MucR from Sinorhizobium meliloti: New Insights into Its DNA Targets and Its Ability to Oligomerize
Abstract
:1. Introduction
2. Results
2.1. Primary Structure Characterization of MucR Purified from S. meliloti by Mass Spectrometry
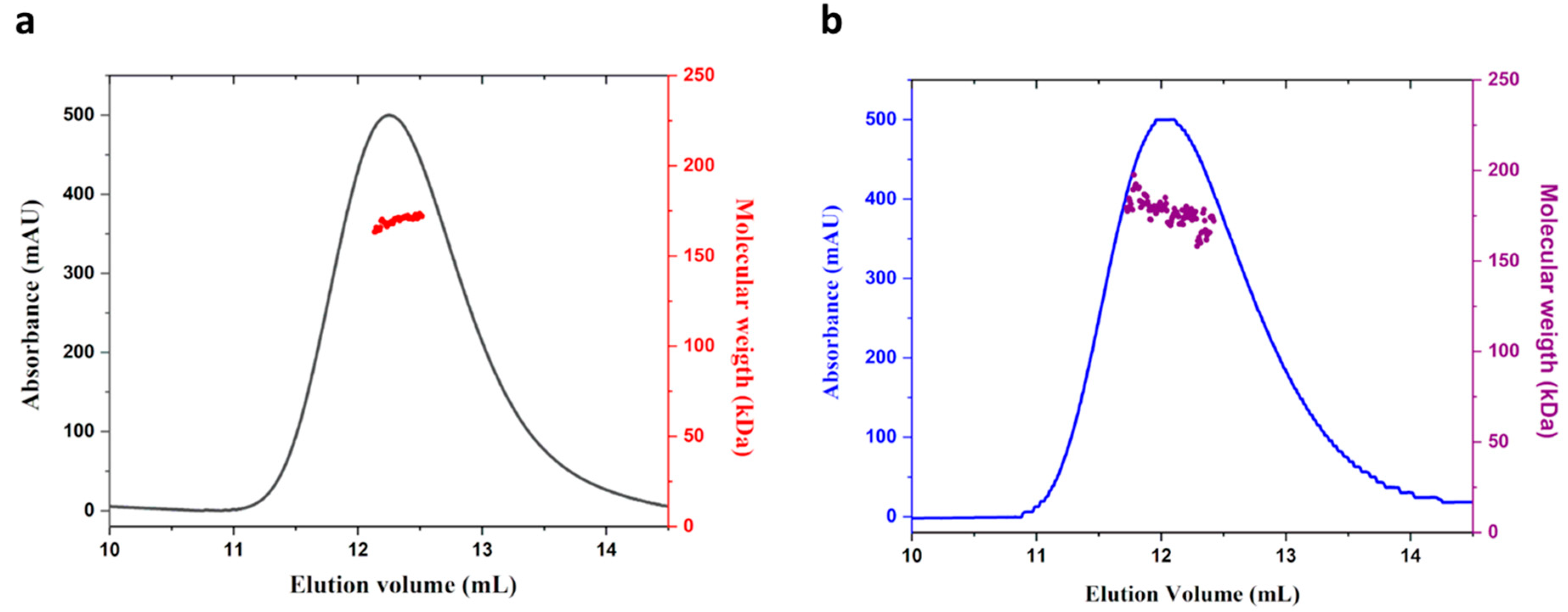
2.2. MucR from S. meliloti Forms High-Order Oligomers
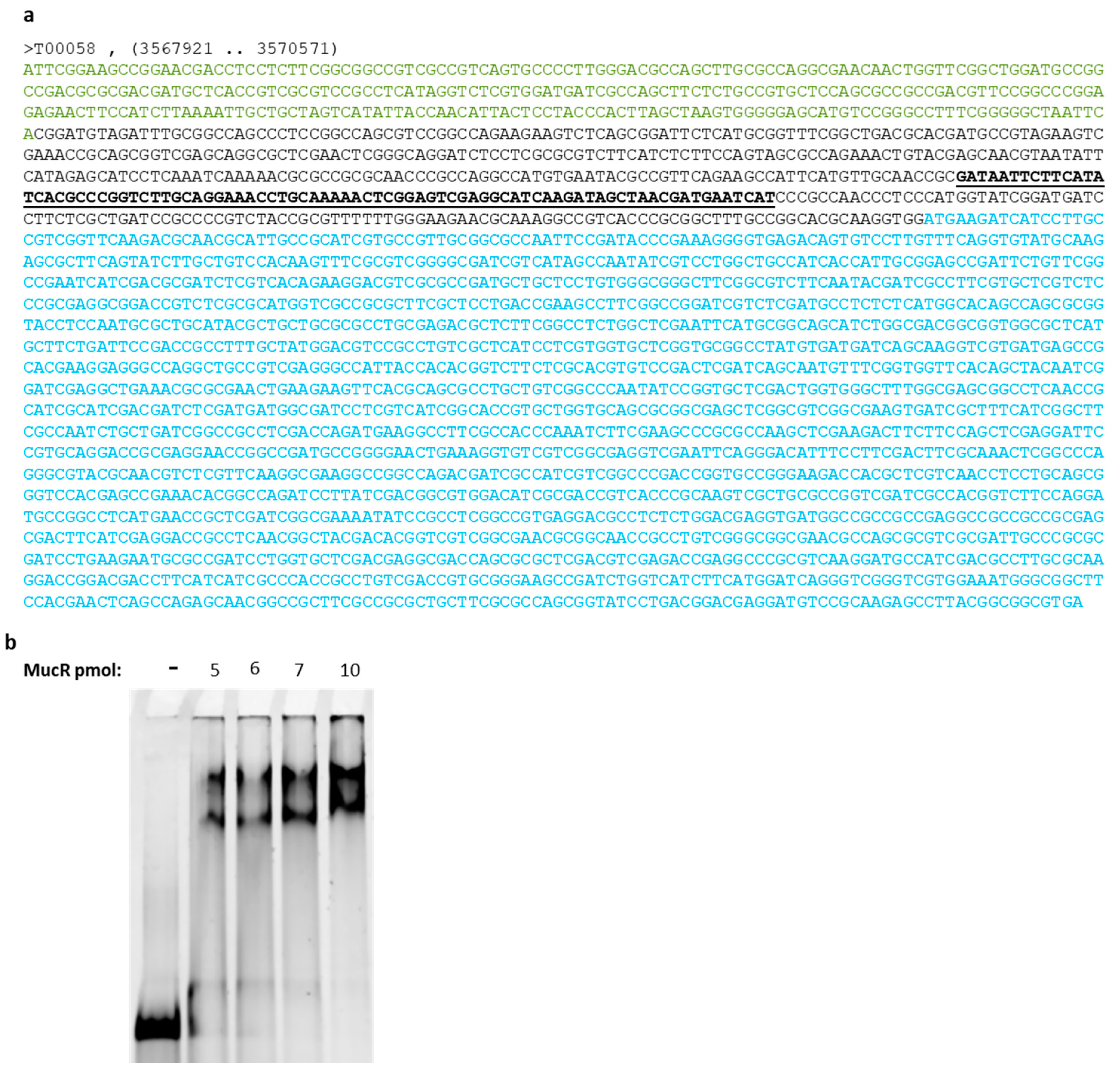
2.3. ndvA Promoter Is a Direct Target of MucR in S. meliloti
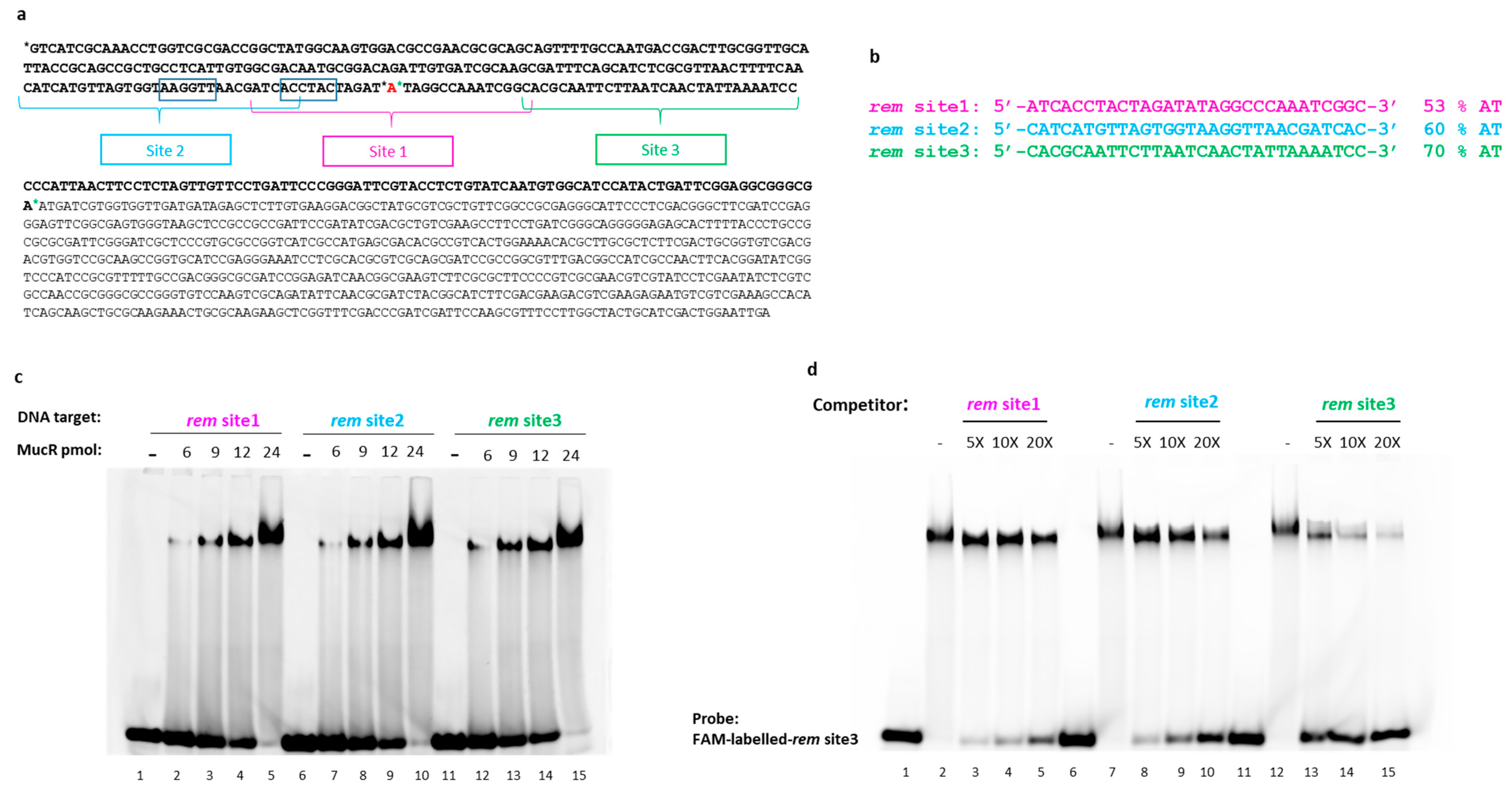
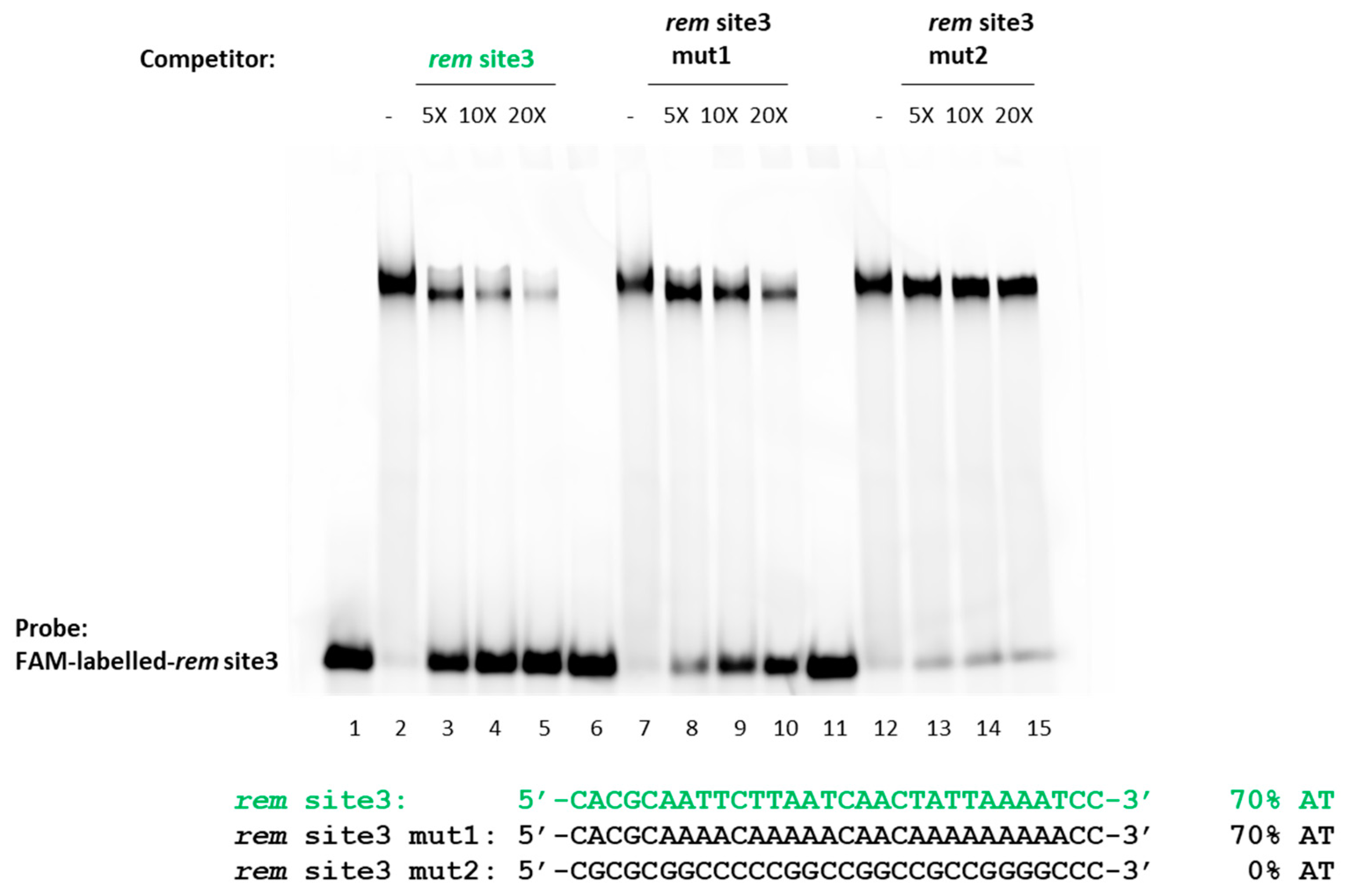
2.4. S. meliloti MucR Binds Three Different AT-Rich Sequences in the Targeted Rem Promoter without any Sequence Specificity
2.5. The Length of DNA Target Sites Affect S. meliloti MucR DNA Binding
3. Discussion
4. Materials and Methods
4.1. Protein Expression and Purification
4.2. High-Resolution NanoLC–Tandem Mass Spectrometry
4.3. MALDI-TOF MS Analysis of MucR Peptides
4.4. Mass Spectrometry Analysis of Intact Protein
4.5. SEC-MALS
4.6. EMSA
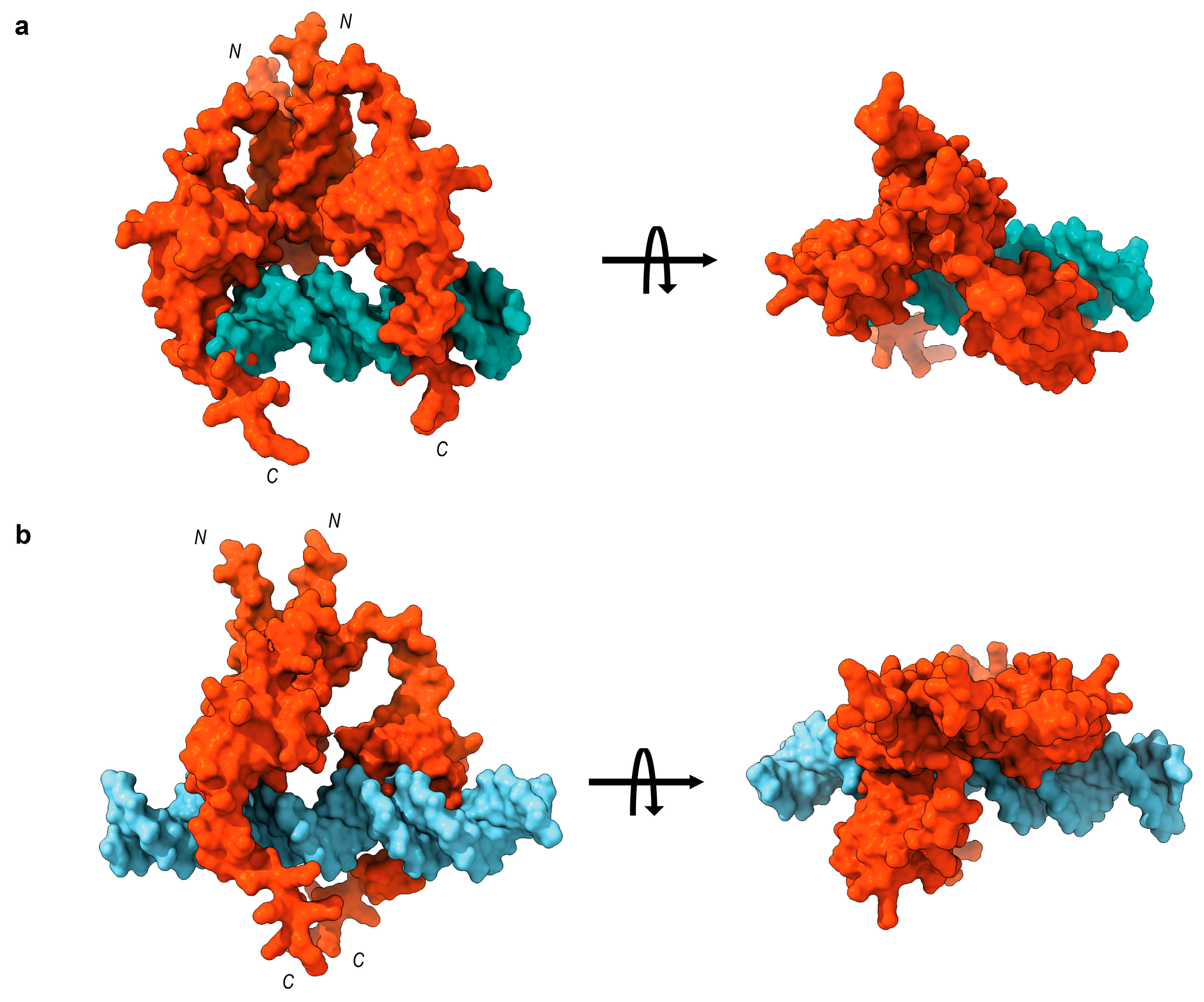
4.7. Molecular Docking
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Janczarek, M. The Ros/MucR Zinc-Finger Protein Family in Bacteria: Structure and Functions. Int. J. Mol. Sci. 2022, 23, 15536. [Google Scholar] [CrossRef]
- Caswell, C.C.; Elhassanny, A.E.; Planchin, E.E.; Roux, C.M.; Weeks-Gorospe, J.N.; Ficht, T.A.; Dunman, P.M.; Roop, R.M., 2nd. Diverse genetic regulon of the virulence-associated transcriptional regulator MucR in Brucella abortus 2308. Infect. Immun. 2013, 81, 1040–1051. [Google Scholar] [CrossRef]
- Baglivo, I.; Russo, L.; Esposito, S.; Malgieri, G.; Renda, M.; Salluzzo, A.; Di Blasio, B.; Isernia, C.; Fattorusso, R.; Pedone, P.V. The structural role of the zinc ion can be dispensable in prokaryotic zinc-finger domains. Proc. Natl. Acad. Sci. USA 2009, 106, 6933–6938. [Google Scholar] [CrossRef]
- Russo, L.; Palmieri, M.; Caso, J.V.; D’Abrosca, G.; Diana, D.; Malgieri, G.; Baglivo, I.; Isernia, C.; Pedone, P.V.; Fattorusso, R. Towards understanding the molecular recognition process in prokaryotic zinc-finger domain. Eur. J. Med. Chem. 2015, 91, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Esposito, S.; Baglivo, I.; Malgieri, G.; Russo, L.; Zaccaro, L.; D’Andrea, L.D.; Mammucari, M.; Di Blasio, B.; Isernia, C.; Fattorusso, R.; et al. A novel type of zinc finger DNA binding domain in the Agrobacterium tumefaciens transcriptional regulator Ros. Biochemistry 2006, 45, 0394–0405. [Google Scholar] [CrossRef] [PubMed]
- Malgieri, G.; Russo, L.; Esposito, S.; Baglivo, I.; Zaccaro, L.; Pedone, E.M.; Di Blasio, B.; Isernia, C.; Pedone, P.V.; Fattorusso, R. The prokaryotic Cys2His2 zinc-finger adopts a novel fold as revealed by the NMR structure of Agrobacterium tumefaciens Ros DNA-binding domain. Proc. Natl. Acad. Sci. USA 2007, 104, 17341–17346. [Google Scholar] [CrossRef] [PubMed]
- Netti, F.; Malgieri, G.; Esposito, S.; Palmieri, M.; Baglivo, I.; Isernia, C.; Omichinski, J.G.; Pedone, P.V.; Lartillot, N.; Fattorusso, R. An experimentally tested scenario for the structural evolution of eukaryotic Cys2His2 zinc fingers from eubacterial ros homologs. Mol. Biol. Evol. 2013, 30, 1504–1513. [Google Scholar] [CrossRef]
- Baglivo, I.; Palmieri, M.; Rivellino, A.; Netti, F.; Russo, L.; Esposito, S.; Iacovino, R.; Farina, B.; Isernia, C.; Fattorusso, R.; et al. Molecular strategies to replace the structural metal site in the prokaryotic zinc finger domain. Biochim. Biophys. Acta 2014, 1844, 497–504. [Google Scholar] [CrossRef]
- Baglivo, I.; Pirone, L.; Pedone, E.M.; Pitzer, J.E.; Muscariello, L.; Marino, M.M.; Malgieri, G.; Freschi, A.; Chambery, A.; Roop Ii, R.M.; et al. Ml proteins from Mesorhizobium loti and MucR from Brucella abortus: An AT-rich core DNA-target site and oligomerization ability. Sci. Rep. 2017, 7, 15805. [Google Scholar] [CrossRef] [PubMed]
- Pirone, L.; Pitzer, J.E.; D’Abrosca, G.; Fattorusso, R.; Malgieri, G.; Pedone, E.M.; Pedone, P.V.; Roop, R.M., 2nd; Baglivo, I. Identifying the region responsible for Brucella abortus MucR higher-order oligomer formation and examining its role in gene regulation. Sci. Rep. 2018, 8, 17238. [Google Scholar] [CrossRef]
- Shi, W.T.; Zhang, B.; Li, M.L.; Liu, K.H.; Jiao, J.; Tian, C.F. The convergent xenogeneic silencer MucR predisposes α-proteobacteria to integrate AT-rich symbiosis genes. Nucleic Acids Res. 2022, 26, 8580–8598. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, M.; Russo, L.; Malgieri, G.; Esposito, S.; Baglivo, I.; Rivellino, A.; Farina, B.; de Paola, I.; Zaccaro, L.; Milardi, D.; et al. Deciphering the zinc coordination properties of the prokaryotic zinc finger domain: The solution structure characterization of Ros87 H42A functional mutant. J. Inorg. Biochem. 2014, 131, 30–36. [Google Scholar] [CrossRef]
- Palmieri, M.; Malgieri, G.; Russo, L.; Baglivo, I.; Esposito, S.; Netti, F.; Del Gatto, A.; de Paola, I.; Zaccaro, L.; Pedone, P.V.; et al. Structural Zn(II) implies a switch from fully cooperative to partly downhill folding in highly homologous proteins. J. Am. Chem. Soc. 2013, 135, 5220–5228. [Google Scholar] [CrossRef]
- Grazioso, R.; García-Viñuales, S.; D’Abrosca, G.; Baglivo, I.; Pedone, P.V.; Milardi, D.; Fattorusso, R.; Isernia, C.; Russo, L.; Malgieri, G. The change of conditions does not affect Ros87 downhill folding mechanism. Sci. Rep. 2020, 10, 21067. [Google Scholar] [CrossRef]
- Sivo, V.; D’Abrosca, G.; Baglivo, I.; Iacovino, R.; Pedone, P.V.; Fattorusso, R.; Russo, L.; Malgieri, G.; Isernia, C. Ni(II), Hg(II), and Pb(II) Coordination in the Prokaryotic Zinc-Finger Ros87. Inorg. Chem. 2019, 58, 1067–1080. [Google Scholar] [CrossRef] [PubMed]
- Dragone, M.; Grazioso, R.; D’Abrosca, G.; Baglivo, I.; Iacovino, R.; Esposito, S.; Paladino, A.; Pedone, P.V.; Russo, L.; Fattorusso, R.; et al. Copper (I) or (II) Replacement of the Structural Zinc Ion in the Prokaryotic Zinc Finger Ros Does Not Result in a Functional Domain. Int. J. Mol. Sci. 2022, 23, 11010. [Google Scholar] [CrossRef] [PubMed]
- Malgieri, G.; Palmieri, M.; Esposito, S.; Maione, V.; Russo, L.; Baglivo, I.; de Paola, I.; Milardi, D.; Diana, D.; Zaccaro, L.; et al. Zinc to cadmium replacement in the prokaryotic zinc-finger domain. Metallomics 2014, 6, 96–104. [Google Scholar] [CrossRef]
- D’Souza-Ault, M.R.; Cooley, M.B.; Kado, C.I. Analysis of the Ros repressor of Agrobacterium virC and virD operons: Molecular intercommunication between plasmid and chromosomal genes. J. Bacteriol. 1993, 175, 3486–3490. [Google Scholar] [CrossRef]
- Kado, C.I. Negative transcriptional regulation of virulence and oncogenes of the Ti plasmid by Ros bearing a conserved C2H2-zinc finger motif. Plasmid 2002, 48, 179–185. [Google Scholar] [CrossRef]
- Mirabella, A.; Terwagne, M.; Zygmunt, M.S.; Cloeckaert, A.; De Bolle, X.; Letesson, J.J. Brucella melitensis MucR, an orthologue of Sinorhizobium meliloti MucR, is involved in resistance to oxidative, detergent, and saline stresses and cell envelope modifications. J. Bacteriol. 2013, 195, 453–465. [Google Scholar] [CrossRef]
- Fumeaux, C.; Radhakrishnan, S.K.; Ardissone, S.; Théraulaz, L.; Frandi, A.; Martins, D.; Nesper, J.; Abel, S.; Jenal, U.; Viollier, P.H. Cell cycle transition from S-phase to G1 in Caulobacter is mediated by ancestral virulence regulators. Nat. Commun. 2014, 5, 4081. [Google Scholar] [CrossRef] [PubMed]
- Rachwał, K.; Matczyńska, E.; Janczarek, M. Transcriptome profiling of a Rhizobium leguminosarum bv. trifolii rosR mutant reveals the role of the transcriptional regulator RosR in motility, synthesis of cell-surface components, and other cellular processes. BMC Genom. 2015, 16, 1111. [Google Scholar] [CrossRef] [PubMed]
- Rachwał, K.; Boguszewska, A.; Kopcińska, J.; Karaś, M.; Tchórzewski, M.; Janczarek, M. The regulatory protein RosR affects Rhizobium leguminosarum bv. trifolii protein profiles, cell surface properties, and symbiosis with clover. Front. Microbiol. 2016, 7, 1302. [Google Scholar] [CrossRef] [PubMed]
- Janczarek, M.; Skorupska, A. The Rhizobium leguminosarum bv. trifolii RosR: Transcriptional regulator involved in exopolysaccharide production. Mol. Plant Microbe Interact. 2007, 20, 867–881. [Google Scholar] [CrossRef]
- Acosta-Jurado, S.; Alias-Villegas, C.; Navarro-Gómez, P.; Zehner, S.; Murdoch, P.D.; Rodríguez-Carvajal, M.A.; Soto, M.J.; Ollero, F.-J.; Ruiz-Sainz, J.E.; Göttfert, M.; et al. The Sinorhizobium fredii HH103 MucR1 Global Regulator Is Connected with the nod Regulon and Is Required for Efficient Symbiosis With Lotus burttii and Glycine max cv. Williams. Mol. Plant Microbe Interact. 2016, 29, 700–712. [Google Scholar] [CrossRef]
- Keller, M.; Roxlau, A.; Weng, W.M.; Schmidt, M.; Quandt, J.; Niehaus, K.; Jording, D.; Arnold, W.; Puhler, A. Molecular analysis of the Rhizobium meliloti mucR gene regulating the biosynthesis of the exopolysaccharides succinoglycan and galactoglucan. Mol. Plant Microbe Interact. 1995, 8, 267–277. [Google Scholar] [CrossRef]
- Becker, A.; Rüberg, S.; Baumgarth, B.; Bertram-Drogatz, P.A.; Quester, I.; Pühler, A. Regulation of succinoglycan and galactoglucan biosynthesis in Sinorhizobium meliloti. J. Mol. Microbiol. Biotechnol. 2002, 4, 187–190. [Google Scholar]
- Rinaudi, L.V.; Sorroche, F.; Zorreguieta, A.; Giordano, W. Analysis of the mucR gene regulating biosynthesis of exopolysaccharides: Implications for biofilm formation in Sinorhizobium meliloti Rm1021. FEMS Microbiol. Lett. 2010, 302, 15–21. [Google Scholar] [CrossRef]
- Bahlawane, C.; McIntosh, M.; Krol, E.; Becker, A. Sinorhizobium meliloti regulator MucR couples exopolysaccharide synthesis and motility. Mol. Plant Microbe Interact. 2008, 21, 1498–1509. [Google Scholar] [CrossRef]
- Bahlawane, C.; Baumgarth, B.; Serrania, J.; Rüberg, S.; Becker, A. Fine-tuning of galactoglucan biosynthesis in Sinorhizobium meliloti by differential WggR (ExpG)-, PhoB-, and MucR-dependent regulation of two promoters. J. Bacteriol. 2008, 190, 3456–3466. [Google Scholar] [CrossRef]
- Battisti, L.; Lara, J.C.; Leigh, J.A. Specific oligosaccharide form of the Rhizobium meliloti exopolysaccharide promotes noduleinvasion in alfalfa. Proc. Natl. Acad. Sci. USA 1992, 89, 5625–5629. [Google Scholar] [CrossRef] [PubMed]
- Gonza´lez, J.E.; Reuhs, B.L.; Walker, G.C. Low molecular weight EPS II of Rhizobium meliloti allows nodule invasion in Medicago sativa. Proc. Natl. Acad. Sci. USA 1996, 93, 8636–8641. [Google Scholar] [CrossRef]
- Wang, L.-X.; Wang, Y.; Pellock, B.; Walker, G.C. Structural characterization of the symbiotically important low-molecularweight succinoglycan of Sinorhizobium meliloti. J. Bacteriol. 1999, 181, 6788–6796. [Google Scholar] [CrossRef] [PubMed]
- Bertram-Drogatz, P.A.; Quester, I.; Becker, A.; Pühler, A. The Sinorhizobium meliloti MucR protein, which is essential for the production of high-molecular-weight succinoglycan exopolysaccharide, binds to short DNA regions upstream of exoH and exoY. Mol. Gen. Genet. 1998, 257, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Mueller, K.; González, J.E. Complex regulation of symbiotic functions is coordinated by MucR and quorum sensing in Sinorhizobium meliloti. J. Bacteriol. 2011, 193, 485–496. [Google Scholar] [CrossRef] [PubMed]
- Baglivo, I.; Pirone, L.; Malgieri, G.; Fattorusso, R.; Roop Ii, R.M.; Pedone, E.M.; Pedone, P.V. MucR binds multiple target sites in the promoter of its own gene and is a heat-stable protein: Is MucR a H-NS-like protein? FEBS Open Bio. 2018, 8, 711–718. [Google Scholar] [CrossRef]
- Borriello, G.; Russo, V.; Paradiso, R.; Riccardi, M.G.; Criscuolo, D.; Verde, G.; Marasco, R.; Pedone, P.V.; Galiero, G.; Baglivo, I. Different Impacts of MucR Binding to the babR and virB Promoters on Gene Expression in Brucella abortus 2308. Biomolecules 2020, 10, 788. [Google Scholar] [CrossRef]
- Jiao, J.; Zhang, B.; Li, M.-L.; Zhang, Z.; Tian, C.-F. The zinc-finger bearing xenogeneic silencer MucR in α-proteobacteria balances adaptation and regulatory integrity. ISME J. 2022, 16, 738–749. [Google Scholar] [CrossRef]
- Gordon, B.R.; Li, Y.; Cote, A.; Weirauch, M.T.; Ding, P.; Hughes, T.R.; Navarre, W.W.; Xia, B.; Liu, J. Structural basis for recognition of AT-rich DNA by unrelated xeogeneic silencing proteins. Proc. Natl. Acad. Sci. USA 2011, 108, 10690–10695. [Google Scholar] [CrossRef]
- Navarre, W.W.; Porwollik, S.; Wang, Y.; McClelland, M.; Rosen, H.; Libby, S.J.; Fang, F.C. Selective Silencing of Foreign DNA with Low GC Content by the H-NS Protein in Salmonella. Science 2006, 31, 236–238. [Google Scholar] [CrossRef]
- Dame, R.T.; Noom, M.C.; Wuite, G.J. Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation. Nature 2006, 444, 387–390. [Google Scholar] [CrossRef] [PubMed]
- Dame, R.T.; Wyman, C.; Goosen, N. H-NS mediated compaction of DNA visualised by atomic force microscopy. Nucleic Acids Res. 2000, 28, 3504–3510. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Erkelens, A.M.; Ben Bdira, F.; Dame, R.T. The architects of bacterial DNA bridges: A structurally and functionally conserved family of proteins. Open Biol. 2019, 9, 190223. [Google Scholar] [CrossRef] [PubMed]
- Rashid, F.M.; Dame, R.T. Three-dimensional chromosome re-modelling: The integral mechanism of transcription regulation in bacteria. Mol. Microbiol. 2023, 120, 60–70. [Google Scholar] [CrossRef]
- Dorman, C.J. H-NS, the genome sentinel. Nat. Rev. Microbiol. 2007, 5, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Dorman, C.J. H-NS: A Universal Regulator For a Dynamic genome. Nat. Rev. Microbiol. 2004, 2, 391–399. [Google Scholar] [CrossRef]
- Castang, S.; McManus, H.R.; Turner, K.H.; Dove, S.L. H-NS family members function coordinately in an opportunistic pathogen. Proc. Natl. Acad. Sci. USA 2008, 105, 18947–18952. [Google Scholar] [CrossRef]
- Stanfield, S.W.; Ielpi, L.; O’Brochta, D.; Helinski, D.R.; Ditta, G.S. The ndvA gene product of Rhizobium meliloti is required for beta-(1—2)glucan production and has homology to the ATP-binding export protein HlyB. J. Bacteriol. 1988, 170, 3523–3530. [Google Scholar] [CrossRef]
- Dylan, T.; Ielpi, L.; Stanfield, S.; Kashyap, L.; Douglas, C.; Yanofsky, M.; Nester, E.; Helinski, D.R.; Ditta, G. Rhizobium meliloti genes required for nodule development are related to chromosomal virulence genes in Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. USA 1986, 83, 4403–4407. [Google Scholar] [CrossRef]
- Rotter, C.; Mühlbacher, S.; Salamon, D.; Schmitt, R.; Scharf, B. Rem, a new transcriptional activator of motility and chemotaxis in Sinorhizobium meliloti. J. Bacteriol. 2006, 188, 6932–6942. [Google Scholar] [CrossRef]
- Harteis, S.; Schneider, S. Making the bend: DNA tertiary structure and protein-DNA interactions. Inter. J. Mol. Sci. 2014, 15, 12335–12363. [Google Scholar] [CrossRef] [PubMed]
- Rohs, R.; West, S.M.; Sosinsky, A.; Liu, P.; Mann, R.S.; Honig, B. The role of DNA shape in protein-DNA recognition. Nature 2009, 461, 1248–1253. [Google Scholar] [CrossRef] [PubMed]
- Udvardi, M.; Poole, P.S. Transport and metabolism in legume-rhizobia symbioses. Annu. Rev. Plant Biol. 2013, 64, 781–805. [Google Scholar] [CrossRef] [PubMed]
- Dilweg, I.W.; Dame, R.T. Post-translational modification of nucleoid-associated proteins: An extra layer of functional modulation in bacteria? Biochem. Soc. Trans. 2018, 46, 1381–1392. [Google Scholar] [CrossRef]
- Arold, S.T.; Leonard, P.G.; Parkinson, G.N.; Ladbury, J.E. H-NS forms a superhelical protein scaffold for DNA condensation. Proc. Natl. Acad. Sci. USA 2010, 7, 15728–15732. [Google Scholar] [CrossRef]
- Winardhi, R.S.; Fu, W.; Castang, S.; Li, Y.; Dove, S.L.; Yan, J. Higher order oligomerization is required for H-NS family member MvaT to form gene-silencing nucleoprotein filament. Nucleic Acids Res. 2012, 40, 8942–8952. [Google Scholar] [CrossRef]
- Iglesias, R.; Russo, R.; Landi, N.; Valletta, M.; Chambery, A.; Di Maro, A.; Bolognesi, A.; Ferreras, J.M.; Citores, L. Structure and Biological Properties of Ribosome-Inactivating Proteins and Lectins from Elder (Sambucus nigra L.) Leaves. Toxins 2022, 14, 611. [Google Scholar] [CrossRef]
- Ragucci, S.; Woodrow, P.; Clemente, A.; Russo, R.; Valletta, M.; Landi, N.; Russo, L.; Chambery, A.; Di Maro, A. Myoglobin from Atlantic and Tinker mackerels: Purification, characterization and its possible use as a molecular marker. Int. J. Biol. Macromol. 2022, 214, 459–469. [Google Scholar] [CrossRef]
- Anvar, Z.; Cammisa, M.; Riso, V.; Baglivo, I.; Kukreja, H.; Sparago, A.; Girardot, M.; Lad, S.; De Feis, I.; Cerrato, F.; et al. ZFP57 recognizes multiple and closely spaced sequence motif variants to maintain repressive epigenetic marks in mouse embryonic stem cells. Nucleic Acids Res. 2016, 44, 1118–1132. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Lumbreras, L.A.; Jiménez-García, B.; Giménez-Santamarina, S.; Fernández-Recio, J. pyDockDNA: A new web server for energy-based protein-DNA docking and scoring. Front. Mol. Biosci. 2022, 9, 988996. [Google Scholar] [CrossRef] [PubMed]
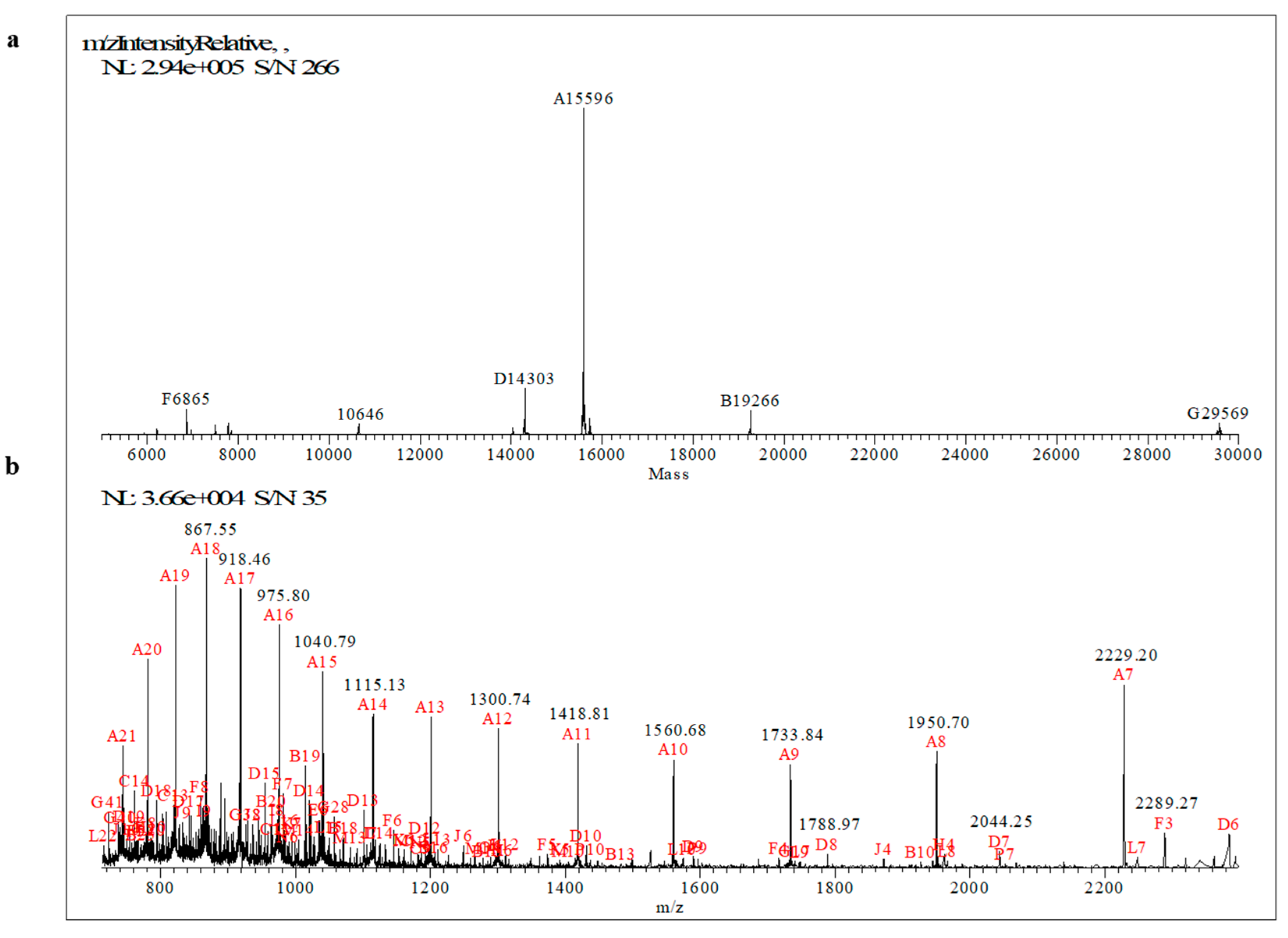

| Tryptic Peptide | Sequence Position | Exp Mr [M+H+]+ | Th Mr [M+H+]+ | Δ (Da) | Missed Cleavages | Modifications |
|---|---|---|---|---|---|---|
| T1 | 2–69 | 7042.55 | 7042.07 | 0.48 | Methionyl residue loss | |
| T2 | 71–87 | 1957.84 | 1957.86 | 0.02 | ||
| T3 | 70–87 | 2085.90 | 2085.96 | 0.06 | K70 | CAM (C79, C82) |
| T4 | 92–105 | 1763.80 | 1763.83 | 0.03 | ||
| T5 | 106–125 | 2278.94 | 2279.08 | 0.14 | K107 | |
| T6 | 131–137 | 790.26 | 790.39 | 0.13 | ||
| T7 | 131–138 | 946.44 | 946.49 | 0.05 |
| Sequence | Modifications | MC | Charge | m/z (Da) | Exp Mr [M+H+]+ | Th Mr [M+H+]+ | Δ (ppm) | RT (min) |
|---|---|---|---|---|---|---|---|---|
| DKWDLPADYPMVAPAYAEAR | 1 | 3 | 760.364 | 2279.078 | 2279.080 | −0.84 | 44.3 | |
| KSVQDDQITcLEcGGTFK | CAM (C10, C13) | 1 | 3 | 695.990 | 2085.957 | 2085.958 | −0.52 | 37.0 |
| SVQDDQITcLEcGGTFK | CAM (C9, C12) | 0 | 2 | 979.434 | 1957.861 | 1957.863 | −1.08 | 40.0 |
| SVQDDQITcLEcGGTFK | CAM (C9, C12) | 0 | 3 | 653.292 | 1957.862 | 1957.863 | −0.53 | 40.0 |
| EMGLGQR | 0 | 2 | 395.697 | 790.387 | 790.388 | −0.44 | 31.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Slapakova, M.; Sgambati, D.; Pirone, L.; Russo, V.; D’Abrosca, G.; Valletta, M.; Russo, R.; Chambery, A.; Malgieri, G.; Pedone, E.M.; et al. MucR from Sinorhizobium meliloti: New Insights into Its DNA Targets and Its Ability to Oligomerize. Int. J. Mol. Sci. 2023, 24, 14702. https://doi.org/10.3390/ijms241914702
Slapakova M, Sgambati D, Pirone L, Russo V, D’Abrosca G, Valletta M, Russo R, Chambery A, Malgieri G, Pedone EM, et al. MucR from Sinorhizobium meliloti: New Insights into Its DNA Targets and Its Ability to Oligomerize. International Journal of Molecular Sciences. 2023; 24(19):14702. https://doi.org/10.3390/ijms241914702
Chicago/Turabian StyleSlapakova, Martina, Domenico Sgambati, Luciano Pirone, Veronica Russo, Gianluca D’Abrosca, Mariangela Valletta, Rosita Russo, Angela Chambery, Gaetano Malgieri, Emilia Maria Pedone, and et al. 2023. "MucR from Sinorhizobium meliloti: New Insights into Its DNA Targets and Its Ability to Oligomerize" International Journal of Molecular Sciences 24, no. 19: 14702. https://doi.org/10.3390/ijms241914702







