Inconspicuous Yet Indispensable: The Coronavirus Spike Transmembrane Domain
Abstract
:1. Introduction

2. Spike TMD Boundaries as Exemplified by SARS-CoV-2 S Protein
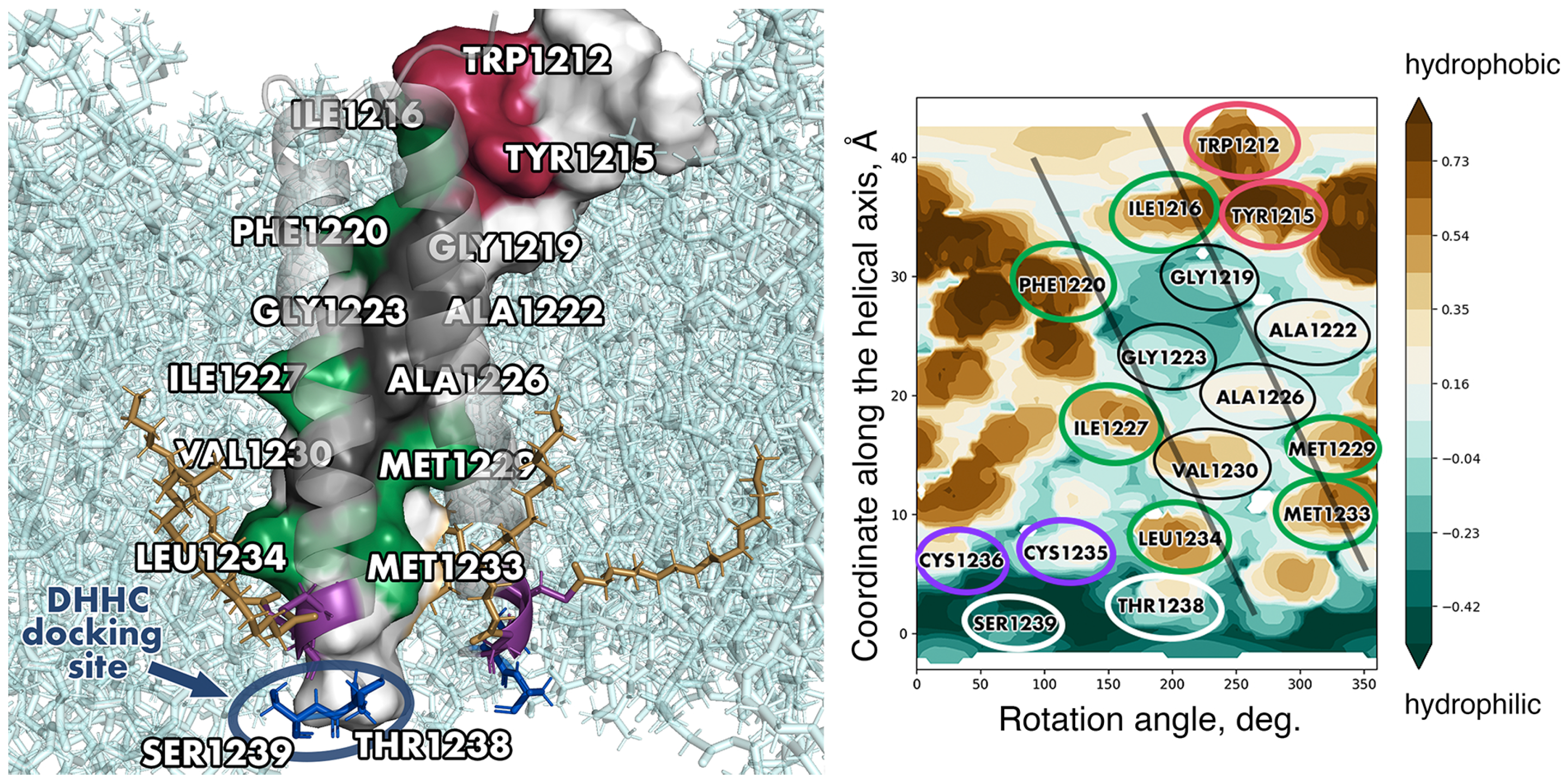
3. Spike TMD Conformation
4. The Aromatic-Rich Region
5. The Hydrophobic Region
6. The Cys-Rich Region
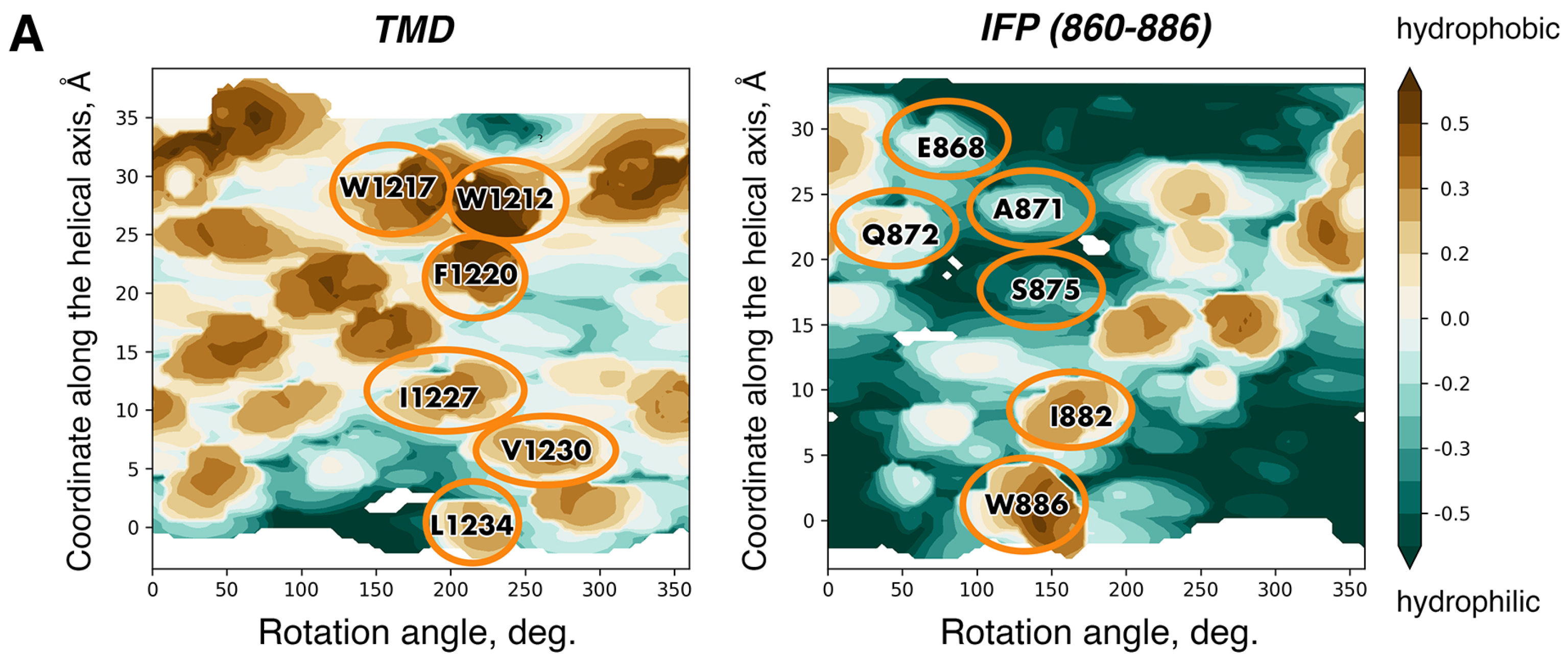
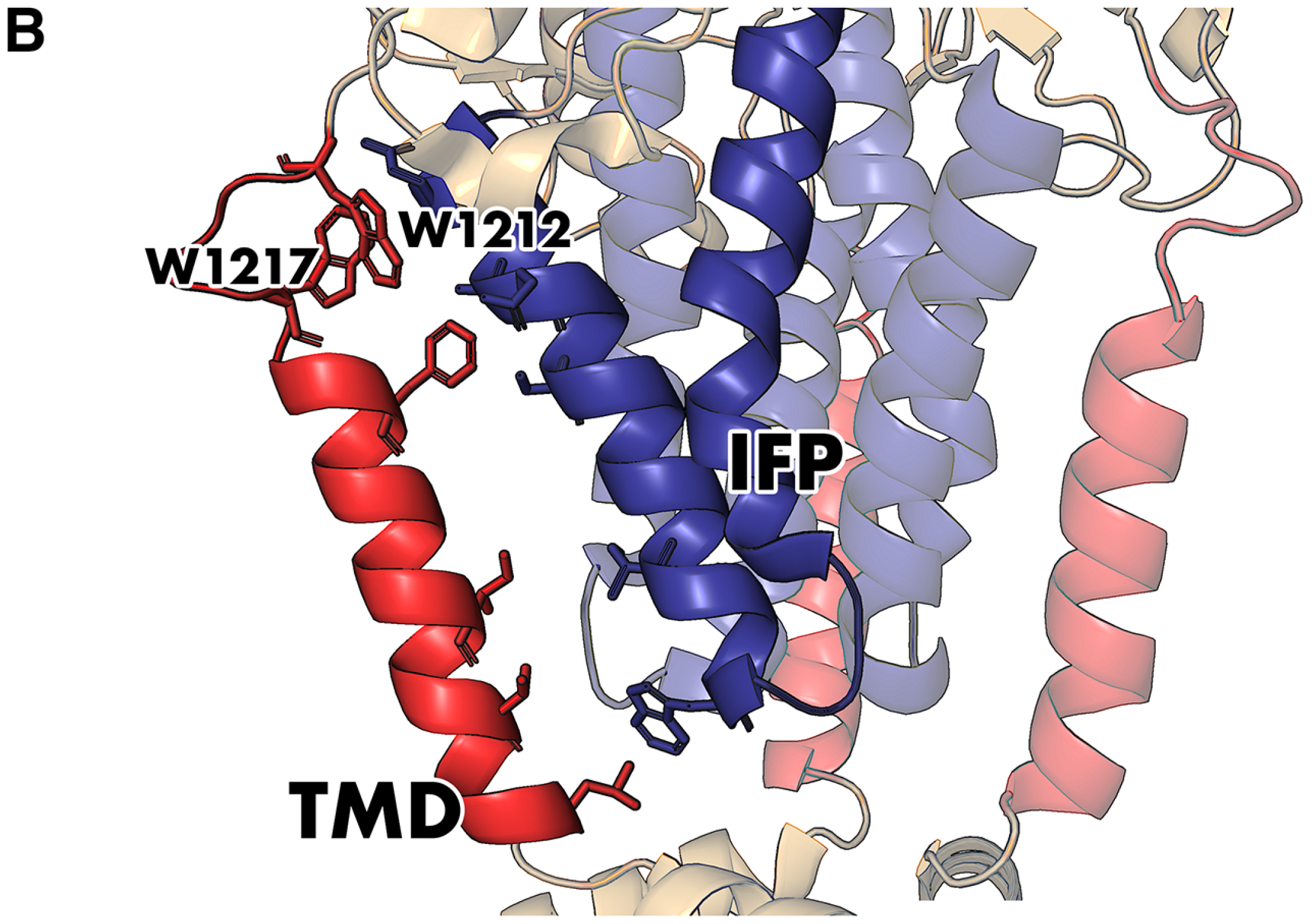
7. Beyond the TMD Trimer
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| Chol | cholesterol |
| Cys-rich | cysteine-rich |
| DH6PC | 1,2-Dihexanoyl-sn-Glycero-3-Phosphocholine |
| DMPC | dimyristoylphosphatidylcholine |
| DMGP | dimyristoylphosphatidylglycerol |
| DPC | dodecylphosphocholine |
| DRM | detergent-resistant membrane |
| FP | fusion peptide |
| IFP | internal fusion peptide |
| JM | juxtamembrane |
| MC | Monte Carlo |
| MD | molecular dynamics |
| MHV | mouse hepatitis virus |
| MHP | molecular hydrophobicity potential |
| PC | phosphatidylcholine |
| POPC | 1-palmitoyl-2-oleoyl-sn-3-glycerophospholcholine |
| pp | pseudotyped particles |
| PTM | pre-transmembrane |
| RBD | receptor-binding domain |
| SM | sphingomyelin |
| TFE | trifluoroethanol |
| TGEV | transmissible gastroenteritis virus |
| TM | transmembrane |
| TMD | transmembrane domain |
| VSV | vesicular stomatitis virus |
Appendix A
| Virus | Modifications Introduced to Spike/ Other Factors Present | Results | Ref. |
|---|---|---|---|
| SARS-CoV | First and second cysteine clusters (ALA) | S-mediated cell fusion levels dropped by 55% and 60%, respectively | [21] |
| Third and fourth cysteine clusters (ALA) | S-mediated cell fusion levels dropped less markedly | idem | |
| C1217, 1218A | 3H-palmitic acid incorporation dropped by 56% | idem | |
| C1222, 1223, 1225A | 3H-palmitic acid incorporation dropped by 49% | idem | |
| C1230, 1232A | 3H-palmitic acid incorporation similar to wild-type | idem | |
| C1235,1236A | 3H-palmitic acid incorporation dropped by 5% | idem | |
| All 9 Cys residues | The mutant spike was capable of interacting with the M protein, but the mutant virus’s capacity for cell–cell fusion was impaired | [62] | |
| Fourth cysteine cluster mutated | The mutant failed to incorporate into virus particles. Mutations of any cysteine did not affect interaction with the M protein | [63] | |
| SARS-CoV-2 | All 10 cysteines (ALA) | Pseudotyped particles bound to ACE2, but lost the ability to fuse target cells. For virus-like particles, fusion levels dropped 20- to 40-fold | [60] |
| Expression in cells where ZDHHC20 and ZDHHC9 were inactivated | Accumulation of the E protein and RdRp dropped by approx. 75% | idem | |
| C1235, 1236A | 3H-palmitic acid incorporation dropped by 80% | idem | |
| C1240, 1241A | 3H-palmitic acid incorporation dropped by 40% | idem | |
| Cys clusters III and IV | 3H-palmitic acid incorporation comparable to wild-type when mutated separately, but dropped more markedly with both III and IV mutated | idem | |
| All 10 cysteines (ALA) | Background levels of infectivity and cell–cell fusion. Palmitoylation not detected | [64] | |
| C1235, C1236, C1240, C1241, C1243 (ALA) | Background levels of infectivity and cell–cell fusion. Palmitoylation levels dropped | idem | |
| C1247, C1248, C1250, C1253, C1254 (ALA) | Infectivity and cell–cell fusion did not alter significantly. Palmitoylation levels dropped less dramatically than for the mutant mentioned above | idem | |
| All 10 cysteines (ALA) | Palmitoylation levels dropped to zero. Pseudoparticles incorporated wild-type levels of the spike, but the level of pp–cell fusion and cell–cell fusion decreased dramatically | [61] | |
| 2-bromopalmitate exposure | Palmitoylation levels dropped; spike fusion was significantly impaired | idem | |
| An array of mutants with all possible variants of Cys clusters mutated to Ala or kept intact | Cys clusters I and II proved to be the ones most significantly affecting the palmitoylation of the spike, especially when mutated simultaneously | [65] | |
| T1238A, S1239A | Both residues are required for palmitoylation | [66] | |
| MHV | A59: C1287, 1288, 1289S | Capacity for cell–cell fusion was lost | [45] |
| A59: C1295, 1296S/F1297N | Capacity for cell–cell fusion was impaired | idem | |
| A59: C1295S/F1297N | Capacity for cell–cell fusion was not affected | idem | |
| A59: C1287, C1292, C1296, C1304 (each with SER, TYR, TRP) | In all cases, fusogenic activity was markedly impaired, i.e., each position contributes to fusion | [46] | |
| A59: C1300, C1303, C1304 (ALA) | Single (1304), double (1303/1304) and triple mutant pseudovirions showed fusion levels 2, 20 and 40 times lower compared with the wild-type, respectively. Affinity to CEACAM was not affected. S2 refolding was shown to slow down significantly. Mutations correlated with failure to associate with the M protein | [67] | |
| A59: deletion of T1290 downstream | Chimaeric protein w/heterologous ectodomain containing the MHV TMD failed to be incorporated into virions, the virus was not viable | [47] | |
| A59: deletion of T1290-N1302 | Spike failed to cause fusion, despite the same deletion not significantly affecting the incorporation of a counterpart chimaera with a heterologous ectodomain into virions | idem | |
| A59: a variety of C/A, C/G, C/S mutants | A minimum of three Cys residues required for viral replication | [68] | |
| 2-bromopalmitate exposure (palmitoyl acyltransferase inhibitor) | Spike accumulated poorly in virions and failed to associate with the M protein. Infectivity was impaired | [69] | |
| JHM: C1347F, C1348F, C1348S (homologous to C1295/C1296 in A59) | Double mutant C1347F/C1348S did not demonstrate lower level of virion incorporation, but a much lower infectivity. Double mutant C1347S/C1348F showed a 10-fold drop in infectivity. Single mutant C1348S showed wild-type-level infectivity. Palmitoylation at these positions is not crucial to spike incorporation into virions | idem | |
| TGEV | 2-bromopalmitate exposure | Affects infectivity | [70] |
| A variety of mutants carrying 3 to 10 C/A substitutions | Mutating all 10 Cys residues affected S incorporation into virions. Three cysteines from any cluster were sufficient for spike incorporation. Interaction with the M protein was not affected | idem |
References
- Wrobel, A.G. Mechanism and evolution of human ACE2 binding by SARS-CoV-2 spike. Curr. Opin. Struct Biol. 2023, 81, 102619. [Google Scholar] [CrossRef]
- Qi, H.; Liu, B.; Wang, X.; Zhang, L. The humoral response and antibodies against SARS-CoV-2 infection. Nat. Immunol. 2022, 23, 1008–1020. [Google Scholar] [CrossRef] [PubMed]
- Kordyukova, L.V.; Shanko, A.V. COVID-19: Myths and Reality. Biochemistry 2021, 86, 800–817. [Google Scholar] [CrossRef]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Basso, L.G.M.; Zeraik, A.E.; Felizatti, A.P.; Costa-Filho, A.J. Membranotropic and biological activities of the membrane fusion peptides from SARS-CoV spike glycoprotein: The importance of the complete internal fusion peptide domain. Biochim. Biophys. Acta Biomembr. 2021, 1863, 183697. [Google Scholar] [CrossRef]
- Bocharov, E.V.; Mineev, K.S.; Pavlov, K.V.; Akimov, S.A.; Kuznetsov, A.S.; Efremov, R.G.; Arseniev, A.S. Helix-helix interactions in membrane domains of bitopic proteins: Specificity and role of lipid environment. Biochim. Biophys. Acta Biomembr. 2017, 1859, 561–576. [Google Scholar] [CrossRef] [PubMed]
- Ellaithy, A.; Gonzalez-Maeso, J.; Logothetis, D.A.; Levitz, J. Structural and Biophysical Mechanisms of Class C G Protein-Coupled Receptor Function. Trends Biochem. Sci. 2020, 45, 1049–1064. [Google Scholar] [CrossRef] [PubMed]
- Westerfield, J.M.; Barrera, F.N. Membrane receptor activation mechanisms and transmembrane peptide tools to elucidate them. J. Biol. Chem. 2020, 295, 1792–1814. [Google Scholar] [CrossRef]
- Bosch, B.J.; van der Zee, R.; de Haan, C.A.; Rottier, P.J. The coronavirus spike protein is a class I virus fusion protein: Structural and functional characterization of the fusion core complex. J. Virol. 2003, 77, 8801–8811. [Google Scholar] [CrossRef]
- Shi, W.; Cai, Y.; Zhu, H.; Peng, H.; Voyer, J.; Rits-Volloch, S.; Cao, H.; Mayer, M.L.; Song, K.; Xu, C.; et al. Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane. Nature 2023, 619, 403–409. [Google Scholar] [CrossRef]
- Bocharov, E.V.; Volynsky, P.E.; Pavlov, K.V.; Efremov, R.G.; Arseniev, A.S. Structure elucidation of dimeric transmembrane domains of bitopic proteins. Cell Adhes. Migr. 2010, 4, 284–298. [Google Scholar] [CrossRef]
- Chiliveri, S.C.; Louis, J.M.; Ghirlando, R.; Bax, A. Transient lipid-bound states of spike protein heptad repeats provide insights into SARS-CoV-2 membrane fusion. Sci. Adv. 2021, 7, eabk2226. [Google Scholar] [CrossRef]
- Tan, Z.W.; Tee, W.V.; Samsudin, F.; Guarnera, E.; Bond, P.J.; Berezovsky, I.N. Allosteric perspective on the mutability and druggability of the SARS-CoV-2 Spike protein. Structure 2022, 30, 590–607. [Google Scholar] [CrossRef] [PubMed]
- Thoma, J.; Burmann, B.M. Fake It ’Till You Make It-The Pursuit of Suitable Membrane Mimetics for Membrane Protein Biophysics. Int. J. Mol. Sci. 2020, 22, 50. [Google Scholar] [CrossRef]
- Bernhofer, M.; Kloppmann, E.; Reeb, J.; Rost, B. TMSEG: Novel prediction of transmembrane helices. Proteins 2016, 84, 1706–1716. [Google Scholar] [CrossRef]
- Hofmann, K.; Stoffel, W. TMbase-a database of membrane spanning proteins segments. Biol. Chem. Hoppe Seyler 1993, 347, 166. [Google Scholar]
- The UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Zhu, Y.; Liu, M.; Lan, Q.; Xu, W.; Wu, Y.; Ying, T.; Liu, S.; Shi, Z.; Jiang, S.; et al. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell. Mol. Immunol. 2020, 17, 765–767. [Google Scholar] [CrossRef]
- Cai, Y.; Zhang, J.; Xiao, T.; Peng, H.; Sterling, S.M.; Walsh, R.M., Jr.; Rawson, S.; Rits-Volloch, S.; Chen, B. Distinct conformational states of SARS-CoV-2 spike protein. Science 2020, 369, 1586–1592. [Google Scholar] [CrossRef]
- Aliper, E.T.; Krylov, N.A.; Nolde, D.E.; Polyansky, A.A.; Efremov, R.G. A Uniquely Stable Trimeric Model of SARS-CoV-2 Spike Transmembrane Domain. Int. J. Mol. Sci. 2022, 23, 9221. [Google Scholar] [CrossRef]
- Petit, C.M.; Chouljenko, V.N.; Iyer, A.; Colgrove, R.; Farzan, M.; Knipe, D.M.; Kousoulas, K.G. Palmitoylation of the cysteine-rich endodomain of the SARS-coronavirus spike glycoprotein is important for spike-mediated cell fusion. Virology 2007, 360, 264–274. [Google Scholar] [CrossRef]
- Li, Q.; Huang, Q.; Kang, C. Secondary Structures of the Transmembrane Domain of SARS-CoV-2 Spike Protein in Detergent Micelles. Int. J. Mol. Sci. 2022, 23, 1040. [Google Scholar] [CrossRef] [PubMed]
- Dev, J.; Park, D.; Fu, Q.; Chen, J.; Ha, H.J.; Ghantous, F.; Herrmann, T.; Chang, W.; Liu, Z.; Frey, G.; et al. Structural basis for membrane anchoring of HIV-1 envelope spike. Science 2016, 353, 172–175. [Google Scholar] [CrossRef] [PubMed]
- Benton, D.J.; Nans, A.; Calder, L.J.; Turner, J.; Neu, U.; Lin, Y.P.; Ketelaars, E.; Kallewaard, N.L.; Corti, D.; Lanzavecchia, A.; et al. Influenza hemagglutinin membrane anchor. Proc. Natl. Acad. Sci. USA 2018, 115, 10112–10117. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Chou, J.J. A Trimeric Hydrophobic Zipper Mediates the Intramembrane Assembly of SARS-CoV-2 Spike. J. Am. Chem. Soc. 2021, 143, 8543–8546. [Google Scholar] [CrossRef]
- Lemmon, M.; Engelman, D.M. Specificity and promiscuity in membrane helix interactions. Quart. Rev. Biophys. 1994, 27, 157–218. [Google Scholar] [CrossRef]
- Efremov, R.G. Dielectric-Dependent Strength of Interlipid Hydrogen Bonding in Biomembranes: Model Case Study. J. Chem. Inf. Model. 2019, 59, 2765–2775. [Google Scholar] [CrossRef] [PubMed]
- Bragin, P.E.; Kuznetsov, A.S.; Bocharova, O.V.; Volynsky, P.E.; Arseniev, A.S.; Efremov, R.G.; Mineev, K.S. Probing the effect of membrane contents on transmembrane protein–protein interaction using solution NMR and computer simulations. Biochim. Biophys. Acta Biomembr. 2018, 1860, 2486–2498. [Google Scholar] [CrossRef]
- Mahajan, M.; Bhattacharjya, S. NMR structures and localization of the potential fusion peptides and the pre-transmembrane region of SARS-CoV: Implications in membrane fusion. Biochim. Biophys. Acta 2015, 1848, 721–730. [Google Scholar] [CrossRef]
- Sainz, B.; Rausch, J.M.; Gallaher, W.R.; Garry, R.F.; Wimley, W.C. The Aromatic Domain of the Coronavirus Class I Viral Fusion Protein Induces Membrane Permeabilization: Putative Role during Viral Entry. Biochemistry 2005, 44, 947–958. [Google Scholar] [CrossRef]
- Guillen, J.; Moreno, M.R.; Perez-Berna, A.J.; Bernabeu, A.; Villalain, J. Interaction of a peptide from the pre-transmembrane domain of the severe acute respiratory syndrome coronavirus spike protein with phospholipid membranes. J. Phys. Chem. B 2007, 111, 13714–13725. [Google Scholar] [CrossRef]
- Liao, Y.; Zhang, S.M.; Neo, T.L.; Tam, J.P. Tryptophan-dependent membrane interaction and heteromerization with the internal fusion peptide by the membrane proximal external region of SARS-CoV spike protein. Biochemistry 2015, 54, 1819–1830. [Google Scholar] [CrossRef]
- Turoňová, B.; Sikora, M.; Schürmann, C.; Hagen, W.J.H.; Welsch, S.; Blanc, F.E.C.; von Bülow, S.; Gecht, M.; Bagola, K.; Hörner, C.; et al. In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science 2020, 370, 203–208. [Google Scholar] [CrossRef]
- Rees, D.C.; DeAntonio, L.; Eisenberg, D. Hydrophobic organization of membrane proteins. Science 1989, 245, 510–513. [Google Scholar] [CrossRef] [PubMed]
- Lemmon, M.; Engelman, D.M. Helix-helix interactions inside lipid bilayers. Curr. Opin. Struct. Biol. 1992, 2, 511–518. [Google Scholar] [CrossRef]
- Donnelly, D.; Overington, J.P.; Ruffle, S.V.; Nugent, J.H.; Blundell, T.L. Modeling α-helical transmembrane domains: The calculation and use of substitution tables for lipid-facing residues. Protein Sci. 1993, 2, 55–70. [Google Scholar]
- Yu, J.; Qiao, S.; Guo, R.; Wang, X. Cryo-EM structures of HKU2 and SADS-CoV spike glycoproteins provide insights into coronavirus evolution. Nat. Commun. 2020, 11, 3070. [Google Scholar] [CrossRef]
- Guillen, J.; Perez-Berna, A.J.; Moreno, M.R.; Villalain, J. Identification of the membrane-active regions of the severe acute respiratory syndrome coronavirus spike membrane glycoprotein using a 16/18-mer Peptide scan: Implications for the viral fusion mechanism. J. Virol. 2005, 79, 1743–1752. [Google Scholar] [CrossRef] [PubMed]
- Guillen, J.; Kinnunen, P.K.; Villalain, J. Membrane insertion of the three main membranotropic sequences from SARS-CoV S2 glycoprotein. Biochim. Biophys. Acta 2008, 1778, 2765–2774. [Google Scholar] [CrossRef] [PubMed]
- Corver, J.; Broer, R.; van Kasteren, P.; Spaan, W. Mutagenesis of the transmembrane domain of the SARS coronavirus spike glycoprotein: Refinement of the requirements for SARS coronavirus cell entry. Virol. J. 2009, 6, 230. [Google Scholar] [CrossRef]
- Howard, M.W.; Travanty, E.A.; Jeffers, S.A.; Smith, M.K.; Wennier, S.T.; Thackray, L.B.; Holmes, K.V. Aromatic amino acids in the juxtamembrane domain of SARS coronavirus spike glycoprotein are important for receptor-dependent virus entry and cell–cell fusion. J. Virol. 2008, 82, 2883–2894. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Neo, T.L.; Liu, D.X.; Tam, J.P. Importance of SARS-CoV spike protein Trp-rich region in viral infectivity. Biochem. Biophys. Res. Commun. 2008, 371, 356–360. [Google Scholar] [CrossRef]
- Barrett, C.T.; Dutch, R.E. Viral Membrane Fusion and the Transmembrane Domain. Viruses 2020, 12, 693. [Google Scholar] [CrossRef] [PubMed]
- Broer, R.; Boson, B.; Spaan, W.; Cosset, F.L.; Corver, J. Important role for the transmembrane domain of severe acute respiratory syndrome coronavirus spike protein during entry. J. Virol. 2006, 80, 1302–1310. [Google Scholar] [CrossRef]
- Bos, E.C.; Heijnen, L.; Luytjes, W.; Spaan, W.J. Mutational analysis of the murine coronavirus spike protein: Effect on cell-to-cell fusion. Virology 1995, 214, 453–463. [Google Scholar] [CrossRef]
- Chang, K.W.; Sheng, Y.; Gombold, J.L. Coronavirus-induced membrane fusion requires the cysteine-rich domain in the spike protein. Virology 2000, 269, 212–224. [Google Scholar] [CrossRef] [PubMed]
- Ye, R.; Montalto-Morrison, C.; and Masters, P.S. Genetic analysis of determinants for spike glycoprotein assembly into murine coronavirus virions: Distinct roles for charge-rich and cysteine-rich regions of the endodomain. J. Virol. 2004, 78, 9904–9917. [Google Scholar] [CrossRef]
- Bosch, B.J.; de Haan, C.A.; Smits, S.L.; Rottier, P.J. Spike protein assembly into the coronavirion: Exploring the limits of its sequence requirements. Virology 2005, 334, 306–318. [Google Scholar] [CrossRef]
- Arbely, E.; Granot, Z.; Kass, I.; Orly, J.; Arkin, I.T. A trimerizing GxxxG motif is uniquely inserted in the severe acute respiratory syndrome (SARS) coronavirus spike protein transmembrane domain. Biochemistry 2006, 45, 11349–11356. [Google Scholar] [CrossRef] [PubMed]
- Corver, J.; Broer, R.; van Kasteren, P.; Spaan, W. GxxxG motif of SARS coronavirus spike glycoprotein transmembrane domain is not involved in trimerization and is not important for entry. J. Virol. 2007, 81, 8352–8355. [Google Scholar] [CrossRef]
- Webb, S.R.; Smith, S.E.; Fried, M.G.; Dutch, R.E. Transmembrane Domains of Highly Pathogenic Viral Fusion Proteins Exhibit Trimeric Association In Vitro. mSphere 2018, 3, e00047–18. [Google Scholar] [CrossRef]
- Azad, T.; Singaravelu, R.; Crupi, M.J.F.; Jamieson, T.; Dave, J.; Brown, E.E.F.; Rezaei, R.; Taha, Z.; Boulton, S.; Martin, N.T.; et al. Implications for SARS-CoV-2 Vaccine Design: Fusion of Spike Glycoprotein Transmembrane Domain to Receptor-Binding Domain Induces Trimerization. Membranes 2020, 10, 215. [Google Scholar] [CrossRef]
- Izvorski, A. Predicted 3D Models of the SARS-CoV-2 Spike Protein Membrane Proximal External Region and Transmembrane Domain. Available online: https://chemrxiv.org/engage/chemrxiv/article-details/60c74f99842e651900db387b (accessed on 17 March 2021).
- Nishima, W.; Kulik, M. Full-Length Computational Model of the SARS-CoV-2 Spike Protein and Its Implications for a Viral Membrane Fusion Mechanism. Viruses 2021, 13, 1126. [Google Scholar] [CrossRef] [PubMed]
- Casalino, L.; Gaieb, Z.; Goldsmith, J.A.; Hjorth, C.K.; Dommer, A.C.; Harbison, A.M.; Fogarty, C.A.; Barros, E.P.; Taylor, B.C.; McLellan, J.S.; et al. Beyond Shielding: The Roles of Glycans in the SARS-CoV-2 Spike Protein. ACS Cent. Sci. 2020, 6, 1722–1734. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.; Park, S.J.; Choi, Y.K.; Park, T.; Tanveer, M.; Cao, Y.; Kern, N.R.; Lee, J.; Yeom, M.S.; Croll, T.I.; et al. Developing a Fully Glycosylated Full-Length SARS-CoV-2 Spike Protein Model in a Viral Membrane. J. Phys. Chem. B. 2020, 124, 7128–7137. [Google Scholar] [CrossRef] [PubMed]
- Efremov, R.G.; Gulyaev, D.I.; Vergoten, G.; Modyanov, N.N. Application of three-dimensional molecular hydrophobicity potential to the analysis of spatial organization of membrane domains in proteins: I. Hydrophobic properties of transmembrane segments of Na+, K+-ATPase. J. Protein Chem. 1992, 11, 665–675. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.F.G. Acylation of virol. spike glycoproteins: A feature of enveloped RNA viruses. Virology 1982, 116, 327–338. [Google Scholar] [CrossRef]
- Gadalla, M.R.; Veit, M. Toward the identification of ZDHHC enzymes required for palmitoylation of viral protein as potential drug targets. Expert Opin. Drug Discov. 2020, 15, 159–177. [Google Scholar] [CrossRef] [PubMed]
- Mesquita, F.S.; Abrami, L.; Sergeeva, O.; Turelli, P.; Qing, E.; Kunz, B.; Raclot, C.; Paz Montoya, J.; Abriata, L.A.; Gallagher, T.; et al. S-acylation controls SARS-CoV-2 membrane lipid organization and enhances infectivity. Dev. Cell 2021, 56, 2790–2807. [Google Scholar] [CrossRef]
- Wu, Z.; Zhang, Z.; Wang, X.; Zhang, J.; Ren, C.; Li, Y.; Gao, L.; Liang, X.; Wang, P.; Ma, C. Palmitoylation of SARS-CoV-2 S protein is essential for viral infectivity. Signal Transduct. Target. Ther. 2021, 6, 231. [Google Scholar] [CrossRef]
- McBride, C.E.; Machamer, C.E. Palmitoylation of SARS-CoV S protein is necessary for partitioning into detergent-resistant membranes and cell–cell fusion but not interaction with M protein. Virology 2010, 405, 139–148. [Google Scholar] [CrossRef]
- Ujike, M.; Huang, C.; Shirato, K.; Matsuyama, S.; Makino, S.; Taguchi, F. Two palmitylated cysteine residues of the severe acute respiratory syndrome coronavirus spike (S) protein are critical for S incorporation into virus-like particles, but not for M-S co-localization. J. Gen. Virol. 2012, 93, 823–828. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Zhang, S.; Wang, Q.; Anang, S.; Wang, J.; Ding, H.; Kappes, J.C.; Sodroski, J. Spike glycoprotein and host cell determinants of SARS-CoV-2 entry and cytopathic effects. J. Virol. 2020, 95, e02304-20. [Google Scholar] [CrossRef]
- Puthenveetil, R.; Lun, C.M.; Murphy, R.E.; Healy, L.B.; Vilmen, G.; Christenson, E.T.; Freed, E.O.; Banerjee, A. S-acylation of SARS-CoV-2 spike protein: Mechanistic dissection, in vitro reconstitution and role in viral infectivity. J. Biol. Chem. 2021, 297, 101112. [Google Scholar] [CrossRef]
- Panina, I.; Krylov, N.; Gadalla, M.R.; Aliper, E.; Kordyukova, L.; Veit, M.; Chugunov, A.; Efremov, R. Molecular Dynamics of DHHC20 Acyltransferase Suggests Principles of Lipid and Protein Substrate Selectivity. Int. J. Mol. Sci. 2022, 23, 5091. [Google Scholar] [CrossRef]
- Shulla, A.; Gallagher, T. Role of spike protein endodomains in regulating coronavirus entry. J. Biol. Chem. 2009, 284, 32725–32734. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Lv, J.; Wang, Y.; Gao, S.; Yao, Q.; Qu, D.; Ye, R. Replication of murine coronavirus requires multiple cysteines in the endodomain of spike protein. Virology 2012, 427, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Thorp, E.B.; Boscarino, J.A.; Logan, H.L.; Goletz, J.T.; Gallagher, T.M. Palmitoylations on murine coronavirus spike proteins are essential for virion assembly and infectivity. J. Virol. 2006, 80, 1280–1289. [Google Scholar] [CrossRef]
- Gelhaus, S.; Thaa, B.; Eschke, K.; Veit, M.; Schwegmann-Weßels, C. Palmitoylation of the Alphacoronavirus TGEV spike protein S is essential for incorporation into virus-like particles but dispensable for S–M interaction. Virology 2014, 464–465, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Polyansky, A.A.; Volynsky, P.E.; Efremov, R.G. Multistate organisation of transmembrane helical protein dimers governed by the host membrane. J. Am. Chem. Soc. 2012, 134, 14390–14400. [Google Scholar] [CrossRef] [PubMed]
- Polyansky, A.A.; Chugunov, A.O.; Volynsky, P.E.; Krylov, N.A.; Nolde, D.E.; Efremov, R.G. PREDDIMER: A web server for prediction of transmembrane helical dimers. Bioinformatics 2014, 30, 889–890. [Google Scholar] [CrossRef]
- Polyansky, A.A.; Efremov, R.G. On a mechanistic impact of transmembrane tetramerization in the pathological activation of RTKs. Comput. Struct Biotechnol. J. 2023, 21, 2837–2844. [Google Scholar] [CrossRef]
- Overduin, M.; Tran, A.; Eekels, D.M.; Overduin, F.; Kervin, T.A. Transmembrane Membrane Readers form a Novel Class of Proteins That Include Peripheral Phosphoinositide Recognition Domains and Viral Spikes. Membranes 2022, 12, 1161. [Google Scholar] [CrossRef] [PubMed]
- de Planque, M.R.; Bonev, B.B.; Demmers, J.A.; Greathouse, D.V.; Koeppe, R.E., 2nd; Separovic, F.; Watts, A.; Killian, J.A. Interfacial anchor properties of tryptophan residues in transmembrane peptides can dominate over hydrophobic matching effects in peptide-lipid interactions. Biochemistry 2003, 42, 5341–5348. [Google Scholar] [CrossRef]
- Epand, R.M. Cholesterol and the interaction of proteins with membrane domains. Prog. Lipid Res. 2006, 45, 279–294. [Google Scholar] [CrossRef]
- Zakany, F.; Mándity, I.M.; Varga, Z.; Panyi, G.; Nagy, P.; Kovacs, T. Effect of the Lipid Landscape on the Efficacy of Cell-Penetrating Peptides. Cells 2023, 12, 1700. [Google Scholar] [CrossRef]
- Yang, S.T.; Kreutzberger, A.J.B.; Lee, J.; Kiessling, V.; Tamm, L.K. The role of cholesterol in membrane fusion. Chem. Phys. Lipids 2016, 199, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Chlanda, P.; Mekhedov, E.; Waters, H.; Sodt, A.; Schwartz, C.; Nair, V.; Blank, P.S.; Zimmerberg, J. Palmitoylation Contributes to Membrane Curvature in Influenza A Virus Assembly and Hemagglutinin-Mediated Membrane Fusion. J. Virol. 2017, 91, e00947-17. [Google Scholar] [CrossRef]
- Yang, S.T.; Kiessling, V.; Simmons, J.A.; White, J.M.; Tamm, L.K. HIV gp41-mediated membrane fusion occurs at edges of cholesterol-rich lipid domains. Nat. Chem. Biol. 2015, 11, 424–431. [Google Scholar] [CrossRef]
- Yang, S.T.; Kiessling, V.; Tamm, L. Line tension at lipid phase boundaries as driving force for HIV fusion peptide-mediated fusion. Nat. Commun. 2016, 7, 11401. [Google Scholar] [CrossRef] [PubMed]
- Chlanda, P.; Mekhedov, E.; Waters, H.; Schwartz, C.L.; Fischer, E.R.; Ryham, R.J.; Cohen, F.S.; Blank, P.S.; Zimmerberg, J. The hemifusion structure induced by influenza virus haemagglutinin is determined by physical properties of the target membranes. Nat. Microbiol. 2016, 18, 16050. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kreutzberger, A.J.B.; Odongo, L.; Nelson, E.A.; Nyenhuis, D.A.; Kiessling, V.; Liang, B.; Cafiso, D.S.; White, J.M.; Tamm, L.K. Ebola virus glycoprotein interacts with cholesterol to enhance membrane fusion and cell entry. Nat. Struct. Mol. Biol. 2021, 28, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Sanders, D.W.; Jumper, C.C.; Ackerman, P.J.; Bracha, D.; Donlic, A.; Kim, H.; Kenney, D.; Castello-Serrano, I.; Suzuki, S.; Tamura, T.; et al. SARS-CoV-2 requires cholesterol for viral entry and pathological syncytia formation. eLife 2021, 10, e65962. [Google Scholar] [CrossRef] [PubMed]
- Bippes, C.A.; Muller, D.J. High-resolution atomic force microscopy and spectroscopy of native membrane proteins. Rep. Prog. Phys. 2011, 74, 086601. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aliper, E.T.; Efremov, R.G. Inconspicuous Yet Indispensable: The Coronavirus Spike Transmembrane Domain. Int. J. Mol. Sci. 2023, 24, 16421. https://doi.org/10.3390/ijms242216421
Aliper ET, Efremov RG. Inconspicuous Yet Indispensable: The Coronavirus Spike Transmembrane Domain. International Journal of Molecular Sciences. 2023; 24(22):16421. https://doi.org/10.3390/ijms242216421
Chicago/Turabian StyleAliper, Elena T., and Roman G. Efremov. 2023. "Inconspicuous Yet Indispensable: The Coronavirus Spike Transmembrane Domain" International Journal of Molecular Sciences 24, no. 22: 16421. https://doi.org/10.3390/ijms242216421





