Peptoids: Smart and Emerging Candidates for the Diagnosis of Cancer, Neurological and Autoimmune Disorders
Abstract
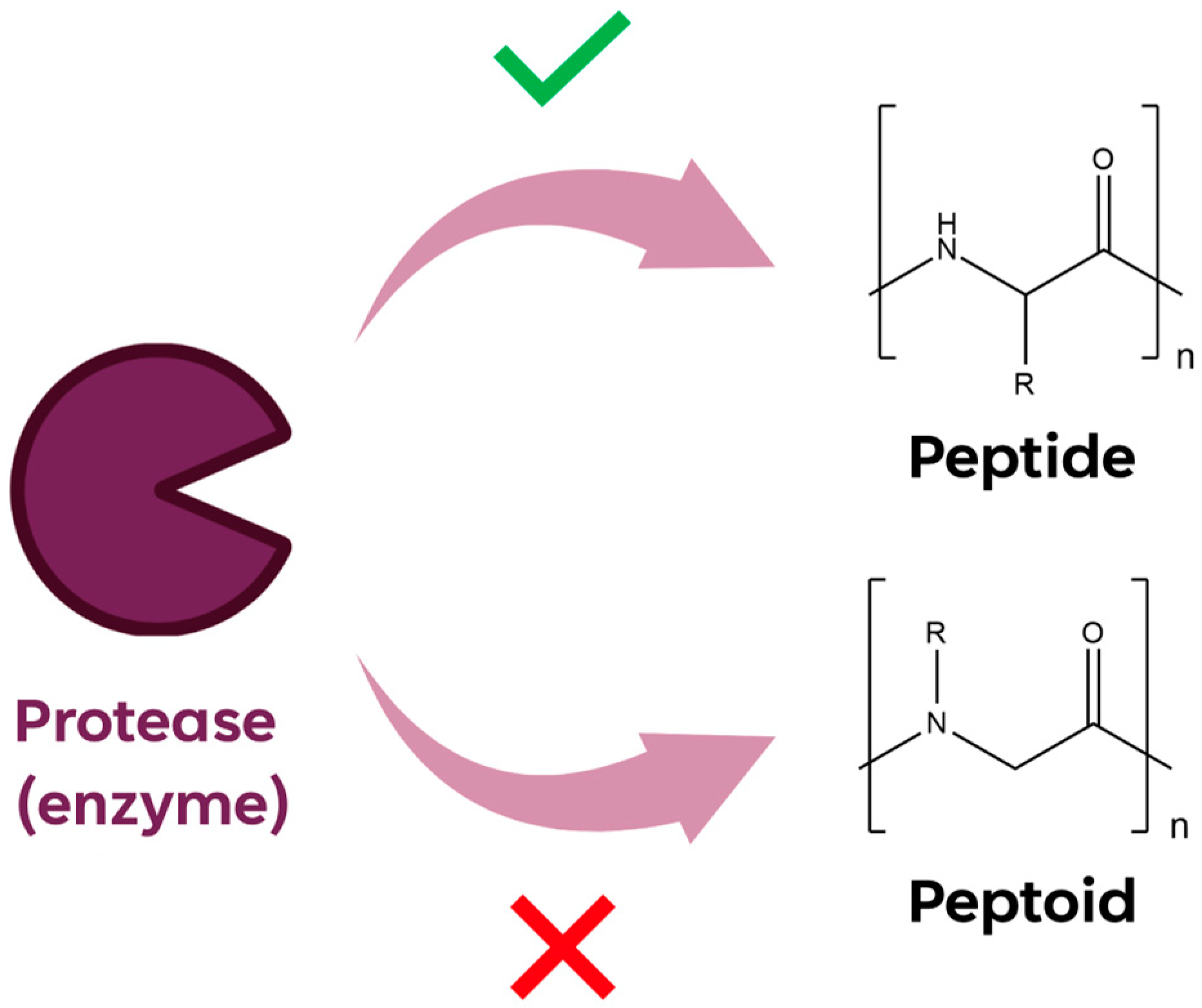
:1. Introduction
2. Radiolabeled Peptoids and Peptoid/Peptide Hybrids for Cancer-Targeted Imaging
2.1. VEGFR2 Biomarker
2.2. NTS1/NTS2 Biomarker
| Name | Sequence | Ref. |
|---|---|---|
| Gd8-dendron-GU40C4 | GU40C * -Ahx-bAla-Lys(GU40C *)-Cys[linker-(Gd8-dendron) **]-NH2 | [22] |
| 64Cu-DOTA-GU40C4 | GU40C *-Ahx-bAla-Lys(GU40C *)-Cys(64Cu-maleimide-monoamide-DOTA derivative)-NH2 | [24] |
| [18F]FGlc-NT4 | Pra *** (218FGlc)-NLys-Lys-Pro-Tyr-Tle-Leu-OH | [32] |
| [68Ga]3 | 68Ga-DOTA-NLys-Lys-Pro-Tyr-Tle-Leu-OH | [34] |
| 18F-4 | Pra *** (618FGlc)-NMeArg-Arg-Pro-NhomoTyr-Ile-Leu-OH | [36] |
3. Peptoids for Blood-Based Screening of Neurological and Autoimmune Diseases
3.1. Alzheimer’s Disease
3.2. Parkinson Disease
3.3. Neuromyelitis Optica
3.4. Systemic Lupus Erythematosus
3.5. Autism Spectrum Disorder
| Name | Sequence | Ref. |
|---|---|---|
| ADP1 | NLeu-NSer-NLys-NLeu-Nspe-Nspe-NLys-NLys-Cys-NH2 | [40] |
| ADP2 | Nall-NSer-Npip-Nall-Nall-Nall-NLys-NLys-Cys-NH2 | |
| ADP3 | NLys-Npip-NLeu-NLys-NLys-Nspe-NLys-NAsp-Cys-NH2 | [40,43] |
| Self-assembled Peptoid nanosheet with ADP3 loops | (…) *-NLys-Npip-NLeu-NLys-NLys-Nspe-NLys-NAsp-NGlu-(…) * | [46,47] |
| PD2 | NLys-NSer-NLys-NLeu-Npyr-Nmea-Ndmpa-Nmea-Cys-NH2 | [49] |
| ASBP-7 | NLys-Na-NGlu-Npip-NLys-NLys-Cys-NH2 | [50] |
| NMOP2 | Napi-Nmba-NLys-Npip-Nbsa-NLys-NLys-Nfur-Met-Cys-NH2 | [57] |
| NMOP5 | Napi-Nmba-Nbsa-Npip-Nbsa-NLys-NLys-Nfur-Met-Cys-NH2 | |
| NMOP6 | NLys-Napi-Npip-Npip-Npip-NLys-NLys-Nfur-Met-Cys-NH2 | |
| NMOP9 | Nbsa-NLys-Npip-NLys-Nmba-NLys-NLys-Nfur-Met-Cys-NH2 | |
| SLE15 | Nmba-NLys-Ndmpa-Npm-Ncha-NLys-Npip-NLys-NLys-Cys-NH2 | [62] |
| ILE2 | Npm-Nfur-Npip-Nmpa-NLeu-NLeu-Nmpa-Nmea-Nmea-Cys-NH2 | |
| ILE7 | Npip-Nfur-Npm-Nall-NSer-Nall-Nfur-Nmea-Nmea-Cys-NH2 | |
| ASDn | (NR1-NR3-NR3-NR4-NR3-NR2-NR1) **-Nmea-Nmea-Met | [66] |
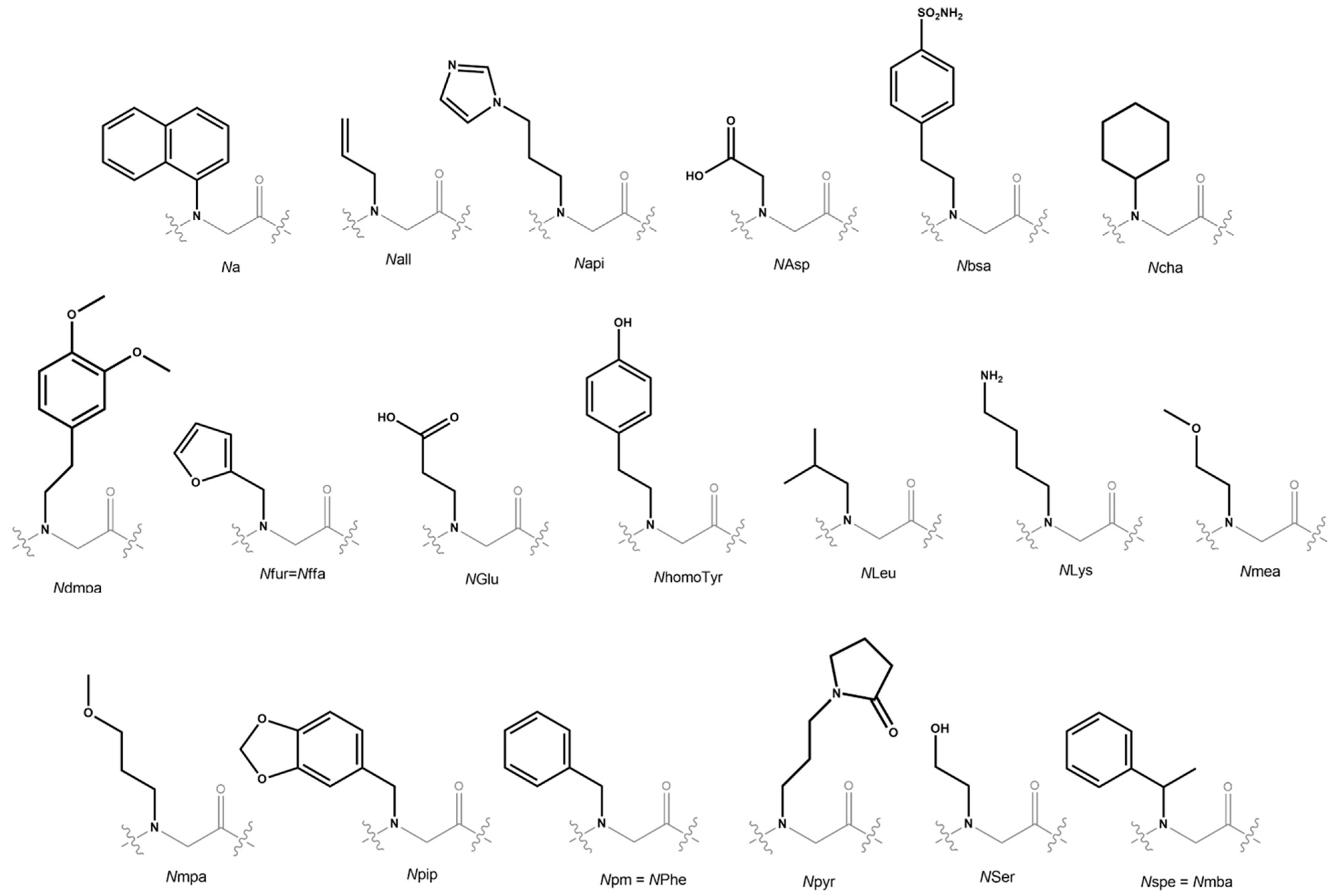
| Monomer Acronym | Amine Precursor |
|---|---|
| Na | 1-naphthylamine |
| Nall | allylamine |
| Napi | 1H-imidazole-4-propylamine |
| NAsp | Glycine |
| Nbsa | 4-(2-aminoethyl)benzenesulfonamide |
| Ncha | cyclohexylamine |
| Ndmpa | 3,4-dimethoxyphenethylamine |
| Nfur = Nffa | furfurylamine |
| NGlu | b-alanine |
| NhomoTyr | p-hydroxyphenylethylamine |
| NLeu | isobutylamine |
| NLys | 1,4-diaminobutane |
| Nmea | 2-methoxyethylamine |
| Nmpa | 3-methoxypropylamine |
| Npip | piperonylamine |
| Npm | benzylamine |
| Npyr | N-(3′-aminopropyl)-2-pyrrolidinone |
| Nser | ethanolamine |
| Nspe = Nmba | (R)-methylbenzylamine |
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pandey, S.; Malviya, G.; Chottova Dvorakova, M. Role of Peptides in Diagnostics. Int. J. Mol. Sci. 2021, 22, 8828. [Google Scholar] [CrossRef] [PubMed]
- Gomari, M.M.; Abkhiz, S.; Pour, T.G.; Lotfi, E.; Rostami, N.; Monfared, F.N.; Ghobari, B.; Mosavi, M.; Alipour, B.; Dokholyan, N.V. Peptidomimetics in Cancer Targeting. Mol. Med. 2022, 28, 146. [Google Scholar] [CrossRef] [PubMed]
- Qvit, N.; Rubin, S.J.S.; Urban, T.J.; Mochly-Rosen, D.; Gross, E.R. Peptidomimetic Therapeutics: Scientific Approaches and Opportunities. Drug Discov. Today 2017, 22, 454–462. [Google Scholar] [CrossRef] [PubMed]
- Zuckermann, R.N.; Kerr, J.M.; Kent, S.B.; Moos, W.H. Efficient Method for the Preparation of Peptoids [Oligo (N-Substituted Glycines)] by Submonomer Solid-Phase Synthesis. J. Am. Chem. Soc. 1992, 114, 10646–10647. [Google Scholar] [CrossRef]
- Sun, J.; Li, Z. Peptoid Applications in Biomedicine and Nanotechnology. In Peptide Applications in Biomedicine, Biotechnology and Bioengineering; Elsevier: Amsterdam, The Netherlands, 2018; pp. 183–213. [Google Scholar]
- Zuckermann, R.N.; Martin, E.J.; Spellmeyer, D.C.; Stauber, G.B.; Shoemaker, K.R.; Kerr, J.M.; Figliozzi, G.M.; Goff, D.A.; Siani, M.A.; Simon, R.J.; et al. Discovery of Nanomolar Ligands for 7-Transmembrane G-Protein-Coupled Receptors from a Diverse N-(Substituted)Glycine Peptoid Library. J. Med. Chem. 1994, 37, 2678–2685. [Google Scholar] [CrossRef]
- Mazaleyrat, J.-P.; Rage, I.; Mouna, A.M.; Šavrda, J.; Wakselman, M.; Boulay, R.; Lelièvre, Y. Peptoid Mimics of a C2-Symmetric Inhibitor of the HIV-1 Protease. Bioorganic Med. Chem. Lett. 1994, 4, 1281–1284. [Google Scholar] [CrossRef]
- Sun, J.; Zuckermann, R.N. Peptoid Polymers: A Highly Designable Bioinspired Material. ACS Nano 2013, 7, 4715–4732. [Google Scholar] [CrossRef]
- Tran, H.; Gael, S.L.; Connolly, M.D.; Zuckermann, R.N. Solid-Phase Submonomer Synthesis of Peptoid Polymers and Their Self-Assembly into Highly-Ordered Nanosheets. JoVE (J. Vis. Exp.) 2011, 57, e3373. [Google Scholar] [CrossRef]
- Diamond, G.; Molchanova, N.; Herlan, C.; Fortkort, J.A.; Lin, J.S.; Figgins, E.; Bopp, N.; Ryan, L.K.; Chung, D.; Adcock, R.S. Potent Antiviral Activity against HSV-1 and SARS-CoV-2 by Antimicrobial Peptoids. Pharmaceuticals 2021, 14, 304. [Google Scholar] [CrossRef]
- Huang, W.; Seo, J.; Willingham, S.B.; Czyzewski, A.M.; Gonzalgo, M.L.; Weissman, I.L.; Barron, A.E. Learning from Host-Defense Peptides: Cationic, Amphipathic Peptoids with Potent Anticancer Activity. PLoS ONE 2014, 9, e90397. [Google Scholar] [CrossRef]
- Chongsiriwatana, N.P.; Patch, J.A.; Czyzewski, A.M.; Dohm, M.T.; Ivankin, A.; Gidalevitz, D.; Zuckermann, R.N.; Barron, A.E. Peptoids That Mimic the Structure, Function, and Mechanism of Helical Antimicrobial Peptides. Proc. Natl. Acad. Sci. USA 2008, 105, 2794–2799. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, R.; Wadman, M.W.; Dohm, M.T.; Czyzewski, A.M.; Spormann, A.M.; Barron, A.E. Antimicrobial Peptoids Are Effective against Pseudomonas Aeruginosa Biofilms. Antimicrob. Agents Chemother. 2011, 55, 3054–3057. [Google Scholar] [CrossRef] [PubMed]
- Mojsoska, B.; Zuckermann, R.N.; Jenssen, H. Structure-Activity Relationship Study of Novel Peptoids That Mimic the Structure of Antimicrobial Peptides. Antimicrob. Agents Chemother. 2015, 59, 4112–4120. [Google Scholar] [CrossRef] [PubMed]
- Mojsoska, B.; Carretero, G.; Larsen, S.; Mateiu, R.V.; Jenssen, H. Peptoids Successfully Inhibit the Growth of Gram Negative E. Coli Causing Substantial Membrane Damage. Sci. Rep. 2017, 7, 42332. [Google Scholar] [CrossRef]
- Nyembe, P.L.; Ntombela, T.; Makatini, M.M. Structure-Activity Relationship of Antimicrobial Peptoids. Pharmaceutics 2023, 15, 1506. [Google Scholar] [CrossRef]
- Gee, A.D.; Herth, M.M.; James, M.L.; Korde, A.; Scott, P.J.; Vasdev, N. Radionuclide Imaging for Neuroscience: Current Opinion and Future Directions. Mol. Imaging 2020, 19, 1536012120936397. [Google Scholar] [CrossRef]
- Brown, R.W.; Cheng, Y.-C.N.; Haacke, E.M.; Thompson, M.R.; Venkatesan, R. Magnetic Resonance Imaging: Physical Principles and Sequence Design; John Wiley & Sons: Hoboken, NJ, USA, 2014; ISBN 978-0-471-72085-0. [Google Scholar]
- Patel, V.; Chisholm, D.; Dua, T.; Laxminarayan, R.; Medina-Mora, M.L.; Vos, T. Disease Control Priorities. In Mental, Neurological, and Substance Use Disorders, 3rd ed.; World Bank Publications: Washinthon, DC, USA, 2016; Volume 4, ISBN 978-1-4648-0428-1. [Google Scholar]
- Pisetsky, D.S. Pathogenesis of Autoimmune Disease. Nat. Rev. Nephrol. 2023, 19, 509–524. [Google Scholar] [CrossRef]
- Zhu, W.; Okollie, B.; Bhujwalla, Z.M.; Artemov, D. PAMAM Dendrimer-Based Contrast Agents for MR Imaging of Her-2/Neu Receptors by a Three-Step Pretargeting Approach. Magn. Reson. Med. 2008, 59, 679–685. [Google Scholar] [CrossRef]
- De León-Rodríguez, L.M.; Lubag, A.; Udugamasooriya, D.G.; Proneth, B.; Brekken, R.A.; Sun, X.; Kodadek, T.; Dean Sherry, A. MRI Detection of VEGFR2 in Vivo Using a Low Molecular Weight Peptoid−(Gd)8-Dendron for Targeting. J. Am. Chem. Soc. 2010, 132, 12829–12831. [Google Scholar] [CrossRef]
- Roland, C.L.; Dineen, S.P.; Lynn, K.D.; Sullivan, L.A.; Dellinger, M.T.; Sadegh, L.; Sullivan, J.P.; Shames, D.S.; Brekken, R.A. Inhibition of Vascular Endothelial Growth Factor Reduces Angiogenesis and Modulates Immune Cell Infiltration of Orthotopic Breast Cancer Xenografts. Mol. Cancer Ther. 2009, 8, 1761–1771. [Google Scholar] [CrossRef]
- Hao, G.; Hajibeigi, A.; León-Rodríguez, L.M.D.; Öz, O.K.; Sun, X. Peptoid-Based PET Imaging of Vascular Endothelial Growth Factor Receptor (VEGFR) Expression. Am. J. Nucl. Med. Mol. Imaging 2011, 1, 65. [Google Scholar] [PubMed]
- Kwon, Y.-U.; Kodadek, T. Quantitative Evaluation of the Relative Cell Permeability of Peptoids and Peptides. J. Am. Chem. Soc. 2007, 129, 1508–1509. [Google Scholar] [CrossRef] [PubMed]
- Udugamasooriya, D.G.; Dineen, S.P.; Brekken, R.A.; Kodadek, T. A Peptoid “Antibody Surrogate” That Antagonizes VEGF Receptor 2 Activity. J. Am. Chem. Soc. 2008, 130, 5744–5752. [Google Scholar] [CrossRef] [PubMed]
- Myers, R.M.; Shearman, J.W.; Kitching, M.O.; Ramos-Montoya, A.; Neal, D.E.; Ley, S.V. Cancer, Chemistry, and the Cell: Molecules That Interact with the Neurotensin Receptors. ACS Chem. Biol. 2009, 4, 503–525. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Q.; Zhou, J.; Yang, W.; Cui, H.; Xu, M.; Yi, L. Oncogenic Role of Neurotensin and Neurotensin Receptors in Various Cancers. Clin. Exp. Pharmacol. Physiol. 2017, 44, 841–846. [Google Scholar] [CrossRef]
- Kokko, K.P.; Hadden, M.K.; Orwig, K.S.; Mazella, J.; Dix, T.A. In Vitro Analysis of Stable, Receptor-Selective Neurotensin[8−13] Analogues. J. Med. Chem. 2003, 46, 4141–4148. [Google Scholar] [CrossRef]
- Previti, S.; Vivancos, M.; Rémond, E.; Beaulieu, S.; Longpré, J.-M.; Ballet, S.; Sarret, P.; Cavelier, F. Insightful Backbone Modifications Preventing Proteolytic Degradation of Neurotensin Analogs Improve NT S1-Induced Protective Hypothermia. Front. Chem. 2020, 8, 406. [Google Scholar] [CrossRef]
- Maschauer, S.; Prante, O. Radiopharmaceuticals for Imaging and Endoradiotherapy of Neurotensin Receptor-positive Tumors. J. Label. Compd. Radiopharm. 2018, 61, 309–325. [Google Scholar] [CrossRef]
- Maschauer, S.; Einsiedel, J.; Haubner, R.; Hocke, C.; Ocker, M.; Hübner, H.; Kuwert, T.; Gmeiner, P.; Prante, O. Labeling and Glycosylation of Peptides Using Click Chemistry: A General Approach to 18F-Glycopeptides as Effective Imaging Probes for Positron Emission Tomography. Angew. Chem. Int. Ed. 2010, 49, 976–979. [Google Scholar] [CrossRef]
- Einsiedel, J.; Hübner, H.; Hervet, M.; Härterich, S.; Koschatzky, S.; Gmeiner, P. Peptide Backbone Modifications on the C-Terminal Hexapeptide of Neurotensin. Bioorganic Med. Chem. Lett. 2008, 18, 2013–2018. [Google Scholar] [CrossRef]
- Maschauer, S.; Einsiedel, J.; Hocke, C.; Hübner, H.; Kuwert, T.; Gmeiner, P.; Prante, O. Synthesis of a 68Ga-Labeled Peptoid−Peptide Hybrid for Imaging of Neurotensin Receptor Expression in Vivo. ACS Med. Chem. Lett. 2010, 1, 224–228. [Google Scholar] [CrossRef] [PubMed]
- Swift, S.L.; Burns, J.E.; Maitland, N.J. Altered Expression of Neurotensin Receptors Is Associated with the Differentiation State of Prostate Cancer. Cancer Res. 2010, 70, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Maschauer, S.; Greff, C.; Einsiedel, J.; Ott, J.; Tripal, P.; Hübner, H.; Gmeiner, P.; Prante, O. Improved Radiosynthesis and Preliminary in Vivo Evaluation of a 18F-Labeled Glycopeptide–Peptoid Hybrid for PET Imaging of Neurotensin Receptor 2. Bioorganic Med. Chem. 2015, 23, 4026–4033. [Google Scholar] [CrossRef]
- Held, C.; Plomer, M.; Hübner, H.; Meltretter, J.; Pischetsrieder, M.; Gmeiner, P. Development of a Metabolically Stable Neurotensin Receptor 2 (NTS2) Ligand. ChemMedChem 2013, 8, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Held, C.; Hübner, H.; Kling, R.; Nagel, Y.A.; Wennemers, H.; Gmeiner, P. Impact of the Proline Residue on Ligand Binding of Neurotensin Receptor 2 (NTS2)-Selective Peptide–Peptoid Hybrids. ChemMedChem 2013, 8, 772–778. [Google Scholar] [CrossRef] [PubMed]
- Einsiedel, J.; Held, C.; Hervet, M.; Plomer, M.; Tschammer, N.; Hübner, H.; Gmeiner, P. Discovery of Highly Potent and Neurotensin Receptor 2 Selective Neurotensin Mimetics. J. Med. Chem. 2011, 54, 2915–2923. [Google Scholar] [CrossRef]
- Reddy, M.M.; Wilson, R.; Wilson, J.; Connell, S.; Gocke, A.; Hynan, L.; German, D.; Kodadek, T. Identification of Candidate IgG Biomarkers for Alzheimer’s Disease via Combinatorial Library Screening. Cell 2011, 144, 132–142. [Google Scholar] [CrossRef]
- Robinson, W.H.; Steinman, L.; Utz, P.J. Protein and Peptide Array Analysis of Autoimmune Disease. Biotechniques 2002, 33, S66–S69. [Google Scholar] [CrossRef]
- Wang, X.; Yu, J.; Sreekumar, A.; Varambally, S.; Shen, R.; Giacherio, D.; Mehra, R.; Montie, J.E.; Pienta, K.J.; Sanda, M.G. Autoantibody Signatures in Prostate Cancer. N. Engl. J. Med. 2005, 353, 1224–1235. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhu, L.; Bu, X.; Ma, H.; Yang, S.; Yang, Y.; Hu, Z. Label-Free Detection of Alzheimer’s Disease through the ADP3 Peptoid Recognizing the Serum Amyloid-Beta42 Peptide. Chem. Commun. 2015, 51, 718–721. [Google Scholar] [CrossRef]
- Kang, J.; Lemaire, H.-G.; Unterbeck, A.; Salbaum, J.M.; Masters, C.L.; Grzeschik, K.-H.; Multhaup, G.; Beyreuther, K.; Müller-Hill, B. The Precursor of Alzheimer’s Disease Amyloid A4 Protein Resembles a Cell-Surface Receptor. Nature 1987, 325, 733–736. [Google Scholar] [CrossRef]
- Zhu, L.; Zhao, Z.; Cheng, P.; He, Z.; Cheng, Z.; Peng, J.; Wang, H.; Wang, C.; Yang, Y.; Hu, Z. Antibody-mimetic Peptoid Nanosheet for Label-free Serum-based Diagnosis of Alzheimer’s Disease. Adv. Mater. 2017, 29, 1700057. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Liu, M.; Zhao, Z.; Yang, C.; Zhu, L.; Cai, Y.; Yang, Y.; Hu, Z. Diagnosis of Mild Cognitive Impairment and Alzheimer’s Disease by the Plasma and Serum Amyloid-Beta 42 Assay through Highly Sensitive Peptoid Nanosheet Sensor. ACS Appl. Mater. Interfaces 2020, 12, 9693–9700. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Wang, J.; Liu, J.; Ye, S.; Meng, X.; Song, S.; Wang, C.; Yu, X.; Zhu, L.; Wang, H.; et al. Peptoid Nanosheet-Based Sensing System for the Diagnosis and Surveillance of Amnestic Mild Cognitive Impairment and Alzheimer’s Disease. ACS Chem. Neurosci. 2021, 12, 4257–4264. [Google Scholar] [CrossRef] [PubMed]
- Yazdani, U.; Zaman, S.; Hynan, L.S.; Brown, L.S.; Dewey, R.B.; Karp, D.; German, D.C. Blood Biomarker for Parkinson Disease: Peptoids. NPJ Park. Dis. 2016, 2, 16012. [Google Scholar] [CrossRef]
- Feng, N.; Simanski, S.; Islam, K.; Hynan, L.S.; Kodadek, T.; German, D.C. Antibody Biomarker for de Novo Parkinson Disease: Attempted Validation. NPJ Park. Dis. 2018, 4, 28. [Google Scholar] [CrossRef]
- Gao, H.; Zhao, Z.; He, Z.; Wang, H.; Liu, M.; Hu, Z.; Cheng, O.; Yang, Y.; Zhu, L. Detection of Parkinson’s Disease through the Peptoid Recognizing α-Synuclein in Serum. ACS Chem. Neurosci. 2019, 10, 1204–1208. [Google Scholar] [CrossRef]
- Crane, J.M.; Lam, C.; Rossi, A.; Gupta, T.; Bennett, J.L.; Verkman, A.S. Binding Affinity and Specificity of Neuromyelitis Optica Autoantibodies to Aquaporin-4 M1/M23 Isoforms and Orthogonal Arrays. J. Biol. Chem. 2011, 286, 16516–16524. [Google Scholar] [CrossRef]
- Hayakawa, S.; Mori, M.; Okuta, A.; Kamegawa, A.; Fujiyoshi, Y.; Yoshiyama, Y.; Mitsuoka, K.; Ishibashi, K.; Sasaki, S.; Hattori, T. Neuromyelitis Optica and Anti-Aquaporin-4 Antibodies Measured by an Enzyme-Linked Immunosorbent Assay. J. Neuroimmunol. 2008, 196, 181–187. [Google Scholar] [CrossRef]
- Kalluri, S.R.; Illes, Z.; Srivastava, R.; Cree, B.; Menge, T.; Bennett, J.L.; Berthele, A.; Hemmer, B. Quantification and Functional Characterization of Antibodies to Native Aquaporin 4 in Neuromyelitis Optica. Arch. Neurol. 2010, 67, 1201–1208. [Google Scholar] [CrossRef]
- Waters, P.; Vincent, A. Detection of Anti-Aquaporin-4 Antibodies in Neuromyelitis Optica: Current Status of the Assays. Int. MS J. 2008, 15, 99–106. [Google Scholar] [PubMed]
- Waters, P.J.; McKeon, A.; Leite, M.I.; Rajasekharan, S.; Lennon, V.A.; Villalobos, A.; Palace, J.; Mandrekar, J.N.; Vincent, A.; Bar-Or, A. Serologic Diagnosis of NMO: A Multicenter Comparison of Aquaporin-4-IgG Assays. Neurology 2012, 78, 665–671. [Google Scholar] [CrossRef] [PubMed]
- Wingerchuk, D.M.; Lennon, V.A.; Pittock, S.J.; Lucchinetti, C.F.; Weinshenker, B. Revised Diagnostic Criteria for Neuromyelitis Optica. Neurology 2006, 66, 1485–1489. [Google Scholar] [CrossRef]
- Raveendra, B.L.; Wu, H.; Baccala, R.; Reddy, M.M.; Schilke, J.; Bennett, J.L.; Theofilopoulos, A.N.; Kodadek, T. Discovery of Peptoid Ligands for Anti-Aquaporin 4 Antibodies. Chem. Biol. 2013, 20, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Kavanaugh, A.F.; Solomon, D.H.; American College of Rheumatology Ad Hoc Committee on Immunologic Testing Guidelines. Guidelines for Immunologic Laboratory Testing in the Rheumatic Diseases: Anti-DNA Antibody Tests. Arthritis Care Res. 2002, 47, 546–555. [Google Scholar] [CrossRef]
- Benito-Garcia, E.; Schur, P.H.; Lahita, R.; American College of Rheumatology Ad Hoc Committee on Immunologic Testing Guidelines. Guidelines for Immunologic Laboratory Testing in the Rheumatic Diseases: Anti-Sm and anti-RNP Antibody Tests. Arthritis Care Res. 2004, 51, 1030–1044. [Google Scholar] [CrossRef]
- Arbuckle, M.R.; McClain, M.T.; Rubertone, M.V.; Scofield, R.H.; Dennis, G.J.; James, J.A.; Harley, J.B. Development of Autoantibodies before the Clinical Onset of Systemic Lupus Erythematosus. N. Engl. J. Med. 2003, 349, 1526–1533. [Google Scholar] [CrossRef]
- Olsen, N.J.; Li, Q.-Z.; Quan, J.; Wang, L.; Mutwally, A.; Karp, D.R. Autoantibody Profiling to Follow Evolution of Lupus Syndromes. Arthritis Res. Ther. 2012, 14, R174. [Google Scholar] [CrossRef]
- Quan, J.; Lakhanpal, A.; Reddy, M.M.; Zaman, S.; Li, Q.-Z.; German, D.C.; Olsen, N.J.; Kodadek, T.; Karp, D.R. Discovery of Biomarkers for Systemic Lupus Erythematosus Using a Library of Synthetic Autoantigen Surrogates. J. Immunol. Methods 2014, 402, 23–34. [Google Scholar] [CrossRef]
- Astle, J.M.; Simpson, L.S.; Huang, Y.; Reddy, M.M.; Wilson, R.; Connell, S.; Wilson, J.; Kodadek, T. Seamless Bead to Microarray Screening: Rapid Identification of the Highest Affinity Protein Ligands from Large Combinatorial Libraries. Chem. Biol. 2010, 17, 38–45. [Google Scholar] [CrossRef]
- Baio, J. Prevalence of Autism Spectrum Disorder among Children Aged 8 Years—Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2010. Morb. Mortal. Wkly. Rep. 2014, 63, 1–21. [Google Scholar]
- Dawson, G.; Rogers, S.; Munson, J.; Smith, M.; Winter, J.; Greenson, J.; Donaldson, A.; Varley, J. Randomized, Controlled Trial of an Intervention for Toddlers with Autism: The Early Start Denver Model. Pediatrics 2010, 125, e17–e23. [Google Scholar] [CrossRef] [PubMed]
- Zaman, S.; Yazdani, U.; Deng, Y.; Li, W.; Gadad, B.S.; Hynan, L.; Karp, D.; Roatch, N.; Schutte, C.; Nathan Marti, C.; et al. A Search for Blood Biomarkers for Autism: Peptoids. Sci. Rep. 2016, 6, 19164. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giorgio, A.; Del Gatto, A.; Pennacchio, S.; Saviano, M.; Zaccaro, L. Peptoids: Smart and Emerging Candidates for the Diagnosis of Cancer, Neurological and Autoimmune Disorders. Int. J. Mol. Sci. 2023, 24, 16333. https://doi.org/10.3390/ijms242216333
Giorgio A, Del Gatto A, Pennacchio S, Saviano M, Zaccaro L. Peptoids: Smart and Emerging Candidates for the Diagnosis of Cancer, Neurological and Autoimmune Disorders. International Journal of Molecular Sciences. 2023; 24(22):16333. https://doi.org/10.3390/ijms242216333
Chicago/Turabian StyleGiorgio, Anna, Annarita Del Gatto, Simone Pennacchio, Michele Saviano, and Laura Zaccaro. 2023. "Peptoids: Smart and Emerging Candidates for the Diagnosis of Cancer, Neurological and Autoimmune Disorders" International Journal of Molecular Sciences 24, no. 22: 16333. https://doi.org/10.3390/ijms242216333






