Interplays between cis- and trans-Acting Factors for Alternative Splicing in Response to Environmental Changes during Biological Invasions of Ascidians
Abstract
:1. Introduction
2. Results
2.1. AS Profiles in Response to Recurrent High Salinity (HS) Stresses in C. robusta
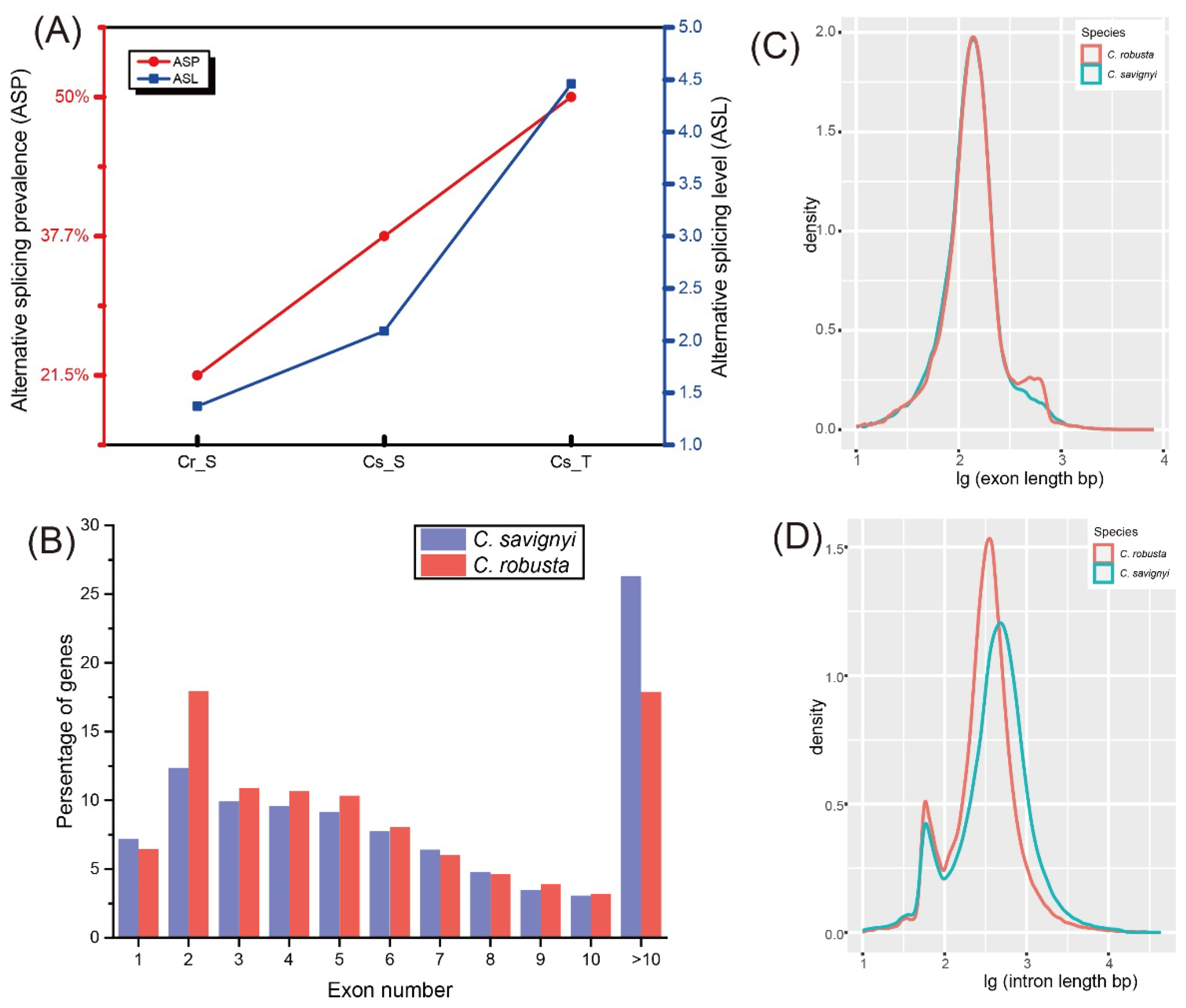
2.2. Comparisons of AS Profiles and Genomic Features between C. robusta and C. savignyi
2.3. Trans-Splicing Factors under AS Regulation
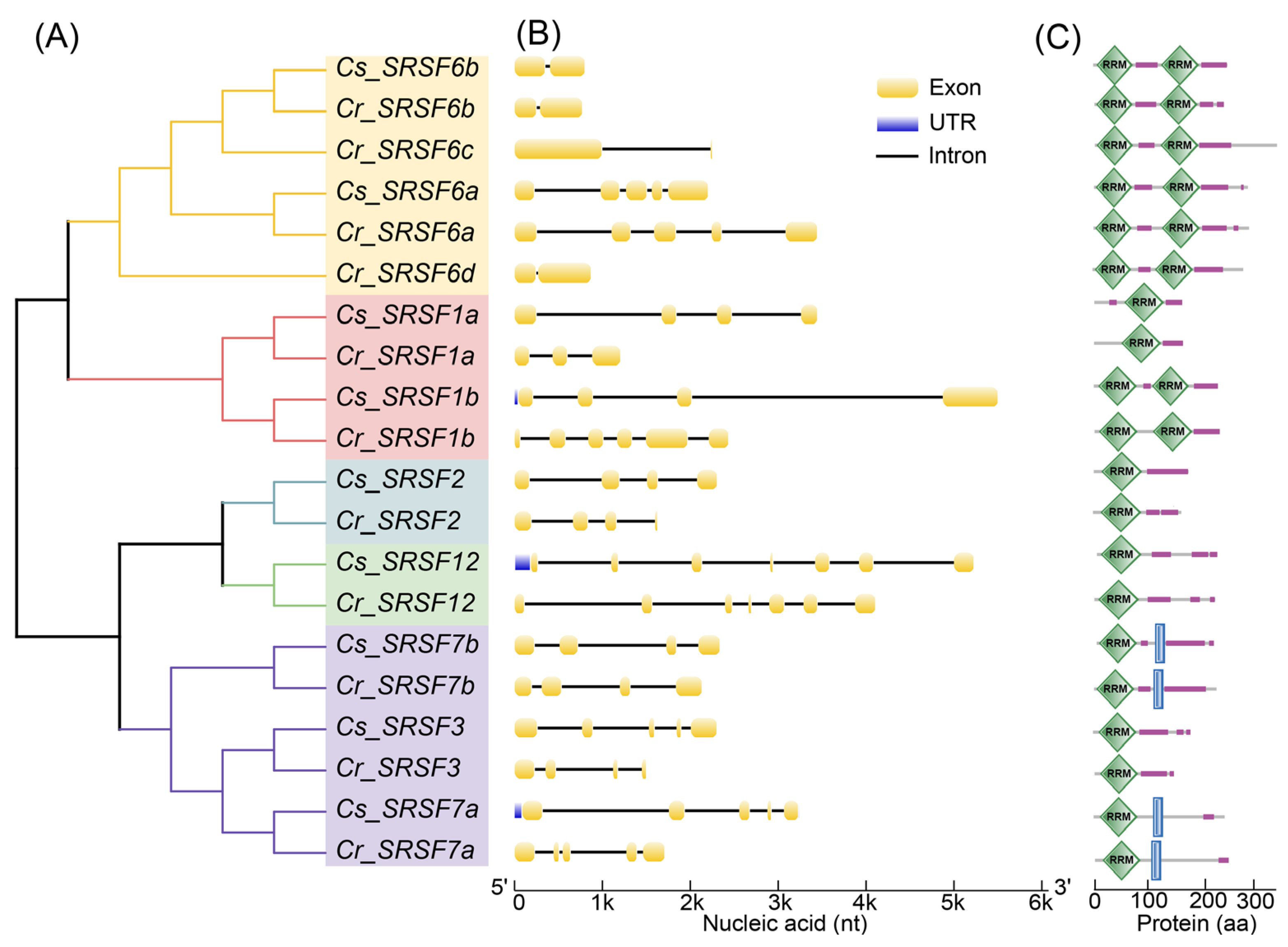
2.4. Genome-Wide Identification of SRSF Members
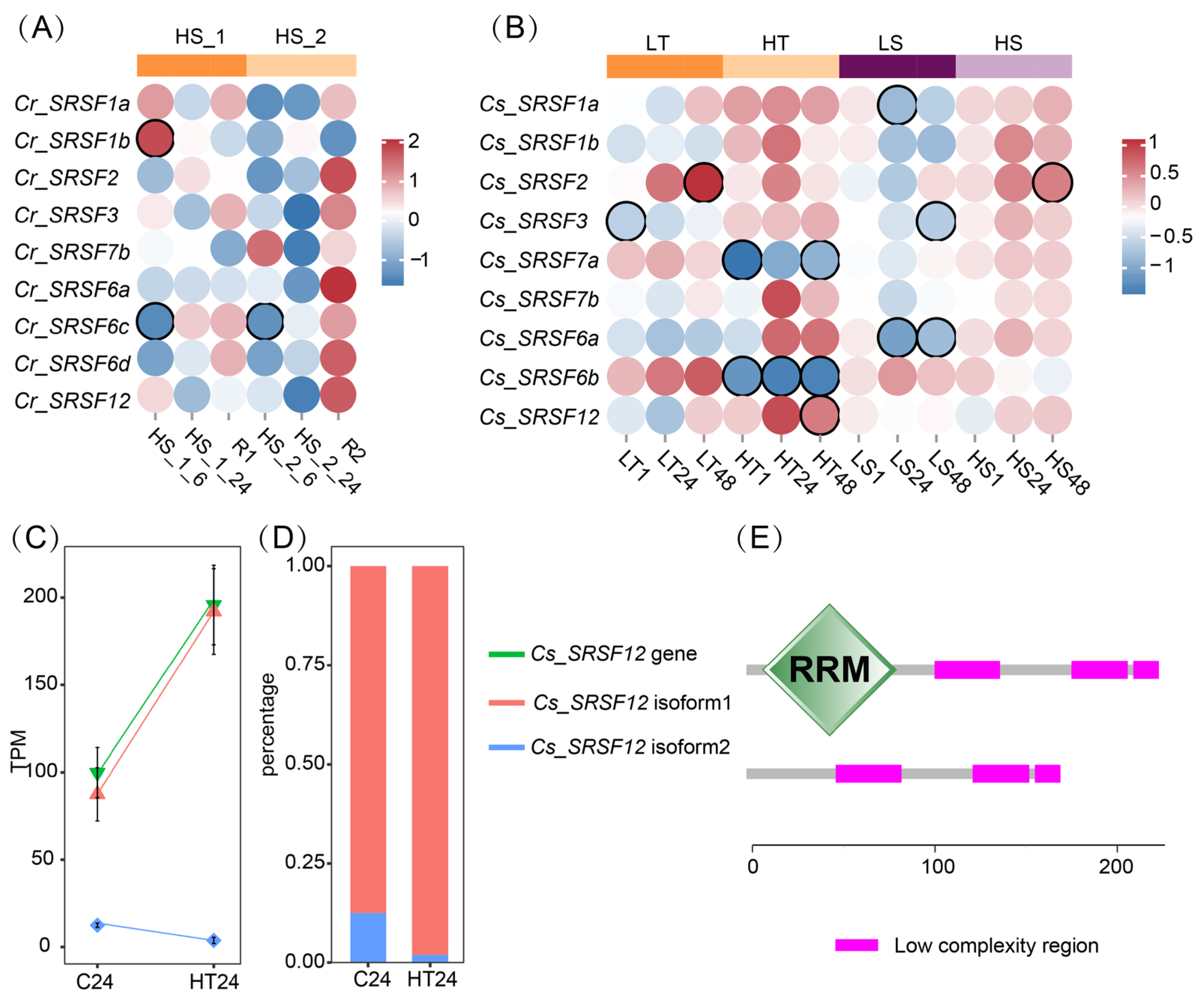
2.5. Gene Expression and AS Responses of SRSF to Environmental Changes
3. Discussion
3.1. Species-Specific and Environmental Context-Dependent AS Patterns
3.2. Genome Architecture Potentially Underlies AS Variation
3.3. Trans-Acting Factors under AS Control in Response to Environmental Stresses
3.4. Future Perspective
4. Materials and Methods
4.1. Sample Collection
4.2. RNA Extraction and Sequencing
4.3. AS Profile Changes of C. robusta to Recurrent High Salinity Stresses
4.4. Transcriptomic Data Collection of C. savignyi
4.5. Genomic Distribution of the Number of Exons and Length of Exons and Introns
4.6. Identification of SRSF Gene Family Members
4.7. Gene Expression and Alternative Splicing Responses to Environmental Changes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wright, C.J.; Smith, C.W.J.; Jiggins, C.D. Alternative splicing as a source of phenotypic diversity. Nat. Rev. Genet. 2022, 23, 697–710. [Google Scholar] [CrossRef]
- Singh, P.; Ahi, E.P. The importance of alternative splicing in adaptive evolution. Mol. Ecol. 2022, 31, 1928–1938. [Google Scholar] [CrossRef]
- Jacobs, A.; Elmer, K.R. Alternative splicing and gene expression play contrasting roles in the parallel phenotypic evolution of a salmonid fish. Mol. Ecol. 2021, 30, 4955–4969. [Google Scholar] [CrossRef] [PubMed]
- Verta, J.P.; Jacobs, A. The role of alternative splicing in adaptation and evolution. Trends Ecol. Evol. 2022, 37, 299–308. [Google Scholar] [CrossRef]
- Merkin, J.; Russell, C.; Chen, P.; Burge, C.B. Evolutionary dynamics of gene and isoform regulation in mammalian tissues. Science 2012, 338, 1593–1599. [Google Scholar] [CrossRef]
- Zhao, F.; Yan, Y.; Wang, Y.; Liu, Y.; Yang, R. Splicing complexity as a pivotal feature of alternative exons in mammalian species. BMC Genomics 2023, 24, 198. [Google Scholar] [CrossRef]
- Griggio, F.; Voskoboynik, A.; Iannelli, F.; Justy, F.; Tilak, M.K.; Turon, X.; Pesole, G.; Douzery, E.J.; Mastrototaro, F.; Gissi, C. Ascidian mitogenomics: Comparison of evolutionary rates in closely related taxa provides evidence of ongoing speciation events. Genome Biol. Evol. 2014, 6, 591–605. [Google Scholar] [CrossRef]
- Zhan, A.; Briski, E.; Bock, D.G.; Ghabooli, S.; MacIsaac, H.J. Ascidians as models for studying invasion success. Mar. Biol. 2015, 162, 2449–2470. [Google Scholar] [CrossRef]
- Tarallo, A.; Yagi, M.; Oikawa, S.; Agnisola, C.; D’Onofrio, G. Comparative morpho-physiological analysis between Ciona robusta and Ciona savignyi. J. Exp. Mar. Biol. Ecol. 2016, 485, 83–87. [Google Scholar] [CrossRef]
- Chen, Y.; Shenkar, N.; Ni, P.; Lin, Y.; Li, S.; Zhan, A. Rapid microevolution during recent range expansion to harsh environments. BMC Evol. Biol. 2018, 18, 187. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Capinha, C.; Karger, D.N.; Turon, X.; MacIsaac, H.J.; Zhan, A. Impacts of climate change on geographical distributions of invasive ascidians. Mar. Environ. Res. 2020, 159, 104993. [Google Scholar] [CrossRef]
- Chen, Y.; Ni, P.; Fu, R.; Murphy, K.J.; Wyeth, R.C.; Bishop, C.D.; Huang, X.; Li, S.; Zhan, A. (Epi)genomic adaptation driven by fine geographical scale environmental heterogeneity after recent biological invasions. Ecol. Appl. 2022, e2772. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Li, S.; Ni, P.; Gao, Y.; Jiang, B.; Zhou, Z.; Zhan, A. Rapid response to changing environments during biological invasions: DNA methylation perspectives. Mol. Ecol. 2017, 26, 6621–6633. [Google Scholar] [CrossRef]
- Ni, P.; Murphy, K.J.; Wyeth, R.C.; Bishop, C.D.; Li, S.; Zhan, A. Significant population methylation divergence and local environmental influence in an invasive ascidian Ciona intestinalis at fine geographical scales. Mar. Biol. 2019, 166, 143. [Google Scholar] [CrossRef]
- Huang, X.; Zhan, A. Highly dynamic transcriptional reprogramming and shorter isoform shifts under acute stresses during biological invasions. RNA Biol. 2021, 18, 340–353. [Google Scholar] [CrossRef]
- Huang, X.; Li, H.; Zhan, A. Multidimensional plasticity jointly contributes to rapid acclimation to environmental challenges during biological invasions. RNA 2023, 29, 675–690. [Google Scholar] [CrossRef]
- Li, H.; Huang, X.; Zhan, A. Stress Memory of Recurrent Environmental Challenges in Marine Invasive Species: Ciona robusta as a Case Study. Front. Physiol. 2020, 11, 94. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, S.; Cheng, J.; Fu, R.; Zhan, A. Proteomic Response to Environmental Stresses in the Stolon of a Highly Invasive Fouling Ascidian. Front. Mar. Sci. 2021, 8, 761628. [Google Scholar] [CrossRef]
- Black, D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003, 72, 291–336. [Google Scholar] [CrossRef]
- Barbosa-Morais, N.L.; Irimia, M.; Pan, Q.; Xiong, H.Y.; Gueroussov, S.; Lee, L.J.; Slobodeniuc, V.; Kutter, C.; Watt, S.; Colak, R.; et al. The evolutionary landscape of alternative splicing in vertebrate species. Science 2012, 338, 1587–1593. [Google Scholar] [CrossRef]
- Busch, A.; Hertel, K.J. Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip Rev. RNA 2012, 3, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Laloum, T.; Martín, G.; Duque, P. Alternative Splicing Control of Abiotic Stress Responses. Trends Plant Sci. 2018, 23, 140–150. [Google Scholar] [CrossRef]
- Long, J.C.; Caceres, J.F. The SR protein family of splicing factors: Master regulators of gene expression. Biochem. J. 2009, 417, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Manley, J.L.; Krainer, A.R. A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes Dev. 2010, 24, 1073–1074. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira Freitas Machado, C.; Schafranek, M.; Bruggemann, M.; Hernandez Canas, M.C.; Keller, M.; Di Liddo, A.; Brezski, A.; Blumel, N.; Arnold, B.; Bremm, A.; et al. Poison cassette exon splicing of SRSF6 regulates nuclear speckle dispersal and the response to hypoxia. Nucleic Acids Res. 2023, 51, 870–890. [Google Scholar] [CrossRef]
- Kataoka, N.; Matsumoto, E.; Masaki, S. Mechanistic Insights of Aberrant Splicing with Splicing Factor Mutations Found in Myelodysplastic Syndromes. Int. J. Mol. Sci. 2021, 22, 7789. [Google Scholar] [CrossRef]
- Yang, P.; Wang, D.; Kang, L. Alternative splicing level related to intron size and organism complexity. BMC Genom. 2021, 22, 853. [Google Scholar] [CrossRef] [PubMed]
- Ling, Y.; Mahfouz, M.M.; Zhou, S. Pre-mRNA alternative splicing as a modulator for heat stress response in plants. Trends Plant Sci. 2021, 26, 1153–1170. [Google Scholar] [CrossRef]
- Schwartz, S.H.; Silva, J.; Burstein, D.; Pupko, T.; Eyras, E.; Ast, G. Large-scale comparative analysis of splicing signals and their corresponding splicing factors in eukaryotes. Genome Res 2008, 18, 88–103. [Google Scholar] [CrossRef]
- Kalyna, M.; Simpson, C.G.; Syed, N.H.; Lewandowska, D.; Marquez, Y.; Kusenda, B.; Marshall, J.; Fuller, J.; Cardle, L.; McNicol, J.; et al. Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res 2012, 40, 2454–2469. [Google Scholar] [CrossRef]
- Chen, L.; Bush, S.J.; Tovar-Corona, J.M.; Castillo-Morales, A.; Urrutia, A.O. Correcting for differential transcript coverage reveals a strong relationship between alternative splicing and organism complexity. Mol. Biol. Evol. 2014, 31, 1402–1413. [Google Scholar] [CrossRef] [PubMed]
- Kannan, S.; Halter, G.; Renner, T.; Waters, E.R. Patterns of alternative splicing vary between species during heat stress. AoB Plants 2018, 10, ply013. [Google Scholar] [CrossRef]
- Bond, U. Heat shock but not other stress inducers leads to the disruption of a sub-set of snRNPs and inhibition of in vitro splicing in HeLa cells. EMBO J. 1988, 7, 3509–3518. [Google Scholar] [CrossRef]
- Martin, G.; Marquez, Y.; Mantica, F.; Duque, P.; Irimia, M. Alternative splicing landscapes in Arabidopsis thaliana across tissues and stress conditions highlight major functional differences with animals. Genome Biol. 2021, 22, 35. [Google Scholar] [CrossRef] [PubMed]
- Tress, M.L.; Abascal, F.; Valencia, A. Alternative Splicing May Not Be the Key to Proteome Complexity. Trends Biochem Sci 2017, 42, 98–110. [Google Scholar] [CrossRef]
- Grau-Bove, X.; Ruiz-Trillo, I.; Irimia, M. Origin of exon skipping-rich transcriptomes in animals driven by evolution of gene architecture. Genome Biol. 2018, 19, 135. [Google Scholar] [CrossRef] [PubMed]
- Delsuc, F.; Philippe, H.; Tsagkogeorga, G.; Simion, P.; Tilak, M.K.; Turon, X.; Lopez-Legentil, S.; Piette, J.; Lemaire, P.; Douzery, E.J.P. A phylogenomic framework and timescale for comparative studies of tunicates. BMC Biol. 2018, 16, 39. [Google Scholar] [CrossRef]
- Satou, Y.; Nakamura, R.; Yu, D.; Yoshida, R.; Hamada, M.; Fujie, M.; Hisata, K.; Takeda, H.; Satoh, N. A Nearly Complete Genome of Ciona intestinalis Type A (C. robusta) Reveals the Contribution of Inversion to Chromosomal Evolution in the Genus Ciona. Genome Biol. Evol. 2019, 11, 3144–3157. [Google Scholar] [CrossRef]
- Kim, N.; Alekseyenko, A.V.; Roy, M.; Lee, C. The ASAP II database: Analysis and comparative genomics of alternative splicing in 15 animal species. Nucleic Acids Res. 2007, 35, D93–D98. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Xiao, J.; Wang, L.; Zhong, J.; Yin, H.; Wu, S.; Zhang, Z.; Yu, J. Systematic analysis of intron size and abundance parameters in diverse lineages. Sci. China Life Sci. 2013, 56, 968–974. [Google Scholar] [PubMed]
- Jakt, L.M.; Dubin, A.; Johansen, S.D. Intron size minimisation in teleosts. BMC Genom. 2022, 23, 628. [Google Scholar] [CrossRef] [PubMed]
- Piovesan, A.; Caracausi, M.; Antonaros, F.; Pelleri, M.C.; Vitale, L. GeneBase 1.1: A tool to summarize data from NCBI gene datasets and its application to an update of human gene statistics. Database 2016, 2016, baw153. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Tan, L.; Wang, S.; Shen, Y.; Guo, L.; Ye, X.; Liu, S.; Feng, Y.; Wu, W. The SR Splicing Factors: Providing Perspectives on Their Evolution, Expression, Alternative Splicing, and Function in Populus trichocarpa. Int. J. Mol. Sci. 2021, 22, 11369. [Google Scholar] [CrossRef]
- Zhang, W.; Du, B.; Liu, D.; Qi, X. Splicing factor SR34b mutation reduces cadmium tolerance in Arabidopsis by regulating iron-regulated transporter 1 gene. Biochem. Biophys. Res. Commun. 2014, 455, 312–317. [Google Scholar] [CrossRef] [PubMed]
- Richardson, D.N.; Rogers, M.F.; Labadorf, A.; Ben-Hur, A.; Guo, H.; Paterson, A.H.; Reddy, A.S.N. Comparative Analysis of Serine/Arginine-Rich Proteins across 27 Eukaryotes: Insights into Sub-Family Classification and Extent of Alternative Splicing. PLoS ONE 2011, 6, e24542. [Google Scholar] [CrossRef]
- Cascarina, S.M.; Ross, E.D. Expansion and functional analysis of the SR-related protein family across the domains of life. RNA 2022, 28, 1298–1314. [Google Scholar] [CrossRef]
- Lareau, L.F.; Brenner, S.E. Regulation of splicing factors by alternative splicing and NMD is conserved between kingdoms yet evolutionarily flexible. Mol. Biol. Evol. 2015, 32, 1072–1079. [Google Scholar] [CrossRef]
- Wei, F.; Chen, P.; Jian, H.; Sun, L.; Lv, X.; Wei, H.; Wang, H.; Hu, T.; Ma, L.; Fu, X.; et al. A Comprehensive Identification and Function Analysis of Serine/Arginine-Rich (SR) Proteins in Cotton (Gossypium spp.). Int. J. Mol. Sci. 2022, 23, 4566. [Google Scholar] [CrossRef]
- Yoon, E.K.; Krishnamurthy, P.; Kim, J.A.; Jeong, M.-J.; Lee, S.I. Genome-wide Characterization of Brassica rapa Genes Encoding Serine/arginine-rich Proteins: Expression and Alternative Splicing Events by Abiotic Stresses. J. Plant Biol. 2018, 61, 198–209. [Google Scholar] [CrossRef]
- Gu, J.; Ma, S.; Zhang, Y.; Wang, D.; Cao, S.; Wang, Z.Y. Genome-Wide Identification of Cassava Serine/Arginine-Rich Proteins: Insights into Alternative Splicing of Pre-mRNAs and Response to Abiotic Stress. Plant Cell Physiol. 2020, 61, 178–191. [Google Scholar] [CrossRef]
- Jin, X. Regulatory Network of Serine/Arginine-Rich (SR) Proteins: The Molecular Mechanism and Physiological Function in Plants. Int. J. Mol. Sci. 2022, 23, 10147. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.; Sinha, S.K.; Maity, U.; Kirti, P.B.; Kumar, K.R.R. Insights into established and emerging roles of SR protein family in plants and animals. Wiley Interdiscip. Rev. RNA 2022, 14, e1763. [Google Scholar] [CrossRef]
- Ding, F.; Su, C.J.; Edmonds, K.K.; Liang, G.; Elowitz, M.B. Dynamics and functional roles of splicing factor autoregulation. Cell Rep. 2022, 39, 110985. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Park, J.W.; Lu, Z.X.; Lin, L.; Henry, M.D.; Wu, Y.N.; Zhou, Q.; Xing, Y. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. USA 2014, 111, E5593–E5601. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Tzioutziou, N.; Stephen, G.; Milne, I.; Calixto, C.; Waugh, R.; Brown, J.W.S.; Zhang, R. 3D RNA-seq—A powerful and flexible tool for rapid and accurate differential expression and alternative splicing analysis of RNA-seq data for biologists. RNA Biol. 2021, 18, 1574–1587. [Google Scholar] [CrossRef] [PubMed]

| Species | Stress | Gene ID a | Gene Name b | Function | Stress-Induced AS Change | Exon Size (bp) |
|---|---|---|---|---|---|---|
| Ciona robusta | High salinity | ENSCING00000001244 | hnRNPLL | Recognition of splicing silencer | Skipping the 7th exon at HS_1_6 and HS_2_6 | 74 |
| ENSCING00000002697 | hnRNPK | Recognition of splicing silencer | Inclusion of the 4th exon at HS_1_24 and R1 | 44 | ||
| ENSCING00000007926 | CLK2 | Phosphorylation of SRSF proteins | Inclusion of the 4th exon at HS_1_6 | 103 | ||
| Ciona savignyi | High salinity | None | ||||
| Low salinity | ENSCSAVG00000000753 | hnRNPM | Recognition of splicing silencer | Inclusion of the 4th exon at LS48 | 8 | |
| ENSCSAVG00000007687 | SNRPA1 | Component of the spliceosome | Skipping the 4th exon at LS1 and LS48 | 80 | ||
| ENSCSAVG00000009055 | BBP | Required for pre-spliceosome formation | Skipping the 3rd exon at LS24 | 76 | ||
| High temperature | ENSCSAVG00000003682 | SRRP53 | Recognition of the 3’ splice site | Skipping the 10th exon at HT1 | 174 | |
| ENSCSAVG00000000753 | hnRNPM | Pre-mRNA binding protein | Skipping the 5th exon at HT1 | 94 | ||
| ENSCSAVG00000004927 | SLU7 | Component of the spliceosome | Skipping the 6th exon at HT1 | 127 | ||
| ENSCSAVG00000007421 | PTBP2 | Negative regulation of exons splicing | Skipping the 8th exon at HT1 | 34 | ||
| ENSCSAVG00000007539 | PHF5A | Component of the minor spliceosome | Skipping the 2nd exon at HT1 | 24 | ||
| ENSCSAVG00000009443 | PRPF38B | Required for pre-mRNA splicing | Skipping the 2nd exon at HT1 | 266 | ||
| Low temperature | ENSCSAVG00000001385 | PUF60 | Binds to the pyrimidine tract and 3’-splice site regions of pre-mRNA | Skipping the 4th exon at LT24 | 84 | |
| ENSCSAVG00000003682 | SRRP53 | Recognition of the 3’ splice site | Skipping the 10th exon at LT48 | 174 | ||
| Species | Family | Gene ID | Gene Name | Genomic Location | Protein | RRM Postion | SR Density % |
|---|---|---|---|---|---|---|---|
| Ciona savignyi | SRSF1/9 | ENSCSAVG00000003567 | Cs_SRSF1a | reftig_15: 65,169–68,621 | 165 | 58–129 | 61.22 |
| ENSCSAVG00000006093 | Cs_SRSF1b | reftig_65: 806,958–812,464 | 233 | 10–79, 111–177 | 59.38 | ||
| SRSF2/8 | ENSCSAVG00000009199 | Cs_SRSF2 | reftig_0: 1,818,675–1,820,985 | 178 | 16–89 | 64.29 | |
| SRSF3/7 | ENSCSAVG00000002676 | Cs_SRSF3 | reftig_11: 177,090–179,396 | 183 | 12–80 | 58.77 | |
| ENSCSAVG00000007369 | Cs_SRSF7a | reftig_26: 994,499–997,745 | 246 | 13–81 | 53.37 | ||
| ENSCSAVG00000010868 | Cs_SRSF7b | reftig_19: 4,084,425–4,086,767 | 220 | 6–74 | 64.08 | ||
| SRSF4/5/6 | ENSCSAVG00000002581 | Cs_SRSF6a | reftig_113: 112,440–114,648 | 289 | 5–70, 130–198 | 69.23 | |
| ENSCSAVG00000011376 | Cs_SRSF6b | reftig_48: 3,543,330–3,544,136 | 252 | 8–73, 129–197 | 75.44 | ||
| SRSF10/12 | ENSCSAVG00000005130 | Cs_SRSF12 | reftig_134: 409,162–425,269 | 226 | 9–82 | 48.55 | |
| Ciona robusta | SRSF1/9 | ENSCING00000000257 | Cr_SRSF1a | Chromosome 8: 137,156–138,366 | 167 | 53–124 | 60.71 |
| ENSCING00000007375 | Cr_SRSF1b | Chromosome 8: 4,297,332–4,299,772 | 235 | 10–79, 111–182 | 57.58 | ||
| SRSF2/8 | ENSCING00000004198 | Cr_SRSF2 | Scaffold HT000041.1: 85,198–86,831 | 166 | 16–89 | 60.81 | |
| SRSF3/7 | ENSCING00000011663 | Cr_SRSF3 | Chromosome 3: 1,159,348–1,160,853 | 149 | 12–80 | 55.41 | |
| ENSCING00000003530 | Cr_SRSF7a | Chromosome 14: 2,961,797–2,963,511 | 251 | 16–84 | 61.9 | ||
| ENSCING00000023459 | Cr_SRSF7b | Chromosome 2: 418,131–420,268 | 230 | 6–74 | 64.65 | ||
| SRSF4/5/6 | ENSCING00000012370 | Cr_SRSF6a | Scaffold HT000150.1: 282,453–285,903 | 292 | 5–70, 130–180 | 68.37 | |
| ENSCING00000020631 | Cr_SRSF6b | Chromosome 5: 3,372,757–3,373,533 | 243 | 5–70, 124–192 | 69.81 | ||
| ENSCING00000024718 | Cr_SRSF6c | Chromosome 5: 3,383,599–3,385,855 | 342 | 5–70, 125–193 | 64.47 | ||
| ENSCING00000011362 | Cr_SRSF6d | Chromosome 2: 6,628,454–6,629,329 | 283 | 5–75, 118–191 | 51.49 | ||
| SRSF10/12 | ENSCING00000018401 | Cr_SRSF12 | Chromosome 8: 1,554,382–1,558,494 | 227 | 9–82 | 49.19 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, X.; Li, H.; Zhan, A. Interplays between cis- and trans-Acting Factors for Alternative Splicing in Response to Environmental Changes during Biological Invasions of Ascidians. Int. J. Mol. Sci. 2023, 24, 14921. https://doi.org/10.3390/ijms241914921
Huang X, Li H, Zhan A. Interplays between cis- and trans-Acting Factors for Alternative Splicing in Response to Environmental Changes during Biological Invasions of Ascidians. International Journal of Molecular Sciences. 2023; 24(19):14921. https://doi.org/10.3390/ijms241914921
Chicago/Turabian StyleHuang, Xuena, Hanxi Li, and Aibin Zhan. 2023. "Interplays between cis- and trans-Acting Factors for Alternative Splicing in Response to Environmental Changes during Biological Invasions of Ascidians" International Journal of Molecular Sciences 24, no. 19: 14921. https://doi.org/10.3390/ijms241914921





