Enhancing Selectivity of Protein Biopharmaceuticals in Ion Exchange Chromatography through Addition of Organic Modifiers
Abstract
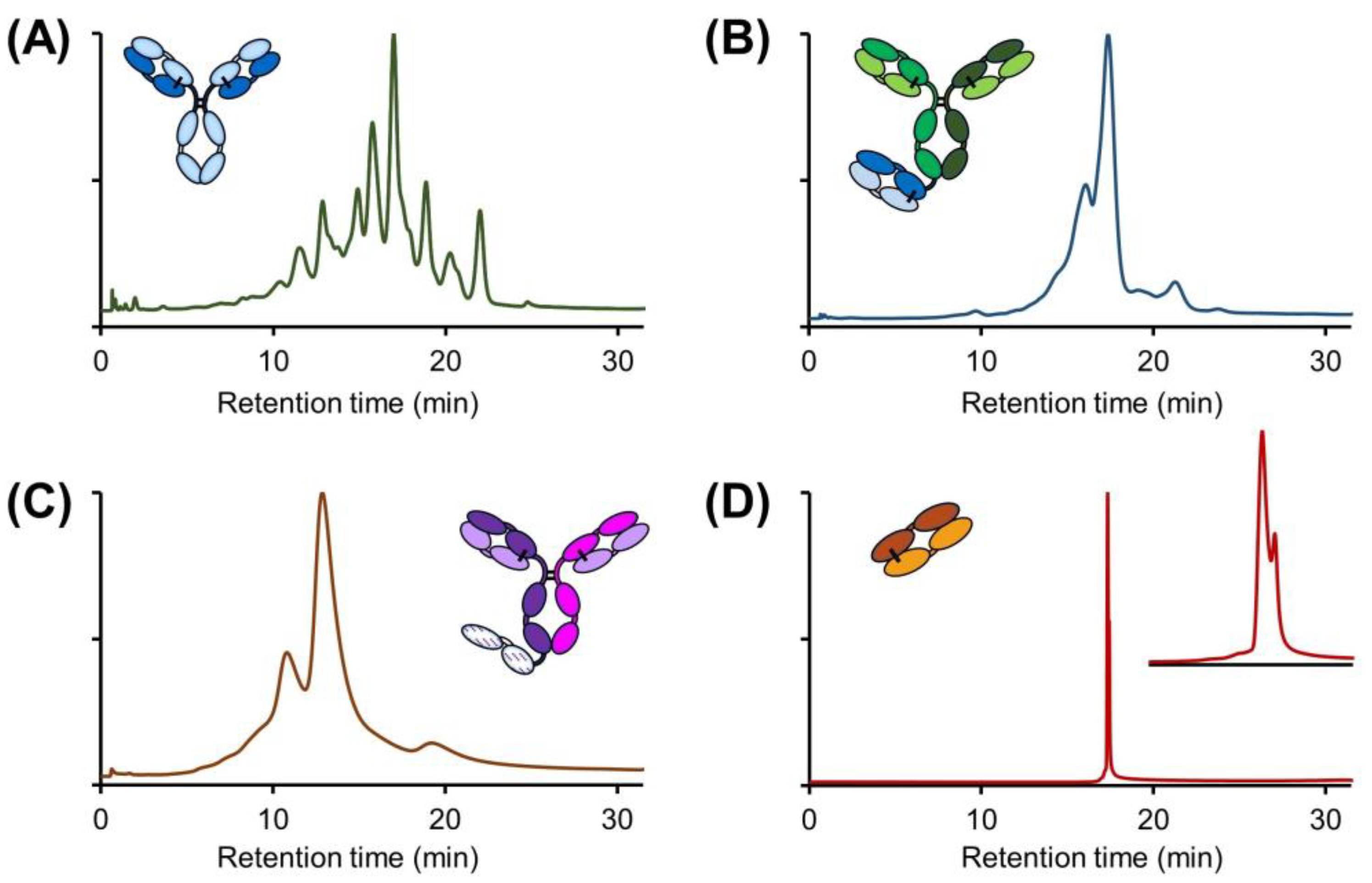
:1. Introduction
2. Results and Discussion
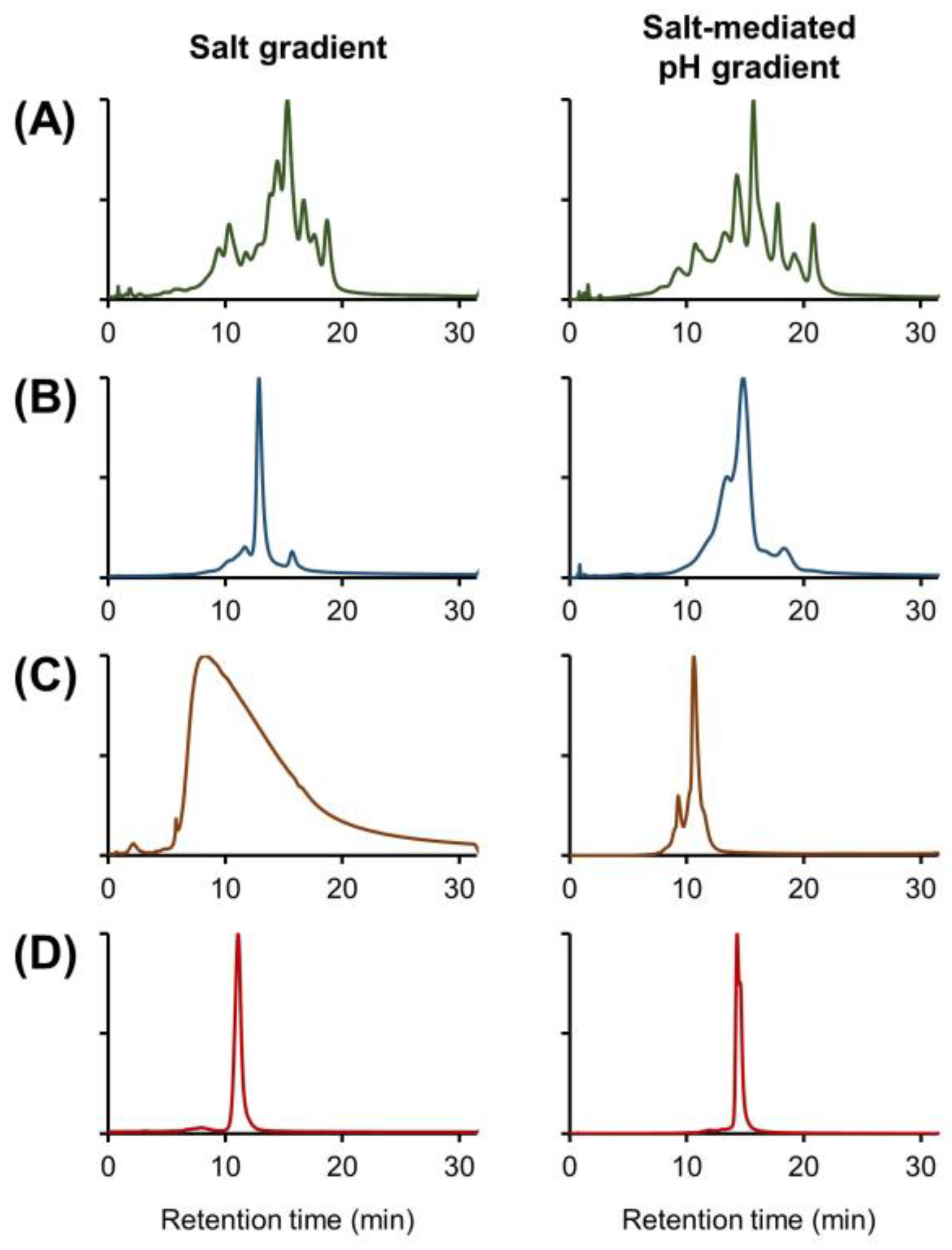
2.1. Evaluation of Buffer Performance
2.2. Evaluation of Solvent Addition in IEX Mobile Phase Buffer Compositions
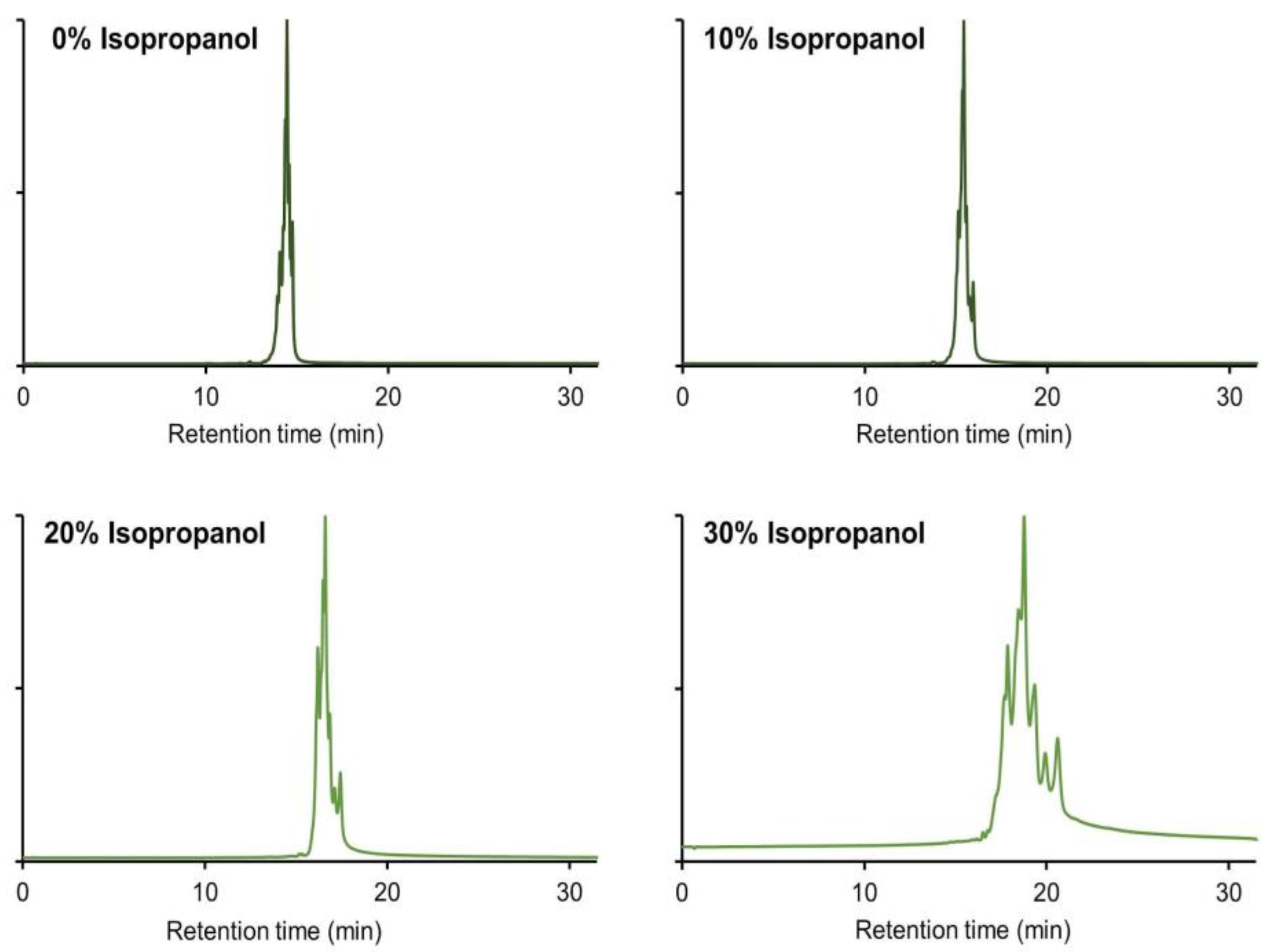
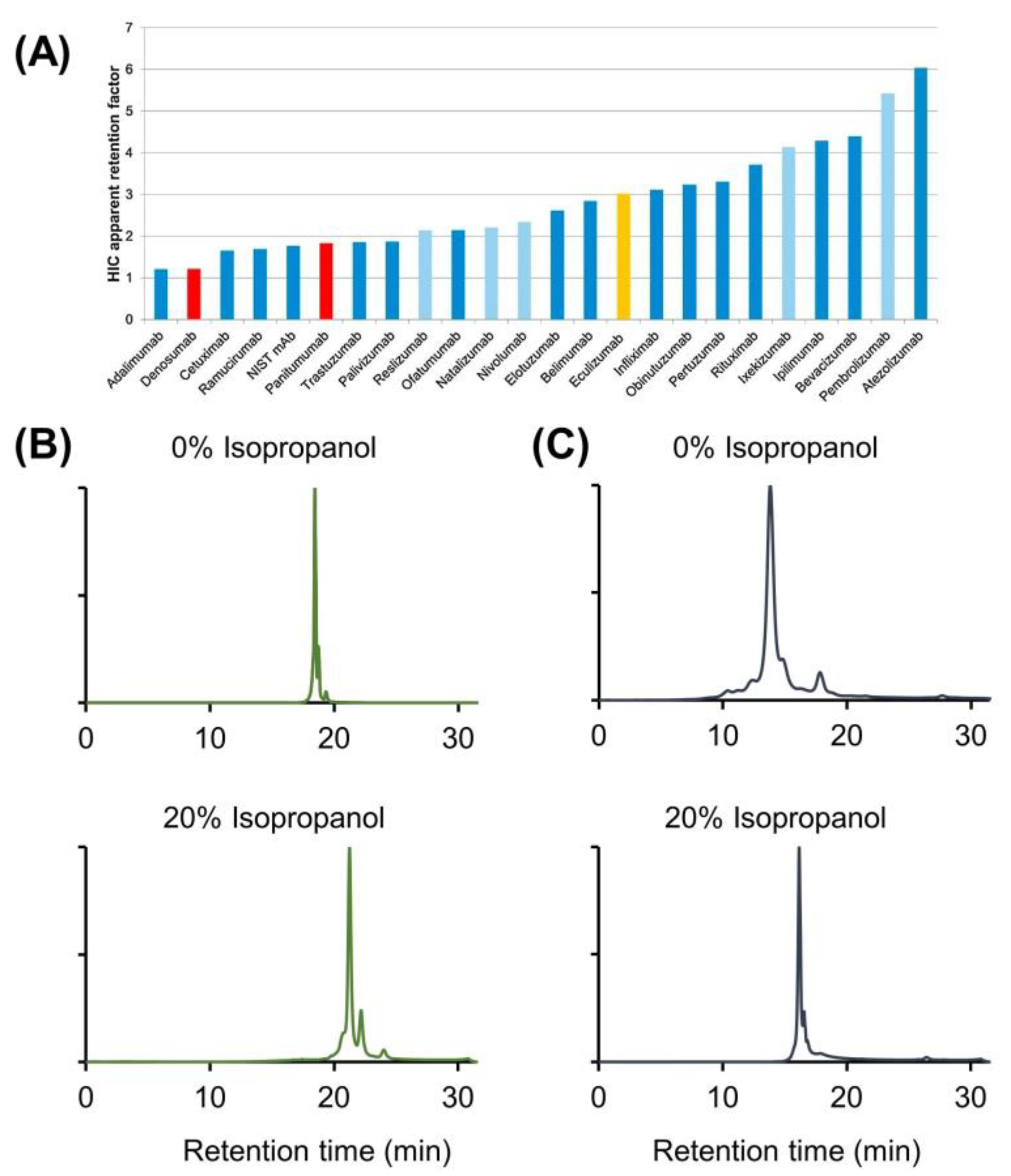
2.3. Use of IPA As Mobile Phase Additive in CEX
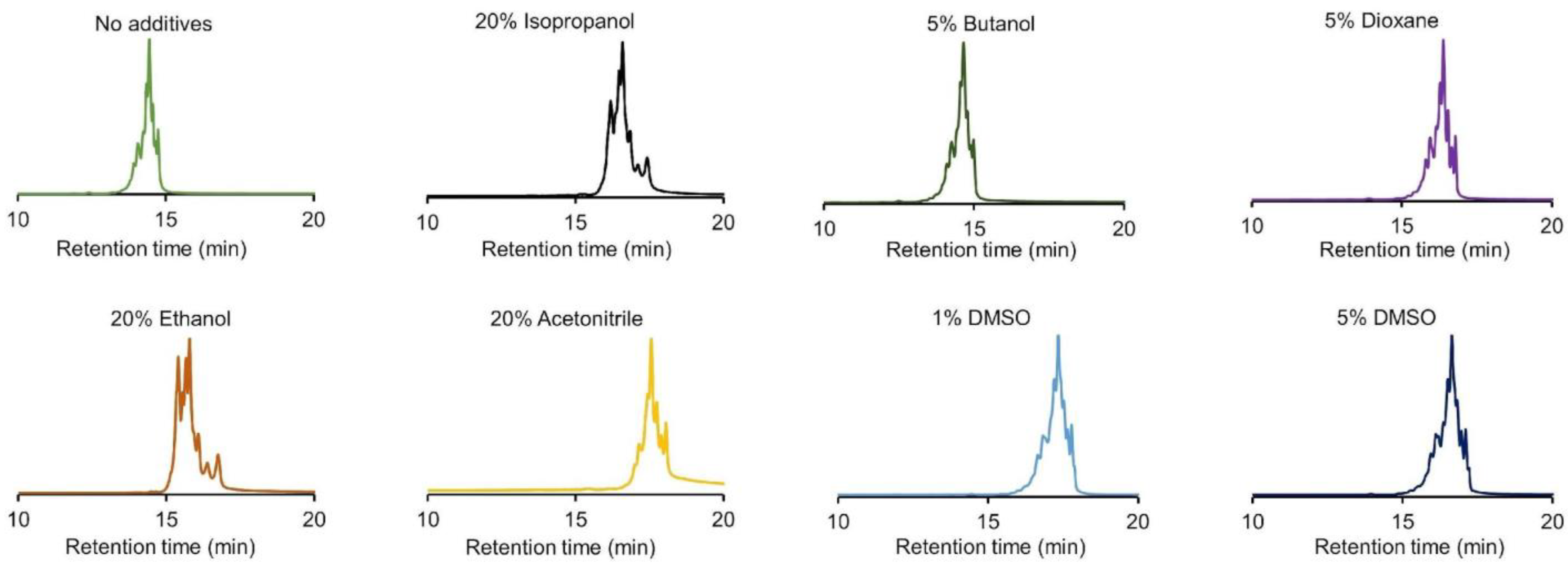
2.4. Addition of Additives to Solvent Enriched Mobile Phases
2.5. Are Solvent- and Additive-Enriched Salt-Mediated pH Gradients Stability Indicating?
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Instrumentation and Columns
3.3. Mobile Phase Preparation
3.4. Chromatographic Conditions
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Castelli, M.S.; McGonigle, P.; Hornby, P.J. The pharmacology and therapeutic applications of monoclonal antibodies. Pharmacol. Res. Perspect. 2019, 7, e00535. [Google Scholar] [CrossRef]
- Li, W.; Pang, I.-H.; Pacheco, M.T.F.; Tian, H. Ligandomics: A paradigm shift in biological drug discovery. Drug. Discov. Today 2018, 23, 636–643. [Google Scholar] [CrossRef]
- Kintzing, J.R.; Filsinger Interrante, M.V.; Cochran, J.R. Emerging Strategies for Developing Next-Generation Protein Therapeutics for Cancer Treatment. Trends Pharmacol. Sci. 2016, 37, 993–1008. [Google Scholar] [CrossRef] [PubMed]
- Jain, T.; Sun, T.; Durand, S.; Hall, A.; Rewa, N.; Nett, J.H.; Sharkey, B. Biophysical properties of the clinical-stage antibody landscape. Proc. Natl. Acad. Sci. USA 2017, 114, 944–949. [Google Scholar] [CrossRef]
- Carrara, S.C.; Ulitzka, M.; Grzeschik, J.; Kornmann, H.; Hock, B.; Kolmar, H. From cell line development to the formulated drug product: The art of manufacturing therapeutic monoclonal antibodies. Int. J. Pharm. 2021, 594, 120164. [Google Scholar] [CrossRef]
- Dumont, J.; Euwart, D.; Mei, B.; Estes, S.; Kshirsagar, R. Human cell lines for biopharmaceutical manufacturing: History, status, and future perspectives. Crit. Rev. Biotechnol. 2016, 36, 1110–1122. [Google Scholar] [CrossRef]
- Liu, J.K.H. The history of monoclonal antibody development–Progress, remaining challenges and future innovations. Ann. Med. Surg. 2014, 3, 113–116. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, D.; Mason, B.; Rossomando, T.; Li, N.; Liu, D.; Cheung, J.K.; Xu, W.; Raghava, S.; Katiyar, A.; et al. Structure, heterogeneity and developability assessment of therapeutic antibodies. MAbs 2019, 11, 239–264. [Google Scholar] [CrossRef]
- Houde, D.; Peng, Y.; Berkowitz, S.A.; Engen, J.R. Post-translational Modifications Differentially Affect IgG1 Conformation and Receptor Binding. Mol. Cell. Proteom. 2010, 9, 1716–1728. [Google Scholar] [CrossRef] [PubMed]
- Walsh, G.; Jefferis, R. Post-translational modifications in the context of therapeutic proteins. Nat. Biotechnol. 2006, 24, 1241–1252. [Google Scholar] [CrossRef]
- Beck, A.; Liu, H. Macro- and Micro-Heterogeneity of Natural and Recombinant IgG Antibodies. Antibodies 2019, 8, 18. [Google Scholar] [CrossRef]
- Millán-Martín, S.; Carillo, S.; Füssl, F.; Sutton, J.; Gazis, P.; Cook, K.; Scheffler, K.; Bones, J. Optimisation of the use of sliding window deconvolution for comprehensive characterisation of trastuzumab and adalimumab charge variants by native high resolution mass spectrometry. Eur. J. Pharm. Biopharm. 2021, 158, 83–95. [Google Scholar] [CrossRef]
- Khawli, L.A.; Goswami, S.; Hutchinson, R.; Kwong, Z.W.; Yang, J.; Wang, X.; Yao, Z.; Sreedhara, A.; Cano, T.; Tesar, D.B.; et al. Charge variants in IgG1. MAbs 2010, 2, 613–624. [Google Scholar] [CrossRef] [PubMed]
- Zupke, C.; Brady, L.J.; Slade, P.G.; Clark, P.; Caspary, R.G.; Livingston, B.; Taylor, L.; Bigham, K.; Morris, A.E.; Bailey, R.W. Real-Time Product Attribute Control to Manufacture Antibodies with Defined N-Linked Glycan Levels. Biotechnol. Prog. 2015, 31, 1433–1441. [Google Scholar] [CrossRef]
- Cho, I.H.; Lee, N.; Song, D.; Jung, S.Y.; Bou-Assaf, G.; Sosic, Z.; Zhang, W.; Lyubarskaya, Y. Evaluation of the structural, physicochemical, and biological characteristics of SB4, a biosimilar of etanercept. MAbs 2016, 8, 1136–1155. [Google Scholar] [CrossRef]
- Goyon, A.; D’Atri, V.; Bobaly, B.; Wagner-Rousset, E.; Beck, A.; Fekete, S.; Guillarme, D. Protocols for the analytical characterization of therapeutic monoclonal antibodies. I—Non-denaturing chromatographic techniques. J. Chromatogr. B 2017, 1058, 73–84. [Google Scholar] [CrossRef]
- Trappe, A.; Füssl, F.; Carillo, S.; Zaborowska, I.; Meleady, P.; Bones, J. Rapid charge variant analysis of monoclonal antibodies to support lead candidate biopharmaceutical development. J. Chromatogr. B 2018, 1095, 166–176. [Google Scholar] [CrossRef]
- Griaud, F.; Denefeld, B.; Lang, M.; Hensinger, H.; Haberl, P.; Berg, M. Unbiased in-depth characterization of CEX fractions from a stressed monoclonal antibody by mass spectrometry. MAbs 2017, 9, 820–830. [Google Scholar] [CrossRef]
- Goyon, A.; Excoffier, M.; Janin Bussat, M.C.; Bobaly, B.; Fekete, S.; Guillarme, D.; Beck, A. Determination of isoelectric points and relative charge variants of 23 therapeutic monoclonal antibodies. J. Chromatogr. B 2017, 1066, 119–128. [Google Scholar] [CrossRef]
- Fekete, S.; Beck, A.; Veuthey, J.-L.; Guillarme, D. Ion-exchange chromatography for the characterization of biopharmaceuticals. J. Pharm. Biomed. Anal. 2015, 113, 43–55. [Google Scholar] [CrossRef]
- Du, Y.; Walsh, A.; Ehrick, R.; Xu, W.; May, K.; Liu, H. Chromatographic analysis of the acidic and basic species of recombinant monoclonal antibodies. MAbs 2012, 4, 578–585. [Google Scholar] [CrossRef]
- Tassi, M.; De Vos, J.; Chatterjee, S.; Sobott, F.; Bones, J.; Eeltink, S. Advances in native high-performance liquid chromatography and intact mass spectrometry for the characterization of biopharmaceutical products. J. Sep. Sci. 2018, 41, 125–144. [Google Scholar] [CrossRef] [PubMed]
- Baek, J.; Schwahn, A.B.; Lin, S.; Pohl, C.A.; De Pra, M.; Tremintin, S.M.; Cook, K. New Insights into the Chromatography Mechanisms of Ion-Exchange Charge Variant Analysis: Dispelling Myths and Providing Guidance for Robust Method Optimization. Anal. Chem. 2020, 92, 13411–13419. [Google Scholar] [CrossRef]
- Füssl, F.; Cook, K.; Scheffler, K.; Farrell, A.; Mittermayr, S.; Bones, J. Charge Variant Analysis of Monoclonal Antibodies Using Direct Coupled pH Gradient Cation Exchange Chromatography to High-Resolution Native Mass Spectrometry. Anal. Chem. 2018, 90, 4669–4676. [Google Scholar] [CrossRef] [PubMed]
- Bailey, A.O.; Han, G.; Phung, W.; Gazis, P.; Sutton, J.; Josephs, J.L.; Sandoval, W. Charge variant native mass spectrometry benefits mass precision and dynamic range of monoclonal antibody intact mass analysis. MAbs 2018, 10, 1214–1225. [Google Scholar] [CrossRef] [PubMed]
- Leblanc, Y.; Ramon, C.; Bihoreau, N.; Chevreux, G. Charge variants characterization of a monoclonal antibody by ion exchange chromatography coupled on-line to native mass spectrometry: Case study after a long-term storage at +5 °C. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2017, 1048, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.L.; Xiao, G.; Dillon, T.M.; Ricci, M.S.; Bondarenko, P.V. Characterization of therapeutic proteins by cation exchange chromatography-mass spectrometry and top-down analysis. MAbs 2020, 12, 1739825. [Google Scholar] [CrossRef] [PubMed]
- Muneeruddin, K.; Nazzaro, M.; Kaltashov, I.A. Characterization of Intact Protein Conjugates and Biopharmaceuticals Using Ion-Exchange Chromatography with Online Detection by Native Electrospray Ionization Mass Spectrometry and Top-Down Tandem Mass Spectrometry. Anal. Chem. 2015, 87, 10138–10145. [Google Scholar] [CrossRef]
- Ma, F.; Raoufi, F.; Bailly, M.A.; Fayadat-Dilman, L.; Tomazela, D. Hyphenation of strong cation exchange chromatography to native mass spectrometry for high throughput online characterization of charge heterogeneity of therapeutic monoclonal antibodies. MAbs 2020, 12, 1763762. [Google Scholar] [CrossRef]
- Zhang, L.; Patapoff, T.; Farnan, D.; Zhang, B. Improving pH gradient cation-exchange chromatography of monoclonal antibodies by controlling ionic strength. J. Chromatogr. A 2013, 1272, 56–64. [Google Scholar] [CrossRef]
- Talebi, M.; Nordborg, A.; Gaspar, A.; Lacher, N.A.; Wang, Q.; He, X.Z.; Haddad, P.R.; Hilder, E.F. Charge heterogeneity profiling of monoclonal antibodies using low ionic strength ion-exchange chromatography and well-controlled pH gradients on monolithic columns. J. Chromatogr. A 2013, 1317, 148–154. [Google Scholar] [CrossRef]
- Murisier, A.; Duivelshof, B.L.; Fekete, S.; Bourquin, J.; Schmudlach, A.; Lauber, M.A.; Nguyen, J.M.; Beck, A.; Guillarme, D.; D’Atri, V. Towards a simple on-line coupling of ion exchange chromatography and native mass spectrometry for the detailed characterization of monoclonal antibodies. J. Chromatogr. A 2021, 1655, 462499. [Google Scholar] [CrossRef]
- Farsang, E.; Murisier, A.; Horváth, K.; Colas, O.; Beck, A.; Guillarme, D.; Fekete, S. Optimization of MS-Compatible Mobile Phases for IEX Separation of Monoclonal Antibodies. LC-GC Eur. 2019, 32, 28–34. [Google Scholar]
- Brinkmann, U.; Kontermann, R. The making of bispecific antibodies. MAbs 2017, 9, 182–212. [Google Scholar] [CrossRef] [PubMed]
- Graf, T.; Heinrich, K.; Grunert, I.; Wegele, H.; Haberger, M.; Bulau, P.; Leiss, M. Recent advances in LC–MS based characterization of protein-based bio-therapeutics—Mastering analytical challenges posed by the increasing format complexity. J. Pharm. Biomed. Anal. 2020, 186, 113251. [Google Scholar] [CrossRef]
- Beck, A.; Goetsch, L.; Dumontet, C.; Corvaïa, N. Strategies and challenges for the next generation of antibody–drug conjugates. Nat. Rev. Drug. Discov. 2017, 16, 315–337. [Google Scholar] [CrossRef]
- Strohl, W.R. Current progress in innovative engineered antibodies. Protein Cell. 2018, 9, 86–120. [Google Scholar] [CrossRef]
- Wang, C.; Vemulapalli, B.; Cao, M.; Wang, J.; Wang, X.; Liu, D.; Gadre, D.; Hunter, A. A systematic approach for analysis and characterization of mispairing in bispecific antibodies with asymmetric architecture. MAbs 2018, 10, 1226–1235. [Google Scholar] [CrossRef]
- Heath, C.; Pettit, D. Fc Fusion Proteins. In Challenges in Protein Product Development; Warne, N., Mahler, H.C., Eds.; Springer: Cham, Switzerland, 2018; pp. 545–558. [Google Scholar]
- Goyon, A.; McDonald, D.; Fekete, S.; Guillarme, D.; Stella, C. Development of an innovative salt-mediated pH gradient cation exchange chromatography method for the characterization of therapeutic antibodies. J. Chromatogr. B 2020, 1160, 122379. [Google Scholar] [CrossRef]
- Farsang, E.; Murisier, A.; Horváth, K.; Beck, A.; Kormány, R.; Guillarme, D.; Fekete, S. Tuning selectivity in cation-exchange chromatography applied for monoclonal antibody separations, part 1: Alternative mobile phases and fine tuning of the separation. J. Pharm. Biomed. Anal. 2019, 168, 138–147. [Google Scholar] [CrossRef]
- Yan, Y.; Xing, T.; Wang, S.; Li, N. Versatile, Sensitive, and Robust Native LC–MS Platform for Intact Mass Analysis of Protein Drugs. J. Am. Soc. Mass. Spectrom. 2020, 31, 2171–2179. [Google Scholar] [CrossRef]
- Goyon, A.; Beck, A.; Colas, O.; Sandra, K.; Guillarme, D.; Fekete, S. Evaluation of size exclusion chromatography columns packed with sub-3 μm particles for the analysis of biopharmaceutical proteins. J. Chromatogr. A 2017, 1498, 80–89. [Google Scholar] [CrossRef]
- Fekete, S.; Fogwill, M.; Lauber, M.A. Pressure-Enhanced Liquid Chromatography, a Proof of Concept: Tuning Selectivity with Pressure Changes and Gradients. Anal. Chem. 2022, 94, 7877–7884. [Google Scholar] [CrossRef] [PubMed]
- Bucciarelli, S.; Sayedi, E.S.; Osella, S.; Trzaskowski, B.; Vissing, K.J.; Vestergaard, B.; Foderà, V. Disentangling the role of solvent polarity and protein solvation in folding and self-assembly of α-lactalbumin. J. Colloid. Interface Sci. 2020, 561, 749–761. [Google Scholar] [CrossRef]
- Goyon, A.; D’Atri, V.; Colas, O.; Fekete, S.; Beck, A.; Guillarme, D. Characterization of 30 therapeutic antibodies and related products by size exclusion chromatography: Feasibility assessment for future mass spectrometry hyphenation. J. Chromatogr. B 2017, 1065–1066, 35–43. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duivelshof, B.L.; Bouvarel, T.; Pirner, S.; Larraillet, V.; Knaupp, A.; Koll, H.; D’Atri, V.; Guillarme, D. Enhancing Selectivity of Protein Biopharmaceuticals in Ion Exchange Chromatography through Addition of Organic Modifiers. Int. J. Mol. Sci. 2023, 24, 16623. https://doi.org/10.3390/ijms242316623
Duivelshof BL, Bouvarel T, Pirner S, Larraillet V, Knaupp A, Koll H, D’Atri V, Guillarme D. Enhancing Selectivity of Protein Biopharmaceuticals in Ion Exchange Chromatography through Addition of Organic Modifiers. International Journal of Molecular Sciences. 2023; 24(23):16623. https://doi.org/10.3390/ijms242316623
Chicago/Turabian StyleDuivelshof, Bastiaan Laurens, Thomas Bouvarel, Sebastian Pirner, Vincent Larraillet, Alexander Knaupp, Hans Koll, Valentina D’Atri, and Davy Guillarme. 2023. "Enhancing Selectivity of Protein Biopharmaceuticals in Ion Exchange Chromatography through Addition of Organic Modifiers" International Journal of Molecular Sciences 24, no. 23: 16623. https://doi.org/10.3390/ijms242316623






