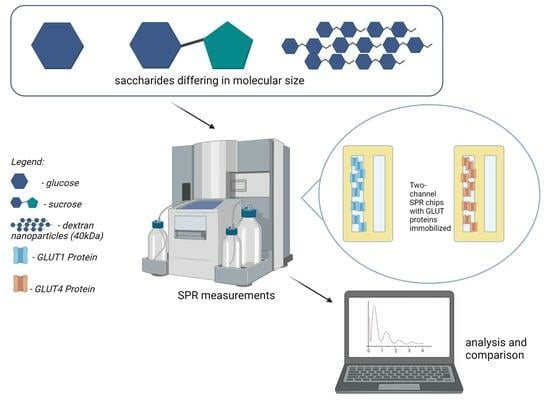
Study on Saccharide–Glucose Receptor Interactions with the Use of Surface Plasmon Resonance
Abstract
:1. Introduction
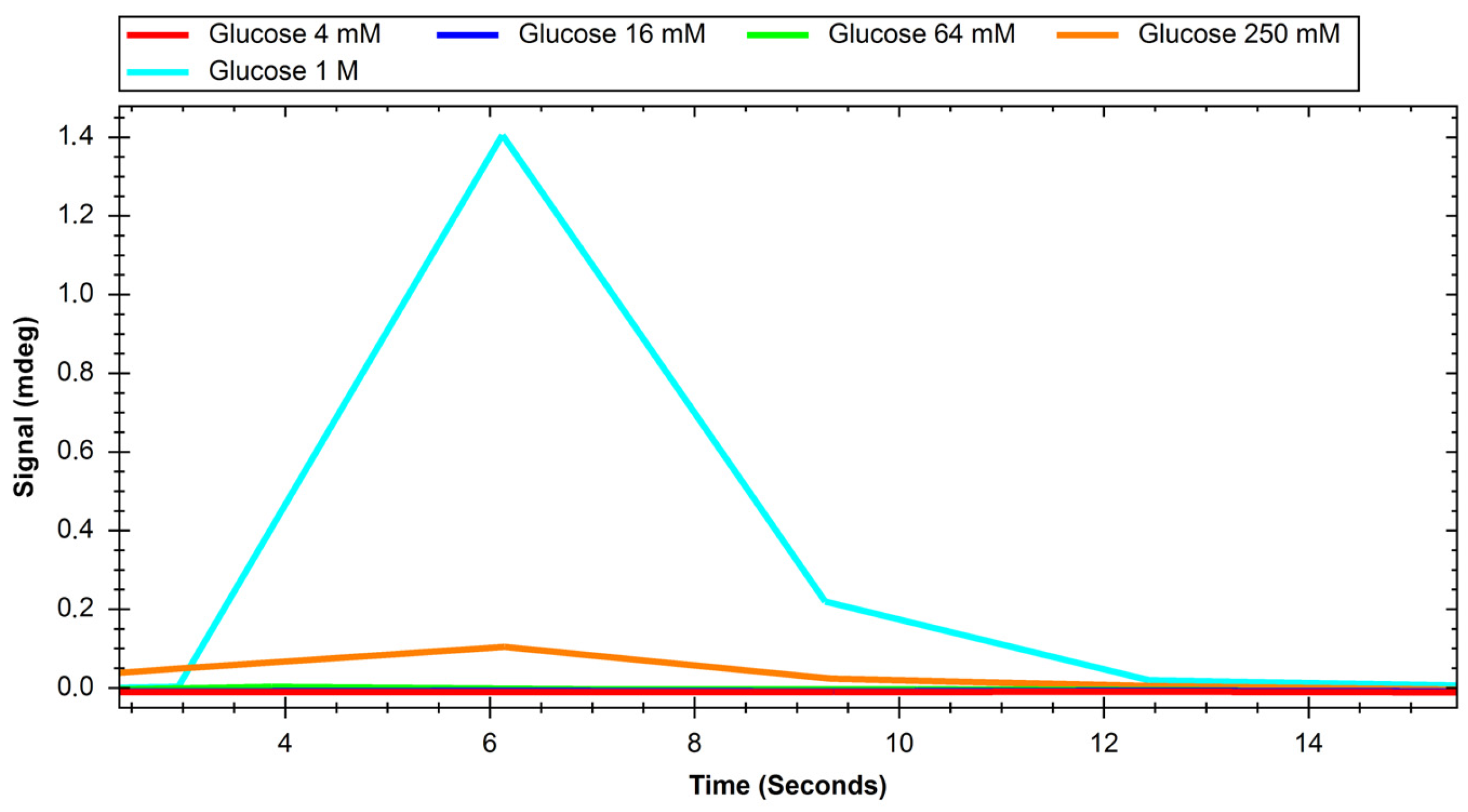
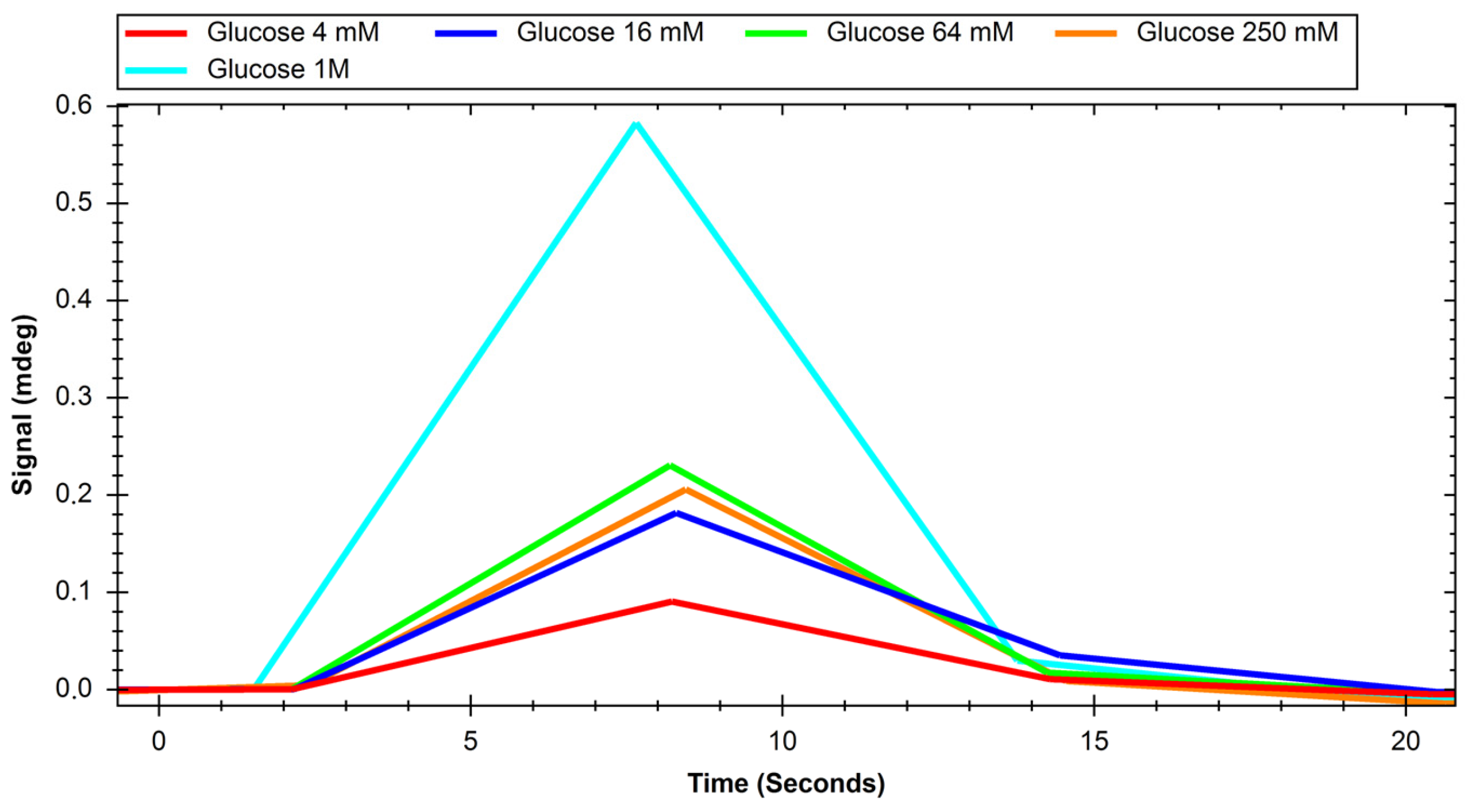
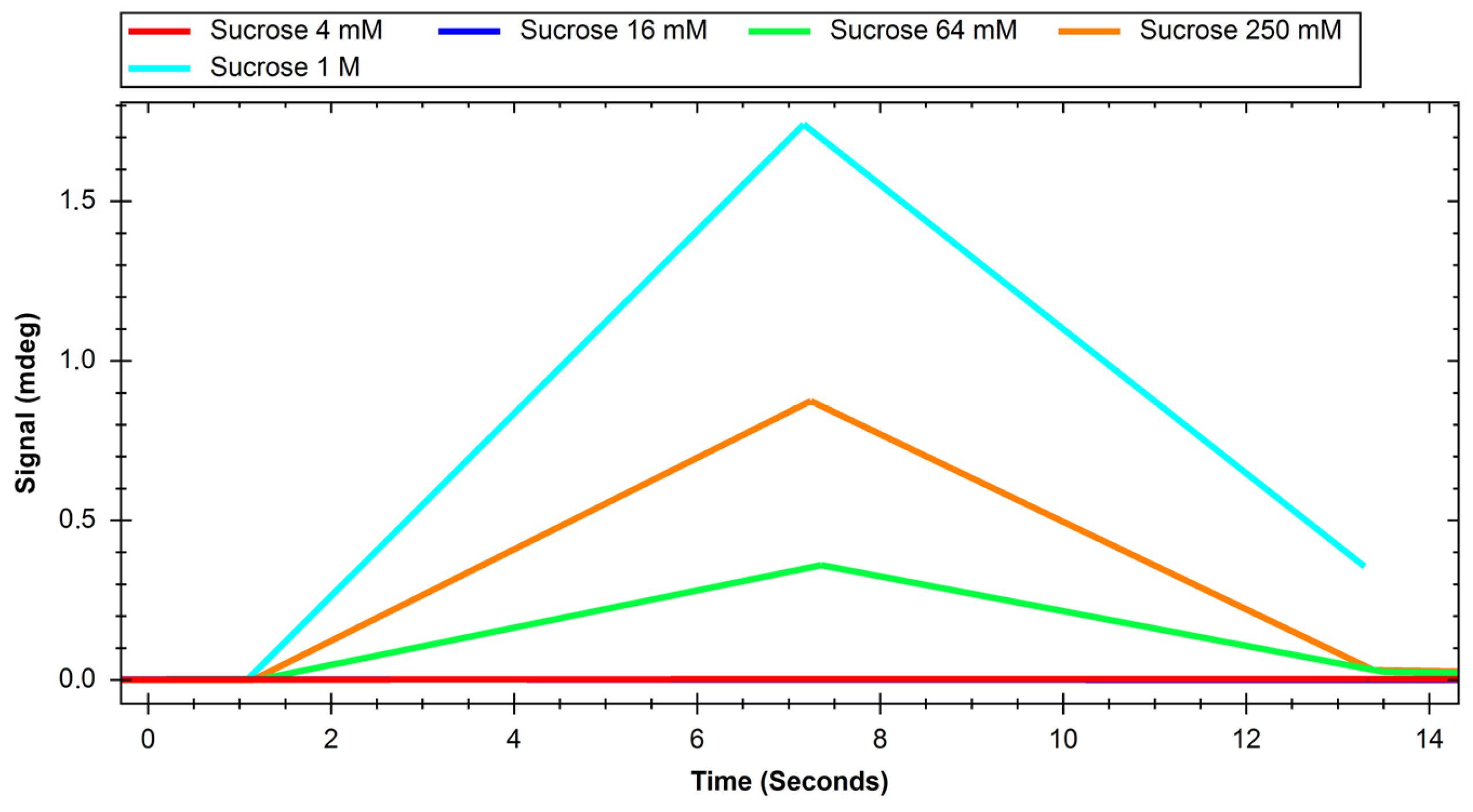
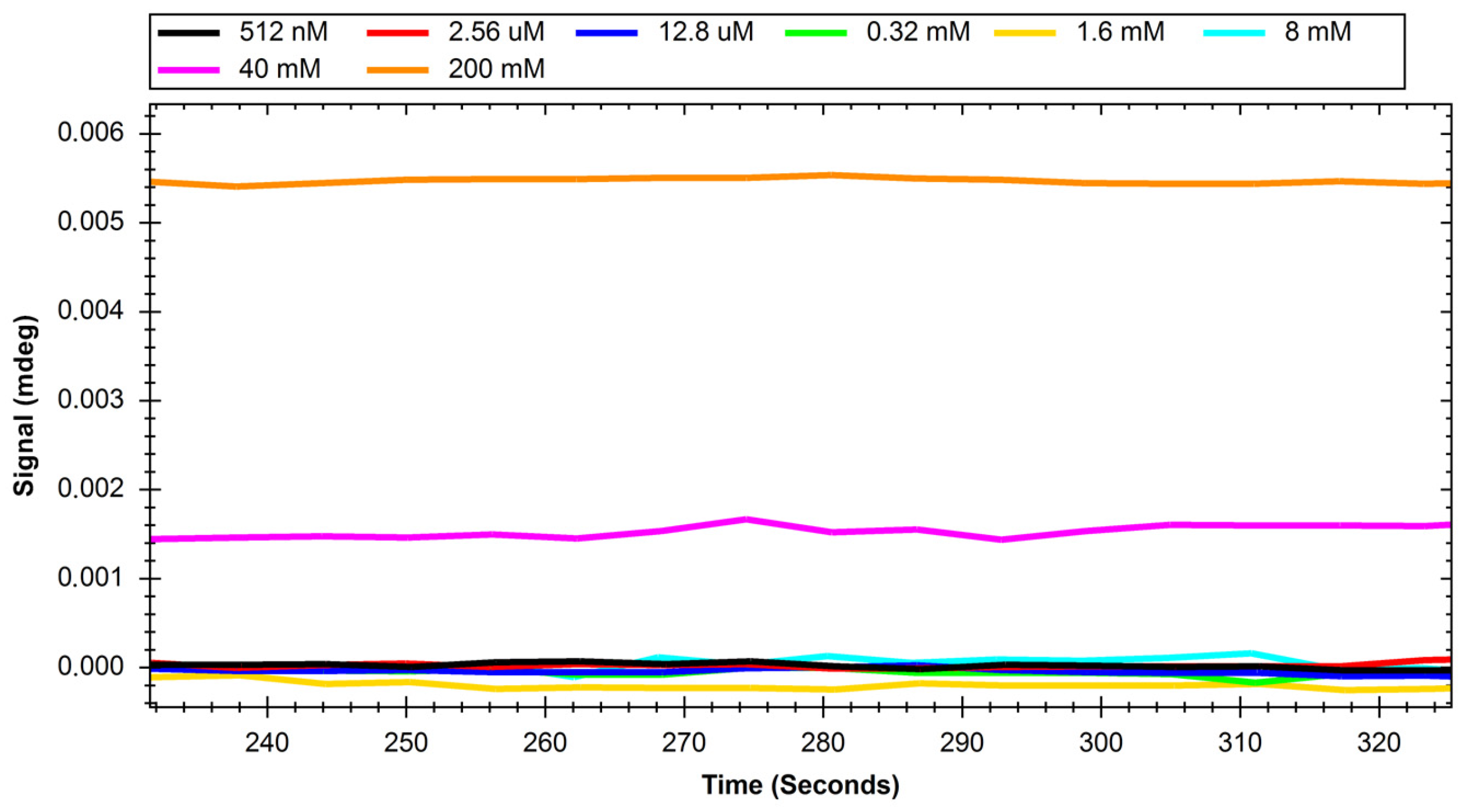
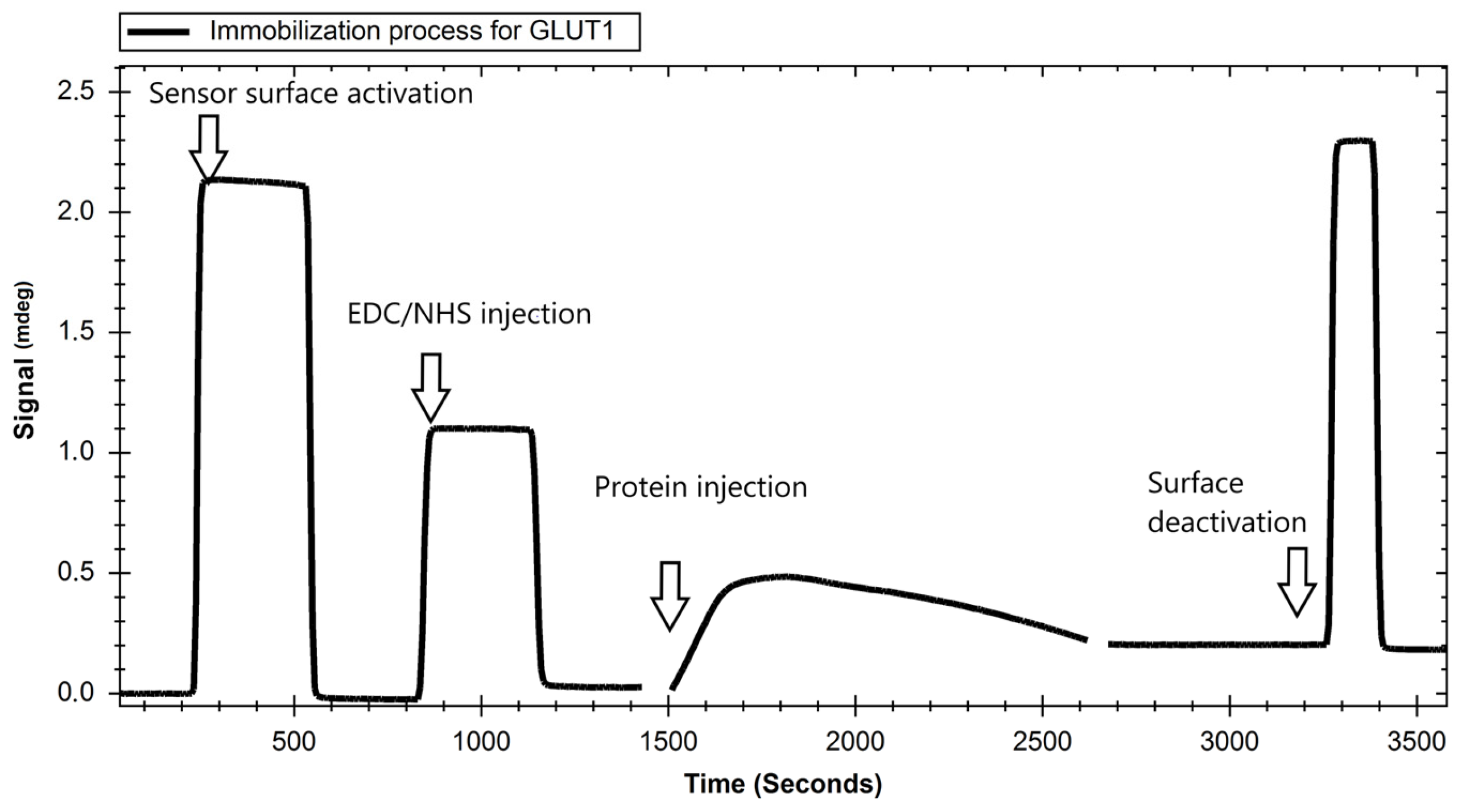
2. Results
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Equipment
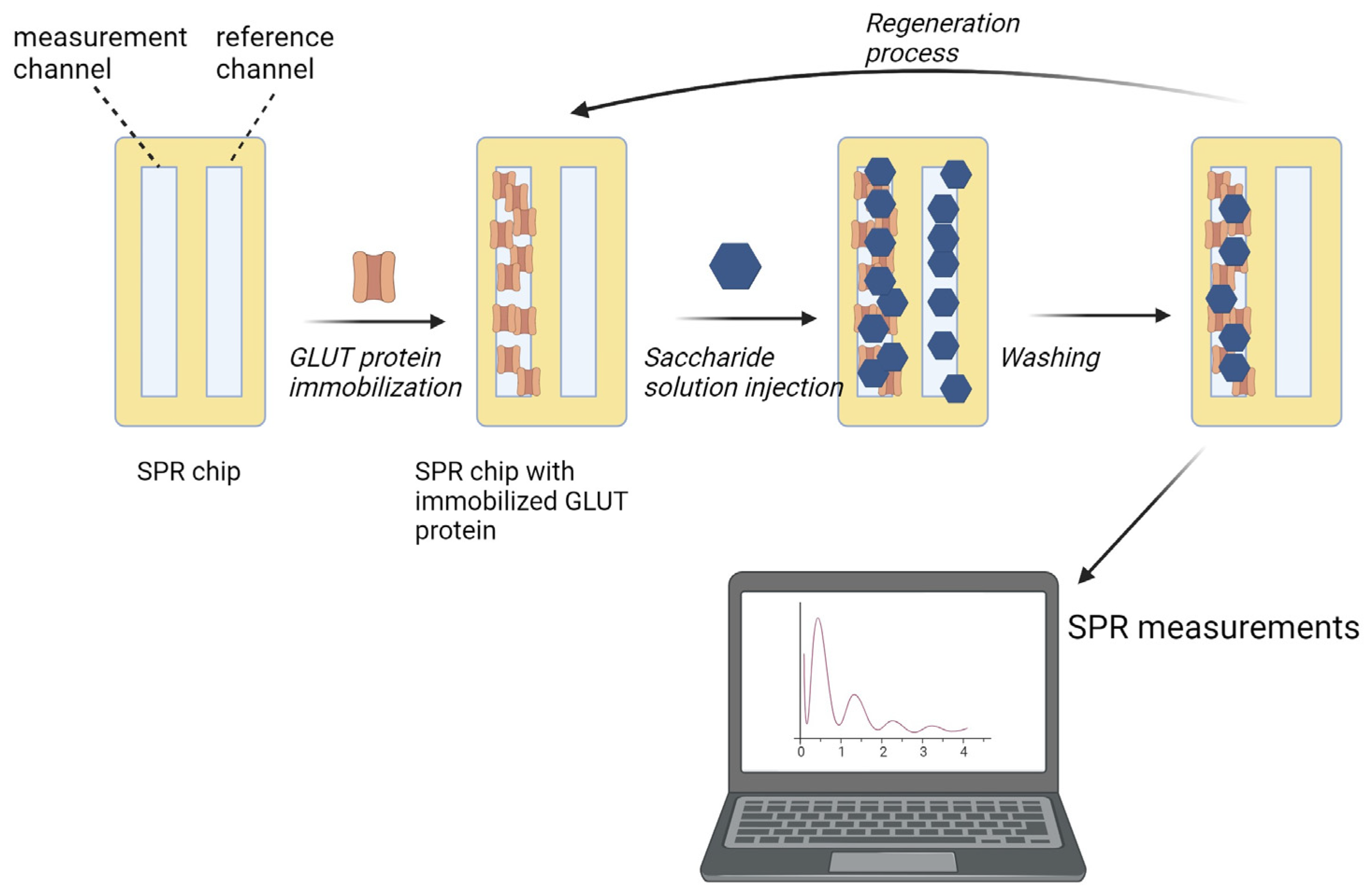
4.3. Methods
4.3.1. Immobilization of GLUT Proteins
4.3.2. Glucose and Sucrose Interaction Experiments
4.3.3. Polysaccharide Nanoparticle Experiments
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guo, X.; Geng, M.; Du, G. Glucose Transporter 1, Distribution in the Brain and in Neural Disorders: Its Relationship with Transport of Neuroactive Drugs through the Blood-Brain Barrier. Biochem. Genet. 2005, 43, 175–187. [Google Scholar] [PubMed]
- Misra, R.; Acharya, S.; Sahoo, S.K. Cancer Nanotechnology: Application of Nanotechnology in Cancer Therapy. Drug Discov. Today 2010, 15, 842–850. [Google Scholar] [CrossRef]
- Nori, A.; Kopecek, J. Intracellular Targeting of Polymer-Bound Drugs for Cancer Chemotherapy. Adv. Drug Deliv. Rev. 2005, 57, 609–636. [Google Scholar] [CrossRef]
- Iversen, T.-G.; Skotland, T.; Sandvig, K. Endocytosis and Intracellular Transport of Nanoparticles: Present Knowledge and Need for Future Studies. Nano Today 2011, 6, 176–185. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Butler, E.B.; Tan, M. Targeting Cellular Metabolism to Improve Cancer Therapeutics. Cell Death Dis. 2013, 4, e532. [Google Scholar] [CrossRef]
- Hamanaka, R.B.; Chandel, N.S. Targeting Glucose Metabolism for Cancer Therapy. J. Exp. Med. 2012, 209, 211–215. [Google Scholar] [CrossRef]
- Calvo, M.B.; Figueroa, A.; Pulido, E.G.; Campelo, R.G.; Aparicio, L.A. Potential Role of Sugar Transporters in Cancer and Their Relationship with Anticancer Therapy. Int. J. Endocrinol. 2010, 2010, 205357. [Google Scholar] [CrossRef]
- Zhu, Y.; Liu, Y.; Jin, K.; Pang, Z. Polysaccharide Nanoparticles for Cancer Drug Targeting. In Polysaccharide Carriers for Drug Delivery; Woodhead Publishing: Sawston, UK, 2019; pp. 365–396. [Google Scholar] [CrossRef]
- Wasiak, I.; Kulikowska, A.; Janczewska, M.; Michalak, M.; Cymerman, I.A.; Nagalski, A.; Kallinger, P.; Szymanski, W.W.; Ciach, T. Dextran Nanoparticle Synthesis and Properties. PLoS ONE 2016, 11, e0146237. [Google Scholar] [CrossRef]
- David, A. Carbohydrate-Based Biomedical Copolymers for Targeted Delivery of Anticancer Drugs. Isr. J. Chem. 2010, 50, 204–219. [Google Scholar] [CrossRef]
- Barclay, T.G.; Day, C.M.; Petrovsky, N.; Garg, S. Review of Polysaccharide Particle-Based Functional Drug Delivery. Carbohydr. Polym. 2019, 221, 94–112. [Google Scholar] [CrossRef] [PubMed]
- Plucinski, A.; Lyu, Z.; Schmidt, B.V.K.J. Polysaccharide Nanoparticles: From Fabrication to Applications. J. Mater. Chem. B 2021, 9, 7030–7062. [Google Scholar] [CrossRef]
- Rammelkamp, D.; Li, W.; Meng, Y. Intracellular Delivery of Fluorescently Labeled Polysaccharide Nanoparticles to Cultured Breast Cancer Cells. Methods Mol. Biol. 2016, 1406, 289–302. [Google Scholar] [PubMed]
- Reshma, P.L.; Unnikrishnan, B.S.; Preethi, G.U.; Syama, H.P.; Archana, M.G.; Remya, K.; Shiji, R.; Sreekutty, J.; Sreelekha, T.T. Overcoming Drug-Resistance in Lung Cancer Cells by Paclitaxel Loaded Galactoxyloglucan Nanoparticles. Int. J. Biol. Macromol. 2019, 136, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Yang, L.; He, L. Overview of the Detection Methods for Equilibrium Dissociation Constant KD of Drug-Receptor Interaction. J. Pharm. Anal. 2018, 8, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Forest-Nault, C.; Gaudreault, J.; Henry, O.; Durocher, Y.; De Crescenzo, G. On the Use of Surface Plasmon Resonance Biosensing to Understand IgG-FcγR Interactions. Int. J. Mol. Sci. 2021, 22, 6616. [Google Scholar] [CrossRef]
- Cho, H.-S.; Ahn, J.-M.; Han, H.-J.; Cho, J.-Y. Glypican 3 Binds to GLUT1 and Decreases Glucose Transport Activity in Hepatocellular Carcinoma Cells. J. Cell. Biochem. 2010, 111, 1252–1259. [Google Scholar] [CrossRef]
- Safina, G. Application of Surface Plasmon Resonance for the Detection of Carbohydrates, Glycoconjugates, and Measurement of the Carbohydrate-Specific Interactions: A Comparison with Conventional Analytical Techniques. A Critical Review. Anal. Chim. Acta 2012, 712, 9–29. [Google Scholar] [CrossRef]
- Gumilar, G.; Chowdhury, S.; Shukri, G.; Patah, A.; Nugraha, N.; Henzie, J.; Anshori, I.; Kaneti, Y.V.; Yuliarto, B. The Revelation of Glucose Adsorption Mechanisms on Hierarchical Metal–Organic Frameworks Using a Surface Plasmon Resonance Sensor. J. Mater. Chem. B 2023, 11, 4428–4444. [Google Scholar] [CrossRef]
- Zeng, Z.; Patel, J.; Lee, S.-H.; McCallum, M.; Tyagi, A.; Yan, M.; Shea, K.J. Synthetic Polymer Nanoparticle–Polysaccharide Interactions: A Systematic Study. J. Am. Chem. Soc. 2012, 134, 2681–2690. [Google Scholar] [CrossRef]
- Zhao, F.-Q.; Keating, A. Functional Properties and Genomics of Glucose Transporters. Curr. Genom. 2007, 8, 113–128. [Google Scholar] [CrossRef] [PubMed]
- Hresko, R.C.; Hruz, P.W. HIV Protease Inhibitors Act as Competitive Inhibitors of the Cytoplasmic Glucose Binding Site of GLUTs with Differing Affinities for GLUT1 and GLUT4. PLoS ONE 2011, 6, e25237. [Google Scholar] [CrossRef] [PubMed]
- Jagdale, S.; Narwade, M.; Sheikh, A.; Md, S.; Salve, R.; Gajbhiye, V.; Kesharwani, P.; Gajbhiye, K.R. GLUT1 Transporter-Facilitated Solid Lipid Nanoparticles Loaded with Anti-Cancer Therapeutics for Ovarian Cancer Targeting. Int. J. Pharm. 2023, 637, 122894. [Google Scholar] [CrossRef] [PubMed]
- Lundqvist, A.; Lundahl, P. Glucose Affinity for the Glucose Transporter Glut1 in Native or Reconstituted Lipid Bilayers: Temperature-Dependence Study by Biomembrane Affinity Chromatography. J. Chromatogr. A 1997, 776, 87–91. [Google Scholar] [CrossRef]
- Tabasi, O.; Falamaki, C. Recent Advancements in the Methodologies Applied for the Sensitivity Enhancement of Surface Plasmon Resonance Sensors. Anal. Methods 2018, 10, 3906–3925. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trzaskowski, M.; Drozd, M.; Ciach, T. Study on Saccharide–Glucose Receptor Interactions with the Use of Surface Plasmon Resonance. Int. J. Mol. Sci. 2023, 24, 16079. https://doi.org/10.3390/ijms242216079
Trzaskowski M, Drozd M, Ciach T. Study on Saccharide–Glucose Receptor Interactions with the Use of Surface Plasmon Resonance. International Journal of Molecular Sciences. 2023; 24(22):16079. https://doi.org/10.3390/ijms242216079
Chicago/Turabian StyleTrzaskowski, Maciej, Marcin Drozd, and Tomasz Ciach. 2023. "Study on Saccharide–Glucose Receptor Interactions with the Use of Surface Plasmon Resonance" International Journal of Molecular Sciences 24, no. 22: 16079. https://doi.org/10.3390/ijms242216079