225Ac-iPSMA-RGD for Alpha-Therapy Dual Targeting of Stromal/Tumor Cell PSMA and Integrins
Abstract
:1. Introduction
2. Results and Discussion
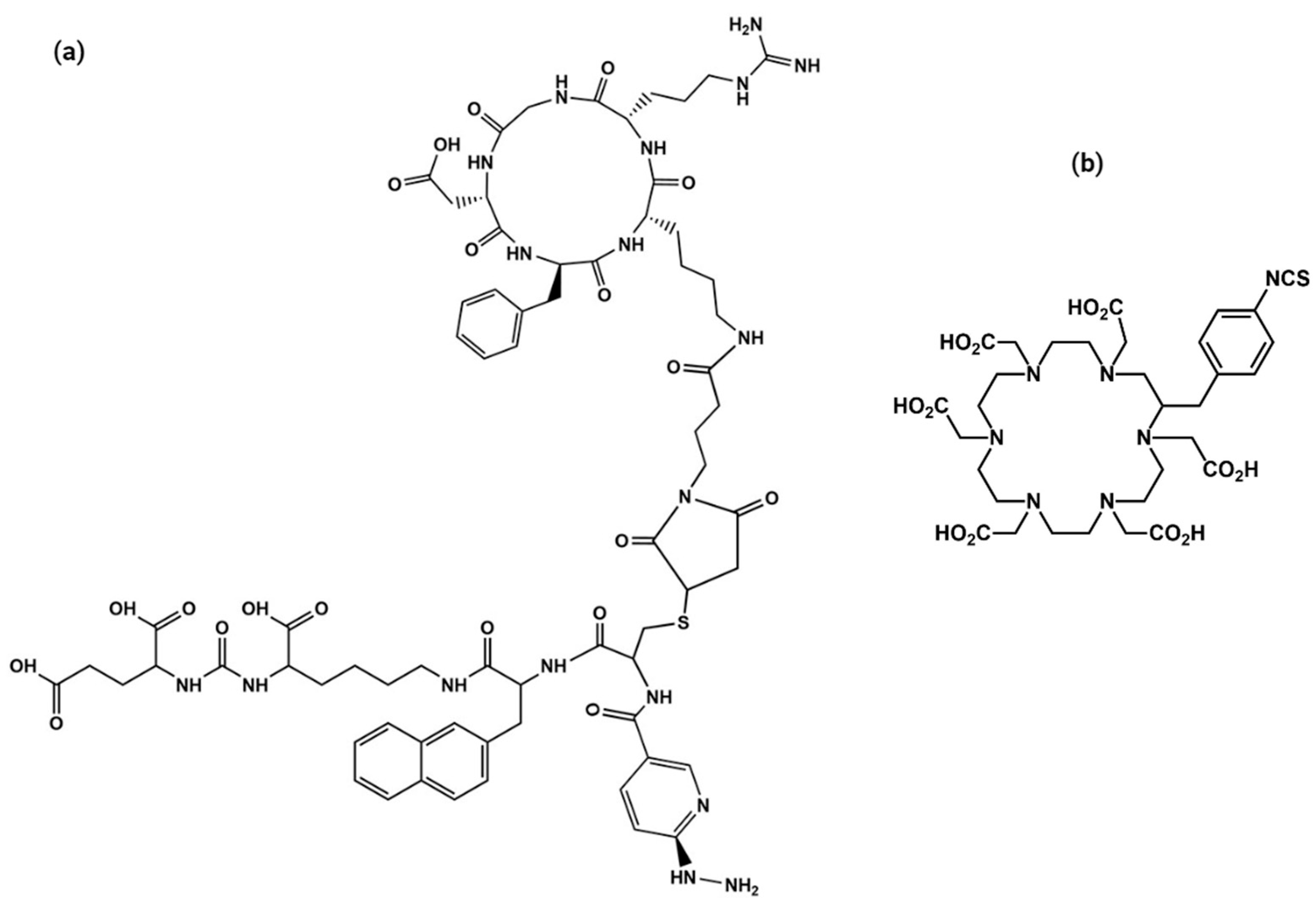
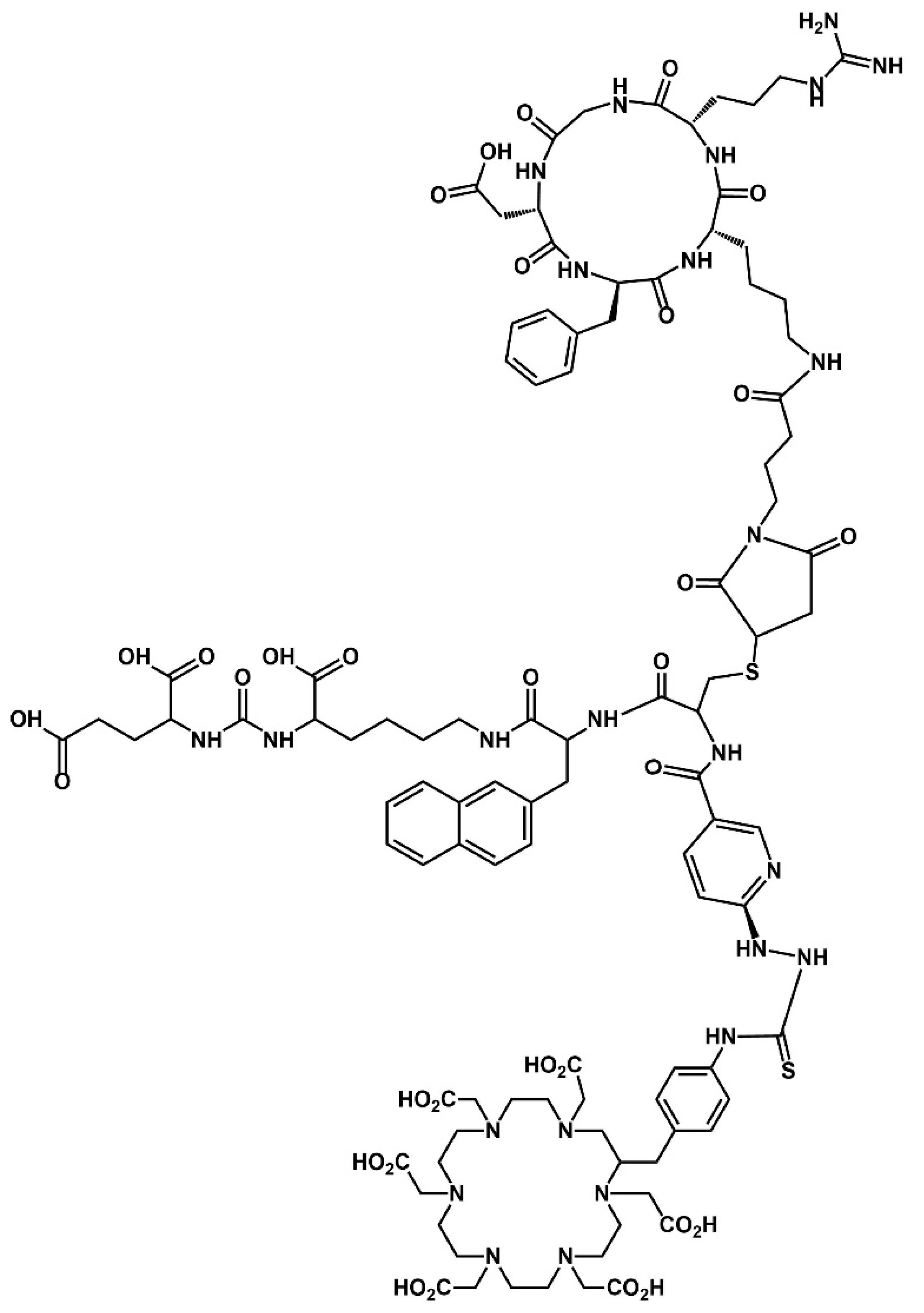
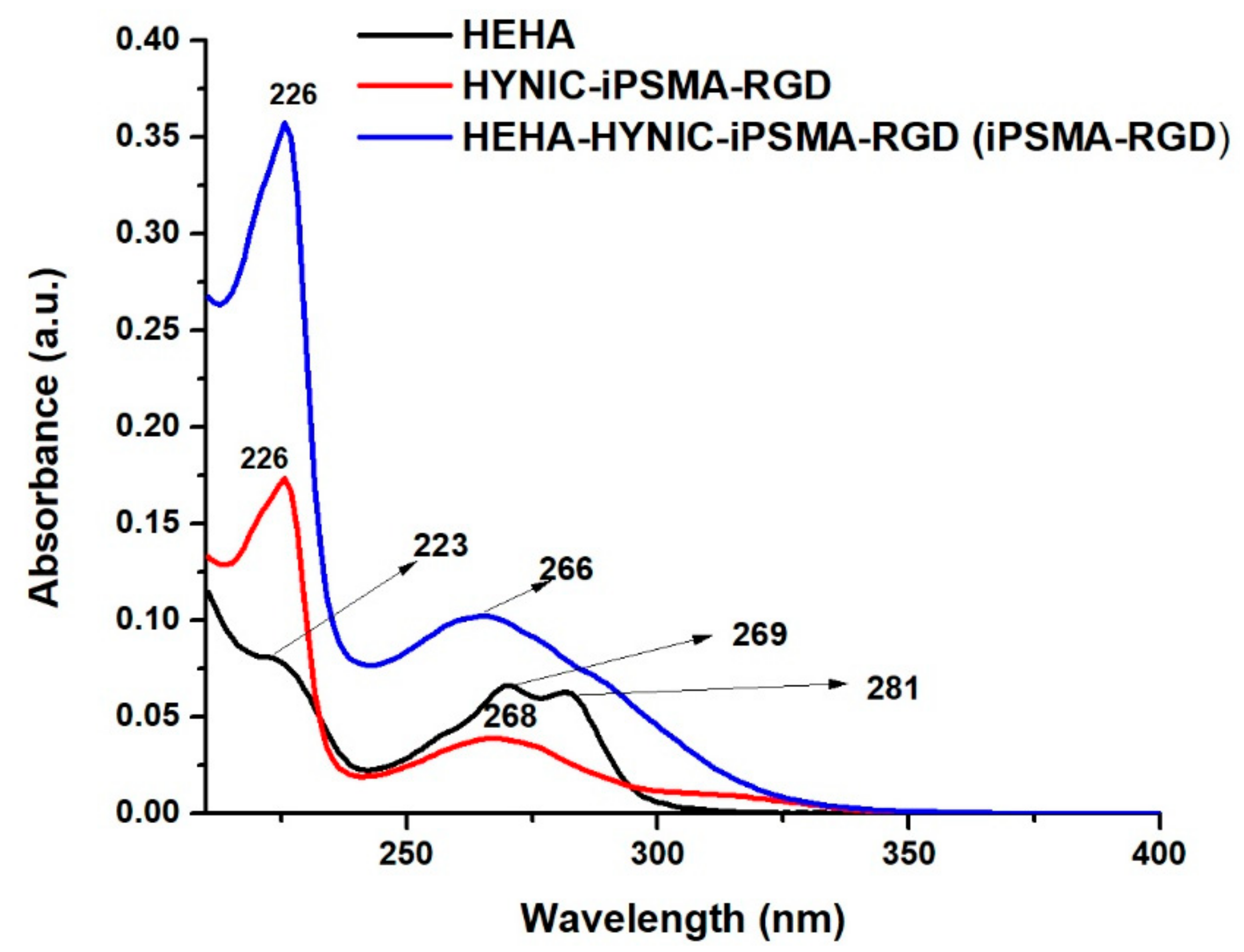
2.1. Synthesis and Chemical Characterization of HEHA-HYNIC-iPSMA-RGD (iPSMA-RGD)
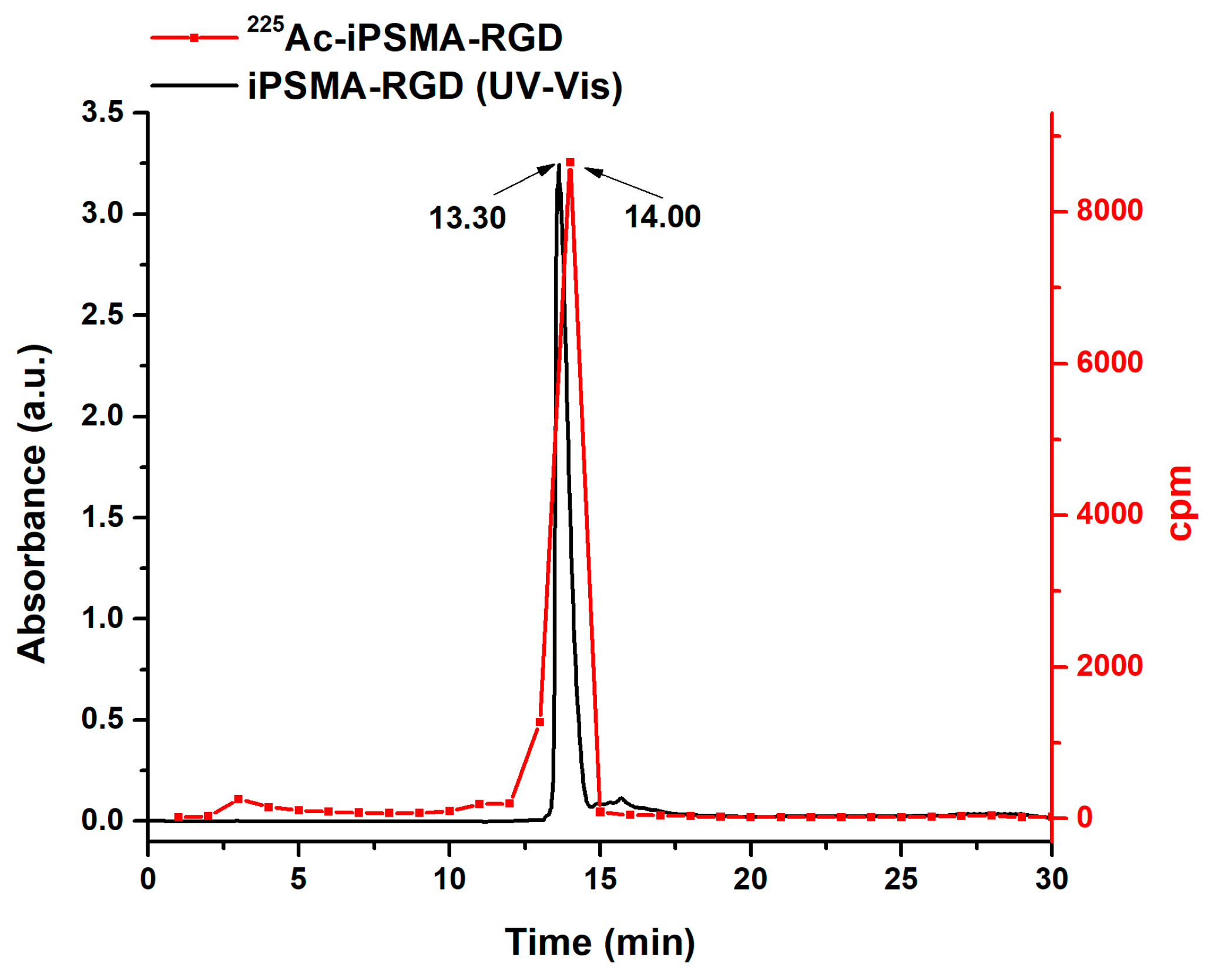
2.2. Preparation of 225Ac-iPSMA-RGD
2.3. In Vitro Evaluation of 225Ac-iPSMA-RGD
2.3.1. Stability in Saline Solution (0.9% NaCl) and Human Serum
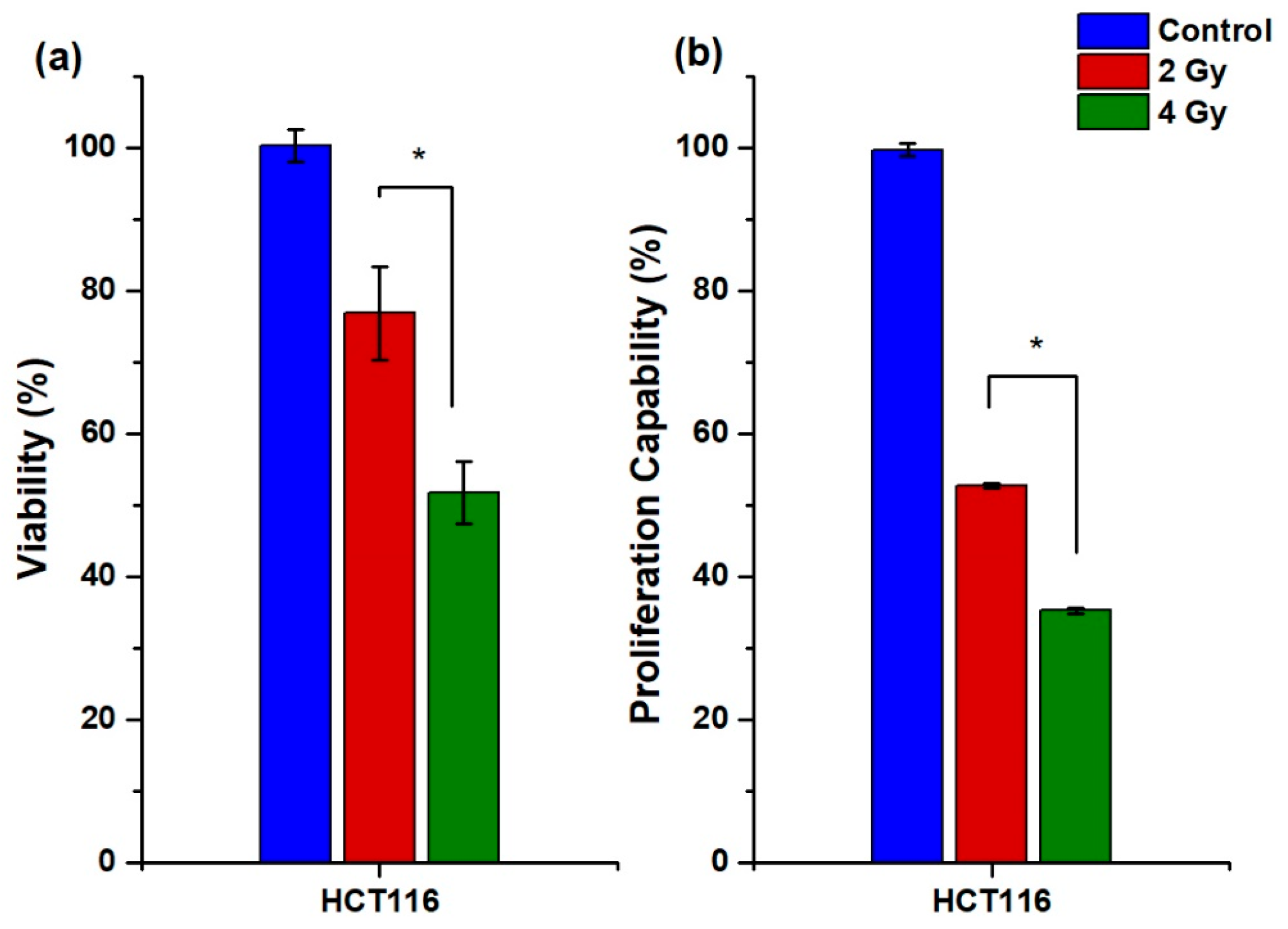
2.3.2. Cellular Uptake, Internalization, and Viability
2.3.3. Proliferation Capability Assay
2.3.4. Apoptosis
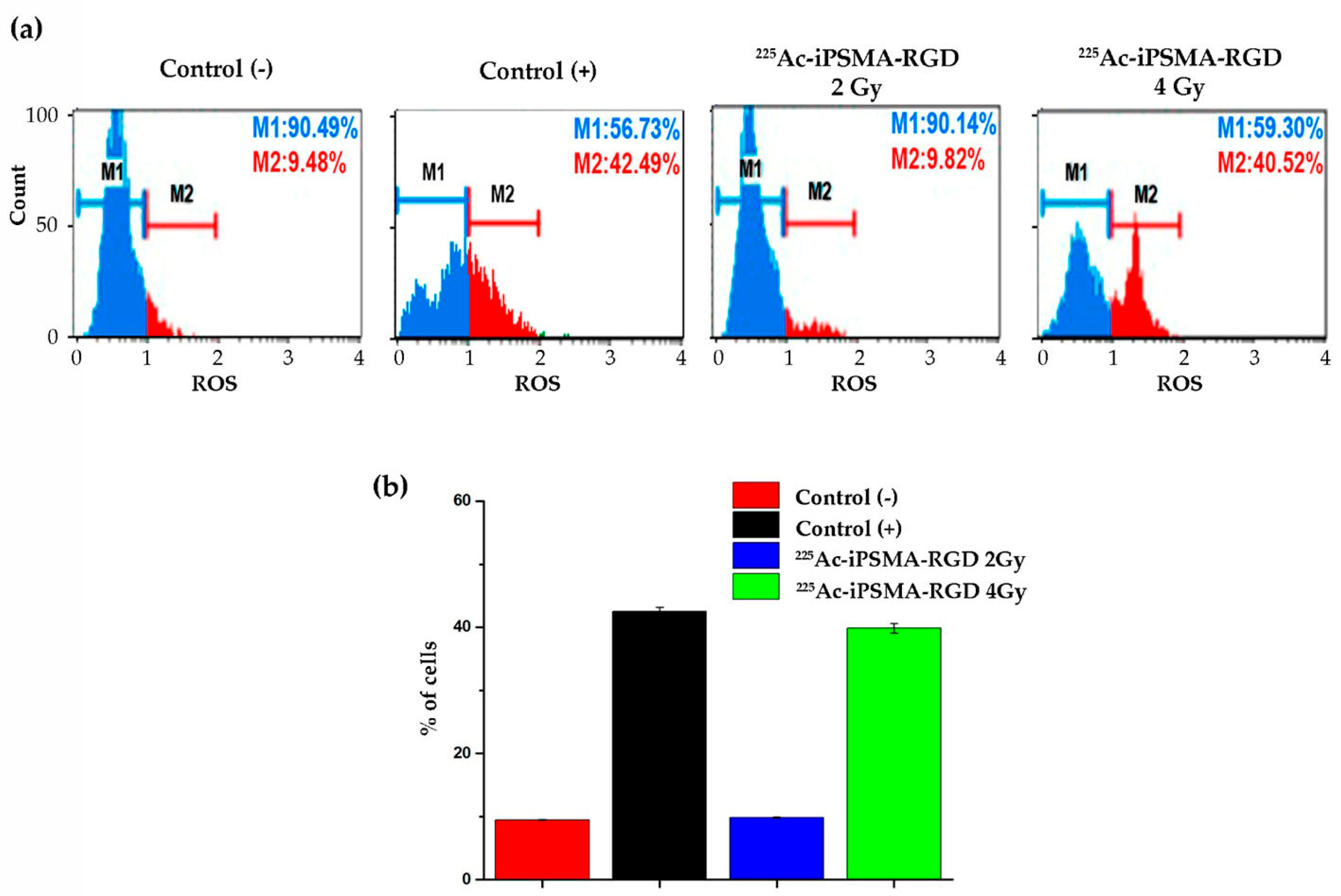
2.3.5. Oxidative Stress
2.3.6. Double-Stranded DNA Breaks
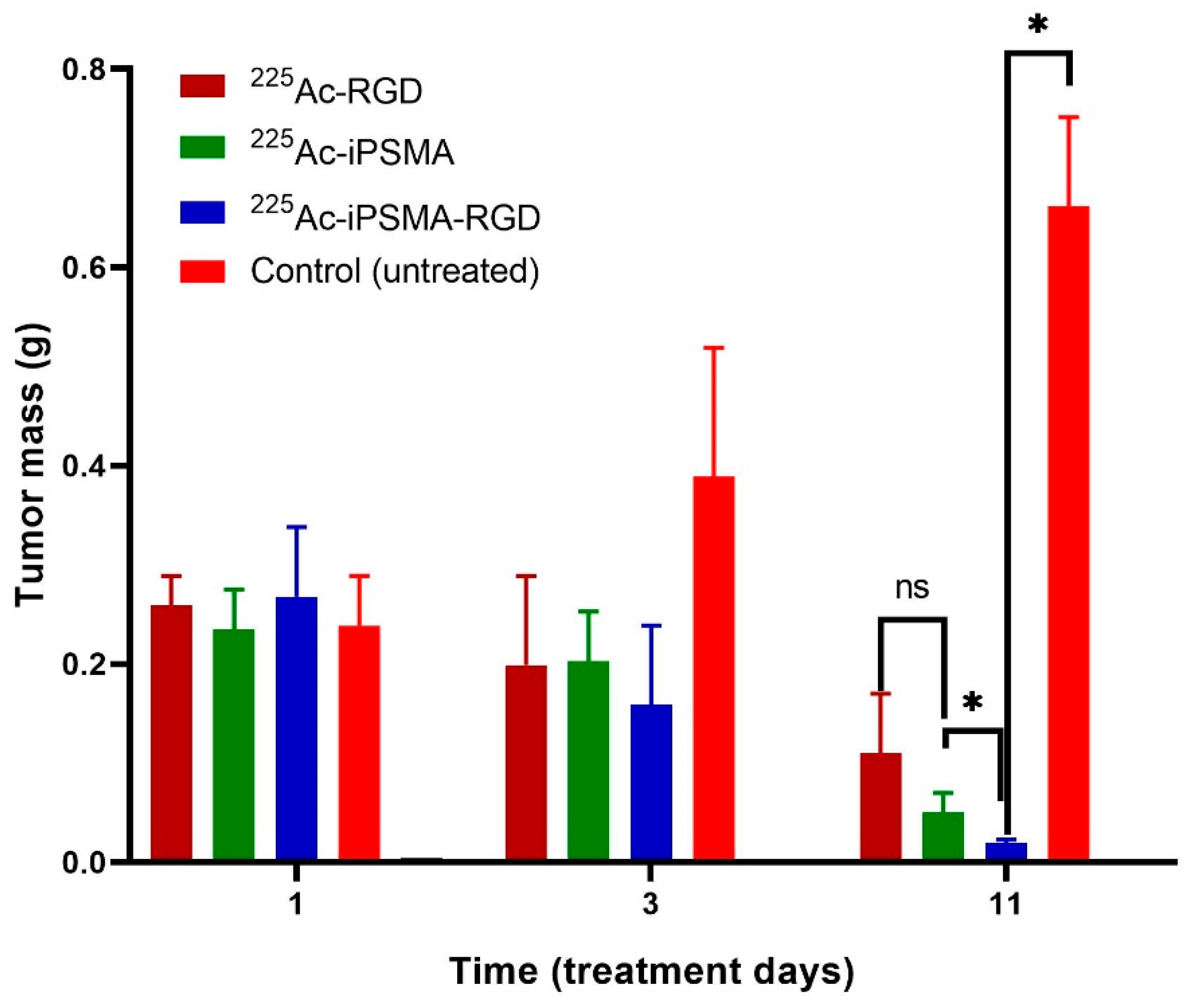
2.4. In Vivo Assessment
2.4.1. Biodistribution and Biokinetics
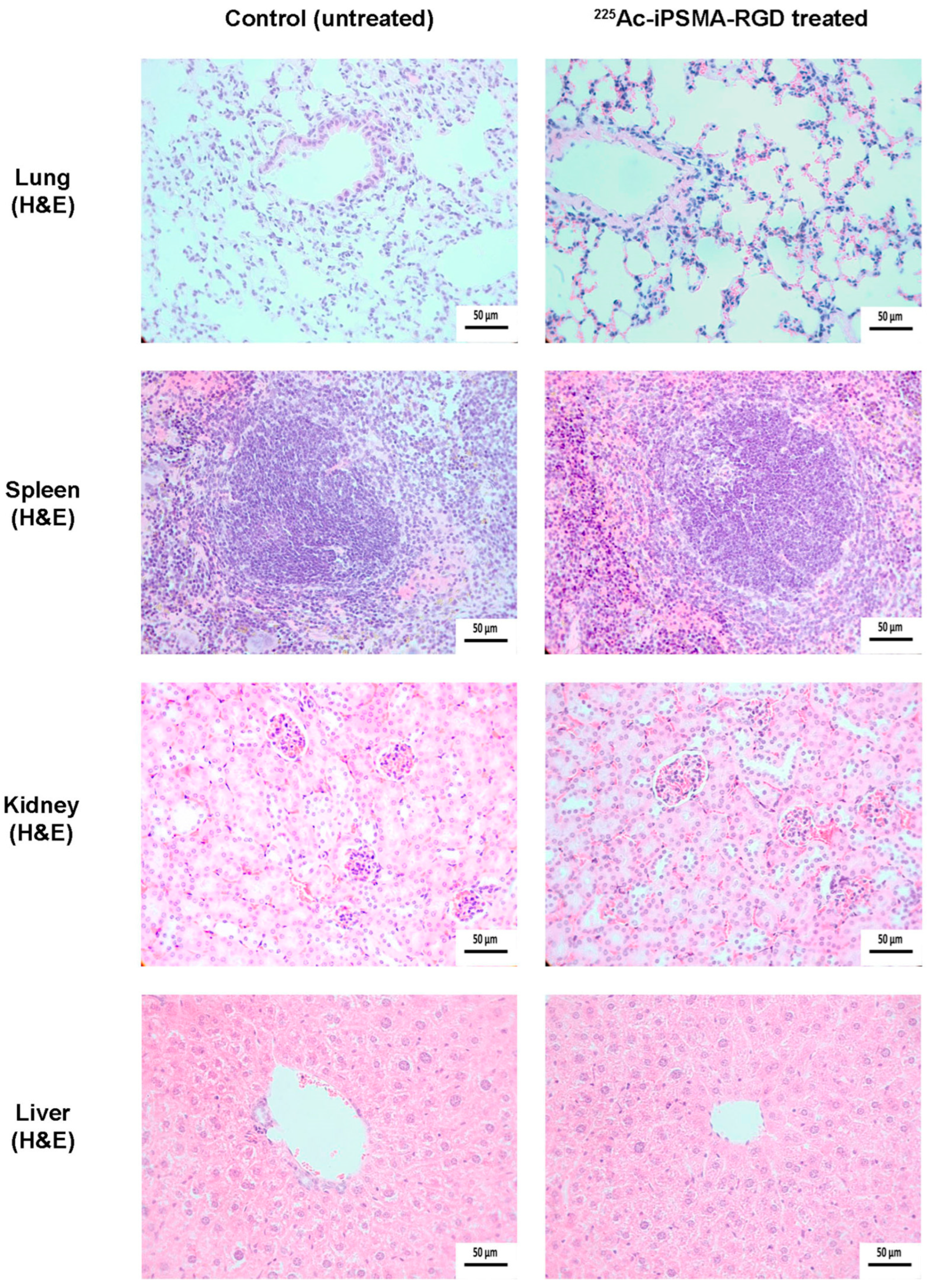
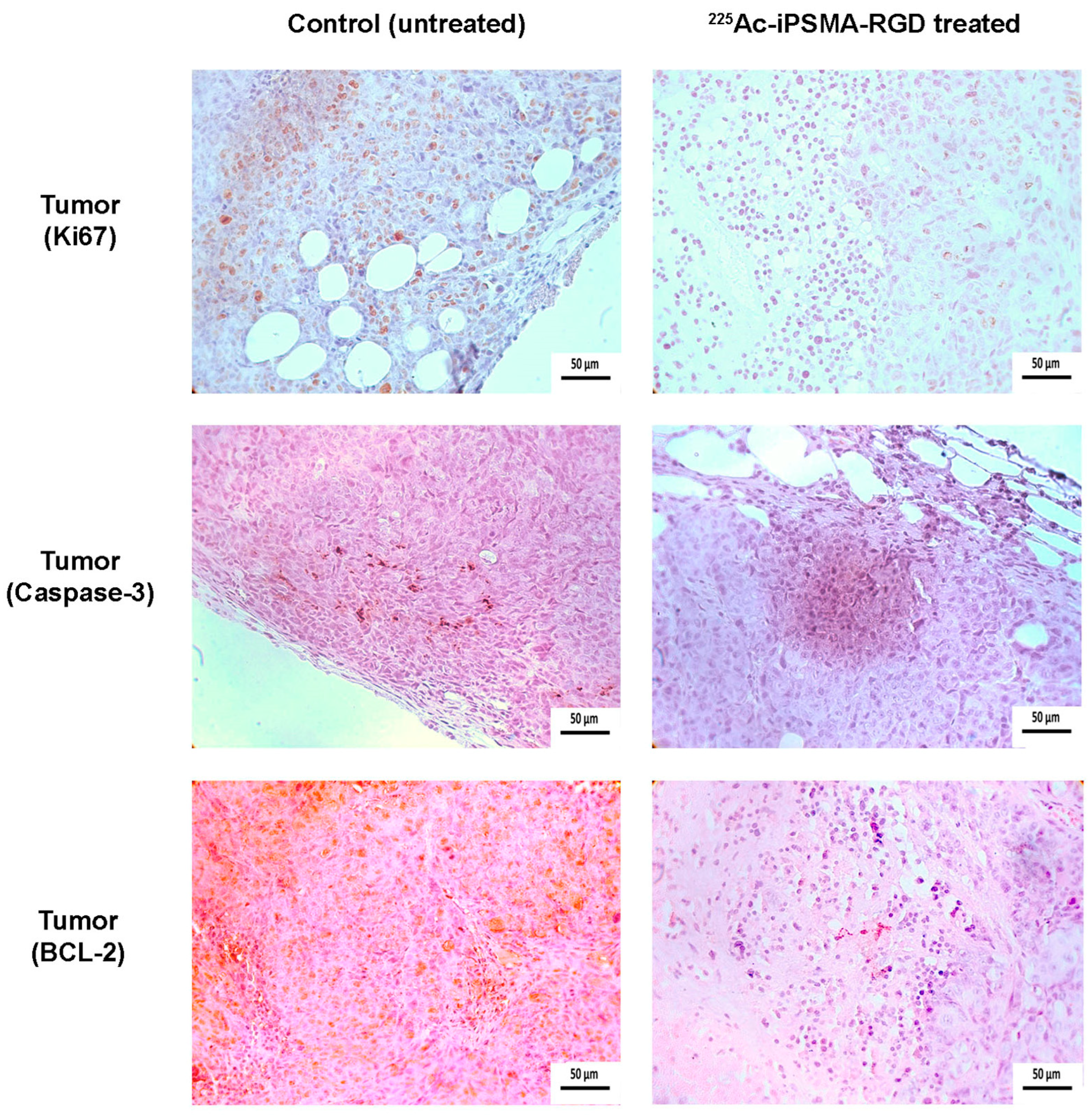
2.4.2. Ex-Vivo Imaging
3. Materials and Methods
3.1. Conjugation of HYNIC-iPSMA-RGD to HEHA-SCN
3.2. Spectroscopic Characterization by FT-IR and UV-Vis Characterization
3.3. Mass Spectrometry Characterization
3.4. Radiolabeling and Radiochemical Purity Evaluation
3.5. Serum Stability
3.6. Cell Culture
3.7. Uptake and Internalization in Cells
3.8. Cytotoxicity after 225Ac-iPSMA-RGD Treatment
3.9. Proliferation Capability Assay
3.10. Apoptosis
3.11. Reactive Oxygen Species
3.12. DNA Double-Stranded Breaks Assay
3.13. Biodistribution
3.14. Absorbed Radiation Dose Estimation
3.15. Histology and Immunohistochemistry Protocol
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Battistini, L.; Bugatti, K.; Sartori, A.; Curti, C.; Zanardi, F. RGD Peptide-Drug Conjugates as Effective Dual Targeting Platforms: Recent Advances. Eur. J. Org. Chem. 2021, 2021, 2506–2528. [Google Scholar] [CrossRef]
- Wernicke, A.G.; Kim, S.; Liu, H.; Bander, N.H.; Pirog, E.C. Prostate-specific Membrane Antigen (PSMA) Expression in the Neovasculature of Gynecologic Malignancies: Implications for PSMA-targeted Therapy. Appl. Immunohistochem. Mol. Morphol. 2017, 25, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Nimmagadda, S.; Pullambhatla, M.; Chen, Y.; Parsana, P.; Lisok, A.; Chatterjee, S.; Mease, R.; Rowe, S.P.; Lupold, S.; Pienta, K.J. Low-level endogenous PSMA expression in nonprostatic tumor xenografts is sufficient for in vivo tumor targeting and imaging. J. Nucl. Med. 2018, 59, 486–493. [Google Scholar] [CrossRef] [PubMed]
- Tolkach, Y.; Goltz, D.; Kremer, A.; Ahmadzadehfar, H.; Bergheim, D.; Essler, M.; Lam, M.; De Keizer, B.; Fischer, H.-P.; Kristiansen, G. Prostate-specific membrane antigen expression in hepatocellular carcinoma: Potential use for prognosis and diagnostic imaging. Oncotarget 2019, 10, 4149. [Google Scholar] [CrossRef] [PubMed]
- Sonni, I.; Caron, J.; Kishan, A.U.; Muthusamy, V.R.; Calais, J. PSMA expression in the neovasculature associated with rectal adenocarcinoma: A potential stromal target for nuclear theranostics. Clin. Nucl. Med. 2020, 45, e309–e310. [Google Scholar] [CrossRef] [PubMed]
- Modest, D.P.; Pant, S.; Sartore-Bianchi, A. Treatment sequencing in metastatic colorectal cancer. Eur. J. Cancer 2019, 109, 70–83. [Google Scholar] [CrossRef] [PubMed]
- Thiele, N.A.; Wilson, J.J. Actinium-225 for targeted α therapy: Coordination chemistry and current chelation approaches. Cancer Biother. Radiopharm. 2018, 33, 336–348. [Google Scholar] [CrossRef] [PubMed]
- Morgenstern, A.; Apostolidis, C.; Kratochwil, C.; Sathekge, M.; Krolicki, L.; Bruchertseifer, F. An overview of targeted alpha therapy with 225Actinium and 213Bismuth. Curr. Radiopharm. 2018, 11, 200–208. [Google Scholar] [CrossRef]
- Castillo Seoane, D.; De Saint-Hubert, M.; Ahenkorah, S.; Saldarriaga Vargas, C.; Ooms, M.; Struelens, L.; Koole, M. Gamma counting protocols for the accurate quantification of 225Ac and 213Bi without the need for a secular equilibrium between parent and gamma-emitting daughter. EJNMMI Radiopharm. Chem. 2022, 7, 28. [Google Scholar] [CrossRef]
- Koniar, H.; Miller, C.; Rahmim, A.; Schaffer, P.; Uribe, C. A GATE simulation study for dosimetry in cancer cell and micrometastasis from the 225Ac decay chain. EJNMMI Phys. 2023, 10, 46. [Google Scholar] [CrossRef]
- Sen, I.; Thakral, P.; Tiwari, P.; Pant, V.; Das, S.S.; Manda, D.; Raina, V. Therapeutic efficacy of 225Ac-PSMA-617 targeted alpha therapy in patients of metastatic castrate resistant prostate cancer after taxane-based chemotherapy. Ann. Nucl. Med. 2021, 35, 794–810. [Google Scholar] [CrossRef] [PubMed]
- Zacherl, M.J.; Gildehaus, F.J.; Mittlmeier, L.; Böning, G.; Gosewisch, A.; Wenter, V.; Unterrainer, M.; Schmidt-Hegemann, N.; Belka, C.; Kretschmer, A. First clinical results for PSMA-targeted α-therapy using 225Ac-PSMA-I&T in advanced-mCRPC patients. J. Nucl. Med. 2021, 62, 669–674. [Google Scholar] [PubMed]
- Pandya, D.N.; Hantgan, R.; Budzevich, M.M.; Kock, N.D.; Morse, D.L.; Batista, I.; Mintz, A.; Li, K.C.; Wadas, T.J. Preliminary therapy evaluation of 225Ac-DOTA-c (RGDyK) demonstrates that Cerenkov radiation derived from 225Ac daughter decay can be detected by optical imaging for in vivo tumor visualization. Theranostics 2016, 6, 698. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, M.; Washiyama, K.; Ohnuki, K.; Kojima, M.; Miller, B.; Yoshii, Y.; Fujii, H. Pre-clinical evaluation of 225Ac-DOTA-E [c (RGDfK)] 2 for targeted alpha therapy in PDCA mice model. J. Nucl. Med. 2023, 64 (Suppl. 1), 276. [Google Scholar]
- Sathekge, M.; Bruchertseifer, F.; Vorster, M.; Lawal, I.O.; Mokoala, K.; Reed, J.; Maseremule, L.; Ndlovu, H.; Hlongwa, K.; Maes, A. 225Ac-PSMA-617 radioligand therapy of de novo metastatic hormone-sensitive prostate carcinoma (mHSPC): Preliminary clinical findings. Eur. J. Nucl. Med. Mol. Imaging 2023, 50, 2210–2218. [Google Scholar] [CrossRef] [PubMed]
- Poty, S.; Francesconi, L.C.; McDevitt, M.R.; Morris, M.J.; Lewis, J.S. α-Emitters for radiotherapy: From basic radiochemistry to clinical studies—Part 1. J. Nucl. Med. 2018, 59, 878–884. [Google Scholar] [CrossRef]
- Poty, S.; Francesconi, L.C.; McDevitt, M.R.; Morris, M.J.; Lewis, J.S. α-emitters for radiotherapy: From basic radiochemistry to clinical studies—Part 2. J. Nucl. Med. 2018, 59, 1020–1027. [Google Scholar] [CrossRef]
- Yilmaz, B.; Nisli, S.; Ergul, N.; Gursu, R.U.; Acikgoz, O.; Çermik, T.F. Effect of external cooling on 177Lu-PSMA uptake by the parotid glands. J. Nucl. Med. 2019, 60, 1388–1393. [Google Scholar] [CrossRef]
- Sarnelli, A.; Belli, M.L.; Di Iorio, V.; Mezzenga, E.; Celli, M.; Severi, S.; Tardelli, E.; Nicolini, S.; Oboldi, D.; Uccelli, L. Dosimetry of 177Lu-PSMA-617 after mannitol infusion and glutamate tablet administration: Preliminary results of EUDRACT/RSO 2016-002732-32 IRST protocol. Molecules 2019, 24, 621. [Google Scholar] [CrossRef]
- Trujillo-Nolasco, M.; Cruz-Nova, P.; Ferro-Flores, G.; Gibbens-Bandala, B.; Morales-Avila, E.; Aranda-Lara, L.; Vargas, M.; Ocampo-García, B. Development of 177Lu-DN (C19)-CXCR4 ligand nanosystem for combinatorial therapy in pancreatic cancer. J. Biomed. Nanotechnol. 2021, 17, 263–278. [Google Scholar] [CrossRef]
- Ferro-Flores, G.; Arteaga de Murphy, C.; Melendez-Alafort, L. Third generation radiopharmaceuticals for imaging and targeted therapy. Curr. Pharm. Anal. 2006, 2, 339–352. [Google Scholar] [CrossRef]
- Basken, N.E.; Mathias, C.J.; Lipka, A.E.; Green, M.A. Species dependence of [64Cu] Cu-Bis (thiosemicarbazone) radiopharmaceutical binding to serum albumins. Nucl. Med. Biol. 2008, 35, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Escudero-Castellanos, A.; Ocampo-García, B.E.; Ferro-Flores, G.; Isaac-Olivé, K.; Santos-Cuevas, C.L.; Olmos-Ortiz, A.; García-Quiroz, J.; García-Becerra, R.; Díaz, L. Preparation and in vitro evaluation of 177Lu-iPSMA-RGD as a new heterobivalent radiopharmaceutical. J. Radioanal. Nucl. Chem. 2017, 314, 2201–2207. [Google Scholar] [CrossRef]
- Ferro-Flores, G.; Luna-Gutiérrez, M.; Ocampo-García, B.; Santos-Cuevas, C.; Azorín-Vega, E.; Jiménez-Mancilla, N.; Orocio-Rodríguez, E.; Davanzo, J.; García-Pérez, F.O. Clinical translation of a PSMA inhibitor for 99mTc-based SPECT. Nucl. Med. Biol. 2017, 48, 36–44. [Google Scholar] [CrossRef]
- Hernández-Jiménez, T.; Cruz-Nova, P.; Ancira-Cortez, A.; Gibbens-Bandala, B.; Lara-Almazán, N.; Ocampo-García, B.; Santos-Cuevas, C.; Morales-Avila, E.; Ferro-Flores, G. Toxicity Assessment of [177Lu] Lu− iFAP/iPSMA Nanoparticles Prepared under GMP-Compliant Radiopharmaceutical Processes. Nanomaterials 2022, 12, 4181. [Google Scholar] [CrossRef]
- Reissig, F.; Zarschler, K.; Novy, Z.; Petrik, M.; Bendova, K.; Kurfurstova, D.; Bouchal, J.; Ludik, M.-C.; Brandt, F.; Kopka, K. Modulating the pharmacokinetic profile of actinium-225-labeled macropa-derived radioconjugates by dual targeting of PSMA and albumin. Theranostics 2022, 12, 7203. [Google Scholar] [CrossRef] [PubMed]
- Majkowska-Pilip, A.; Rius, M.; Bruchertseifer, F.; Apostolidis, C.; Weis, M.; Bonelli, M.; Laurenza, M.; Królicki, L.; Morgenstern, A. In vitro evaluation of 225Ac-DOTA-substance P for targeted alpha therapy of glioblastoma multiforme. Chem. Biol. Drug Des. 2018, 92, 1344–1356. [Google Scholar] [CrossRef]
- Liu, R.; Bian, Y.; Liu, L.; Liu, L.; Liu, X.; Ma, S. Molecular pathways associated with oxidative stress and their potential applications in radiotherapy. Int. J. Mol. Med. 2022, 49, 65. [Google Scholar] [CrossRef]
- Rubira, L.; Deshayes, E.; Santoro, L.; Kotzki, P.O.; Fersing, C. 225Ac-Labeled Somatostatin Analogs in the Management of Neuroendocrine Tumors: From Radiochemistry to Clinic. Pharmaceutics 2023, 15, 1051. [Google Scholar] [CrossRef]
- Garg, R.; Allen, K.J.; Dawicki, W.; Geoghegan, E.M.; Ludwig, D.L.; Dadachova, E. 225Ac-labeled CD33-targeting antibody reverses resistance to Bcl-2 inhibitor venetoclax in acute myeloid leukemia models. Cancer Med. 2021, 10, 1128–1140. [Google Scholar] [CrossRef]
- Graf, F.; Fahrer, J.; Maus, S.; Morgenstern, A.; Bruchertseifer, F.; Venkatachalam, S.; Fottner, C.; Weber, M.M.; Huelsenbeck, J.; Schreckenberger, M. DNA double strand breaks as predictor of efficacy of the alpha-particle emitter Ac-225 and the electron emitter Lu-177 for somatostatin receptor targeted radiotherapy. PLoS ONE 2014, 9, e88239. [Google Scholar] [CrossRef]
- Manohar, S.; Kompotiatis, P.; Halfdanarson, T.R.; Hobday, T.J.; Thorpe, M.; Johnson, G.B.; Kendi, A.T.; Leung, N. 177Lu-dotatate use in chronic kidney disease patients: A single center experience. J. Onco-Nephrol. 2021, 5, 162–171. [Google Scholar] [CrossRef]
- Trujillo-Nolasco, R.M.; Morales-Avila, E.; Ocampo-García, B.E.; Ferro-Flores, G.; Gibbens-Bandala, B.V.; Escudero-Castellanos, A.; Isaac-Olive, K. Preparation and in vitro evaluation of radiolabeled HA-PLGA nanoparticles as novel MTX delivery system for local treatment of rheumatoid arthritis. Mater. Sci. Eng. C Mater. Biol. Appl. 2019, 103, 109766. [Google Scholar] [CrossRef]
- Kratochwil, C.; Bruchertseifer, F.; Giesel, F.L.; Weis, M.; Verburg, F.A.; Mottaghy, F.; Kopka, K.; Apostolidis, C.; Haberkorn, U.; Morgenstern, A. 225Ac-PSMA-617 for PSMA-targeted α-radiation therapy of metastatic castration-resistant prostate cancer. J. Nucl. Med. 2016, 57, 1941–1944. [Google Scholar] [CrossRef]
- Sa-Nongdej, W.; Chongthammakun, S.; Songthaveesin, C. Nutrient starvation induces apoptosis and autophagy in C6 glioma stem-like cells. Heliyon 2021, 7, e06352–e06359. [Google Scholar] [CrossRef]








| Organ | 225Ac-iPSMA | 225Ac-RGD | 225Ac-iPSMA-RGD | |||
|---|---|---|---|---|---|---|
| Radiation Absorbed Dose Gy/37 kBq | Radiation Absorbed Dose Gy/37 kBq | Radiation Absorbed Dose Gy/37 kBq | ||||
| Kidney | 4.41 | 22.04 | 1.78 | 8.90 | 3.52 | 17.63 |
| Liver | 2.10 | 6.38 | 3.92 | 11.89 | 5.23 | 15.85 |
| Tumor | 6.04 | 182.87 | 1.72 | 51.92 | 7.83 | 237.07 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ocampo-García, B.; Cruz-Nova, P.; Jiménez-Mancilla, N.; Luna-Gutiérrez, M.; Oros-Pantoja, R.; Lara-Almazán, N.; Pérez-Velasco, D.; Santos-Cuevas, C.; Ferro-Flores, G. 225Ac-iPSMA-RGD for Alpha-Therapy Dual Targeting of Stromal/Tumor Cell PSMA and Integrins. Int. J. Mol. Sci. 2023, 24, 16553. https://doi.org/10.3390/ijms242316553
Ocampo-García B, Cruz-Nova P, Jiménez-Mancilla N, Luna-Gutiérrez M, Oros-Pantoja R, Lara-Almazán N, Pérez-Velasco D, Santos-Cuevas C, Ferro-Flores G. 225Ac-iPSMA-RGD for Alpha-Therapy Dual Targeting of Stromal/Tumor Cell PSMA and Integrins. International Journal of Molecular Sciences. 2023; 24(23):16553. https://doi.org/10.3390/ijms242316553
Chicago/Turabian StyleOcampo-García, Blanca, Pedro Cruz-Nova, Nallely Jiménez-Mancilla, Myrna Luna-Gutiérrez, Rigoberto Oros-Pantoja, Nancy Lara-Almazán, Diana Pérez-Velasco, Clara Santos-Cuevas, and Guillermina Ferro-Flores. 2023. "225Ac-iPSMA-RGD for Alpha-Therapy Dual Targeting of Stromal/Tumor Cell PSMA and Integrins" International Journal of Molecular Sciences 24, no. 23: 16553. https://doi.org/10.3390/ijms242316553





