Genome-Wide Identification and Characterization of Gibberellic Acid-Stimulated Arabidopsis Gene Family in Pineapple (Ananas comosus)
Abstract
:1. Introduction
2. Results
2.1. Physicochemical Properties of the AcGASA Gene
2.2. Characterization of the AcGASA Genes
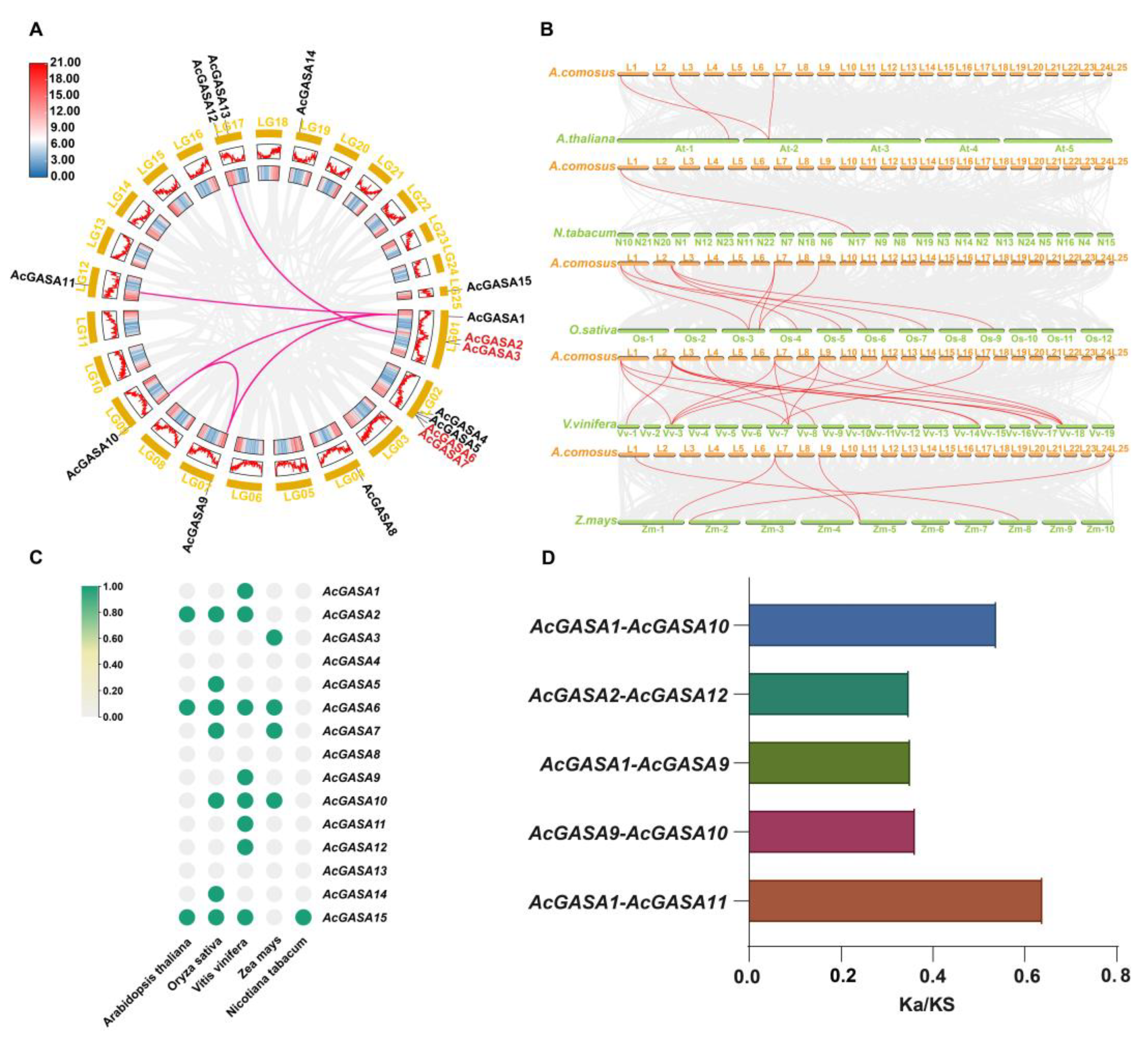
2.3. Developmental Relationships of Different Species of the GASA Family
2.4. Phylogenetic Analysis of the GASA Family Genes
2.5. Origins of the AcGASA Gene
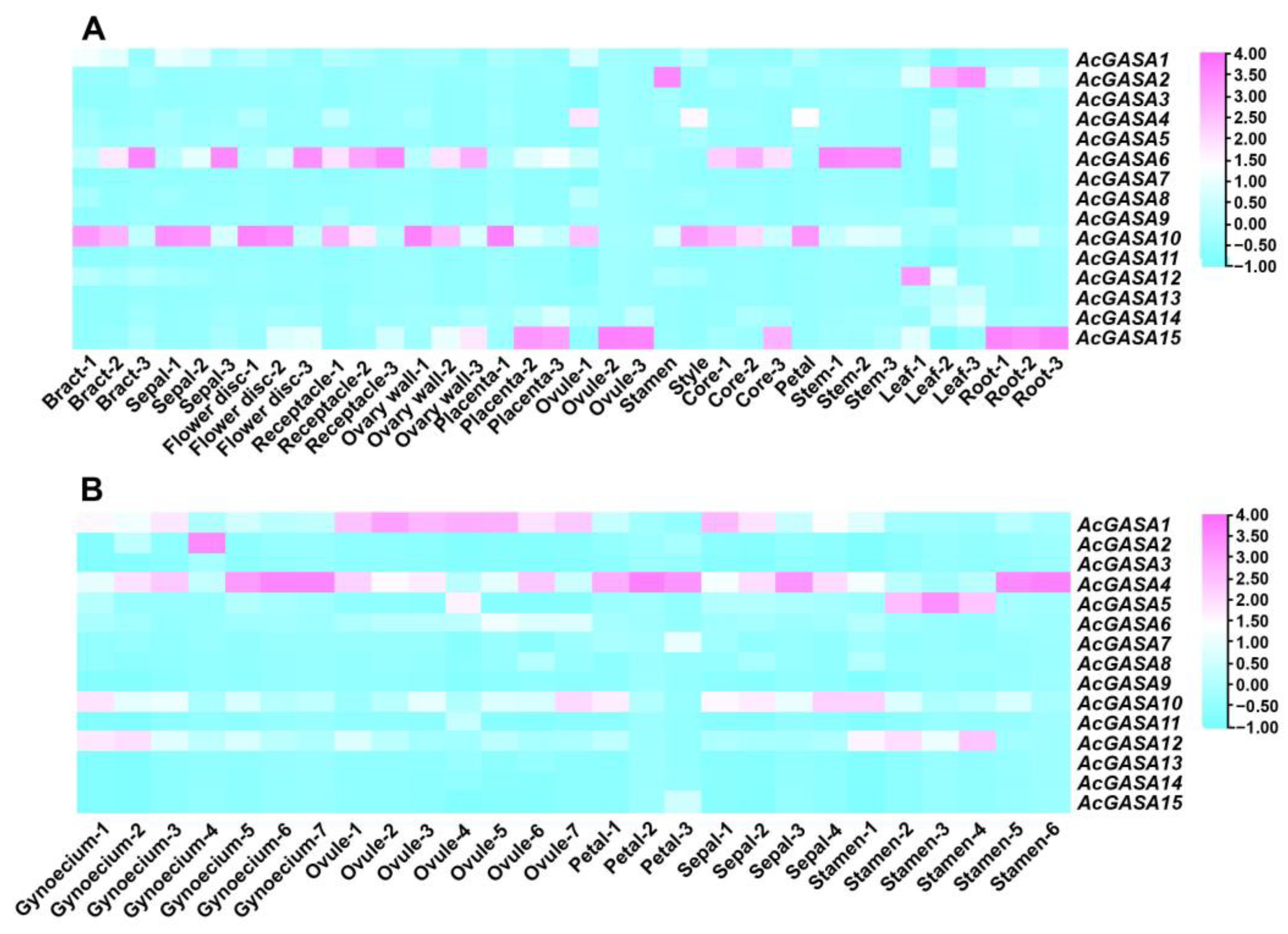
2.6. Expression Profile of AcGASA Gene in Different Tissue Parts of Pineapple
2.7. AcGASA Gene Protein Interactions Prediction and miRNA Target Prediction
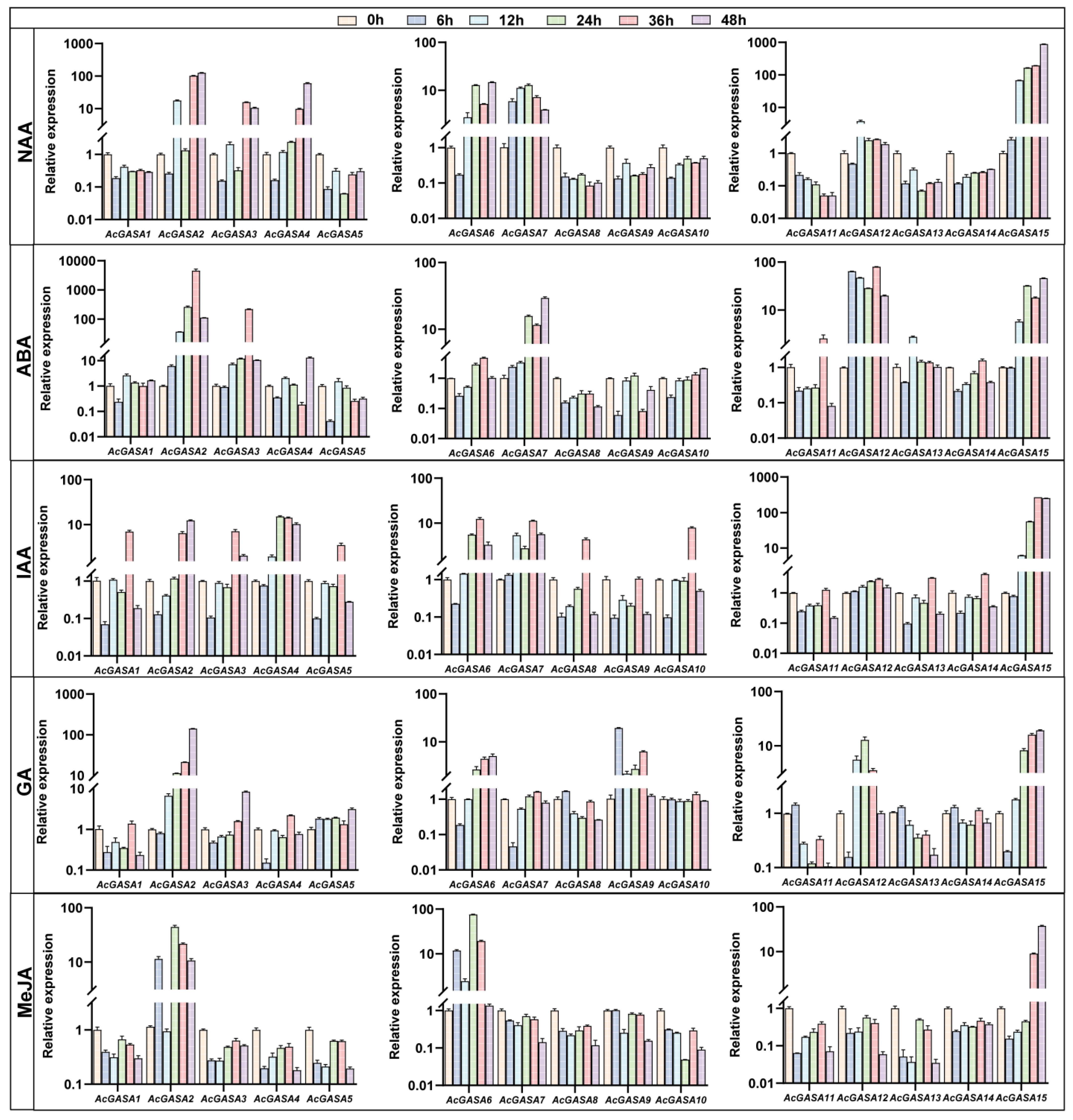
2.8. AcGASA Gene Promoter Analysis and Expression Patterns upon Different Hormone Treatments
3. Discussion
4. Materials and Methods
4.1. Protein Sequence Source
4.2. Identification and Classification of GASA
4.3. Gene Structure and Conserved Motif Analysis of the AcGASA Gene
4.4. Collinear Analysis and Chromosomal Location of the AcGASA Gene
4.5. Analysis of the Cis-Elements of the AcGASA Gene Promoter
4.6. Protein Interaction Network and miRNA Target Prediction of the AcGASA Gene
4.7. Expression Map of the AcGASA Gene in Different Pineapple Tissues
4.8. Hormone Treatment of Pineapple Seedlings and AcGASA Expression Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nahirñak, V.; Almasia, N.I.; Hopp, H.E.; Vazquez-Rovere, C. Snakin/GASA Proteins. Plant Signal. Behav. 2012, 7, 1004–1008. [Google Scholar] [CrossRef]
- Hou, D.; Bai, Q.; Bai, Q.; Li, J.; Xie, L.; Li, X.; Cheng, Z.; Gao, J. The Gibberellic Acid-Stimulated Transcript Gene Family in Moso Bamboo: A Genome-Wide Survey and Expression Profiling during Development and Abiotic Stresses. J. Plant Growth Regul. 2018, 37, 1135–1147. [Google Scholar] [CrossRef]
- Aubert, D.; Chevillard, M.; Dorne, A.M.; Arlaud, G.; Herzog, M. Expression patterns of GASA genes in Arabidopsis thaliana: The GASA4 gene is up-regulated by gibberellins in meristematic regions. Plant Mol. Biol. 1998, 36, 871–883. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Gao, J.; Wang, G.; Wang, S.; Chen, K.; Pu, W.; Wang, Y.; Xia, Q.; Fan, X. Genome-Wide Identification and Characterization of GASA Gene Family in Nicotiana tabacum. Front. Genet. 2022, 12, 768942. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, T.; Sakaguchi, N.; Shimada, H. Two OsGASR genes, rice GAST homologue genes that are abundant in proliferating tissues, show different expression patterns in developing panicles. Genes Genet. Syst. 2006, 81, 171–180. [Google Scholar] [CrossRef]
- Ahmad, B.; Yao, J.; Zhang, S.; Li, X.; Zhang, X.; Yadav, V.; Wang, X. Genome-Wide Characterization and Expression Profiling of GASA Genes during Different Stages of Seed Development in Grapevine (Vitis vinifera L.) Predict Their Involvement in Seed Development. Int. J. Mol. Sci. 2020, 21, 1088. [Google Scholar] [CrossRef]
- Nahirñak, V.; Rivarola, M.; Gonzalez de Urreta, M.; Paniego, N.; Hopp, H.E.; Almasia, N.I.; Vazquez-Rovere, C. Genome-wide Analysis of the Snakin/GASA Gene Family in Solanum tuberosum cv. Kennebec. Am. J. Potato Res. 2016, 93, 172–188. [Google Scholar] [CrossRef]
- Han, S.; Jiao, Z.; Niu, M.X.; Yu, X.; Huang, M.; Liu, C.; Wang, H.L.; Zhou, Y.; Mao, W.; Wang, X.; et al. Genome-Wide Comprehensive Analysis of the GASA Gene Family in Populus. Int. J. Mol. Sci. 2021, 22, 12336. [Google Scholar] [CrossRef]
- García, A.N.; Ayub, N.D.; Fox, A.R.; Gómez, M.C.; Diéguez, M.J.; Pagano, E.M.; Berini, C.A.; Muschietti, J.P.; Soto, G. Alfalfa snakin-1 prevents fungal colonization and probably coevolved with rhizobia. BMC Plant Biol. 2014, 14, 248. [Google Scholar] [CrossRef]
- Wu, T.; Zhong, Y.; Chen, M.; Wu, B.; Wang, T.; Jiang, B.; Zhong, G. Analysis of CcGASA family members in Citrus clementina (Hort. ex Tan.) by a genome-wide approach. BMC Plant Biol. 2021, 21, 565. [Google Scholar] [CrossRef]
- Moyano-Canete, E.; Bellido, M.L.; García-Caparrós, N.; Medina-Puche, L.; Amil-Ruiz, F.; González-Reyes, J.A.; Caballero, J.L.; Munoz-Blanco, J.; Blanco-Portales, R. FaGAST2, a strawberry ripening-related gene, acts together with FaGAST1 to determine cell size of the fruit receptacle. Plant Cell Physiol. 2013, 54, 218–236. [Google Scholar] [CrossRef]
- Nahirñak, V.; Almasia, N.I.; Fernandez, P.V.; Hopp, H.E.; Estevez, J.M.; Carrari, F.; Vazquez-Rovere, C. Potato Snakin-1 Gene Silencing Affects Cell Division, Primary Metabolism, and Cell Wall Composition. Plant Physiol. 2012, 158, 252–263. [Google Scholar] [CrossRef] [PubMed]
- de la Fuente, J.I.; Amaya, I.; Castillejo, C.; Sánchez-Sevilla, J.F.; Quesada, M.A.; Botella, M.A.; Valpuesta, V. The strawberry gene FaGAST affects plant growth through inhibition of cell elongation. J. Exp. Bot. 2006, 57, 2401–2411. [Google Scholar] [CrossRef]
- Zimmermann, R.; Sakai, H.; Hochholdinger, F. The Gibberellic Acid Stimulated-Like gene family in maize and its role in lateral root development. Plant Physiol. 2010, 152, 356–365. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Yang, C.; Peng, J.; Sun, S.; Wang, X. GASA5, a regulator of flowering time and stem growth in Arabidopsis thaliana. Plant Mol. Biol. 2009, 69, 745–759. [Google Scholar] [CrossRef] [PubMed]
- Roxrud, I.; Lid, S.E.; Fletcher, J.C.; Schmidt, E.D.L.; Opsahl-Sorteberg, H.G. GASA4, one of the 14-member Arabidopsis GASA family of small polypeptides, regulates flowering and seed development. Plant Cell Physiol. 2007, 48, 471–483. [Google Scholar] [CrossRef] [PubMed]
- Ben-Nissan, G.; Lee, J.Y.; Borohov, A.; Weiss, D. GIP, a Petunia hybrida GA-induced cysteine-rich protein: A possible role in shoot elongation and transition to flowering. Plant J. 2003, 37, 229–238. [Google Scholar] [CrossRef]
- Rezaee, S.; Ahmadizadeh, M.; Heidari, P. Genome-wide characterization, expression profiling, and post-transcriptional study of GASA gene family. Gene Rep. 2020, 20, 100795. [Google Scholar] [CrossRef]
- Li, X.; Shi, S.; Tao, Q.; Tao, Y.; Miao, J.; Peng, X.; Li, C.; Yang, Z.; Zhou, Y.; Liang, G. OsGASR9 positively regulates grain size and yield in rice (Oryza sativa). Plant Sci. 2019, 286, 17–27. [Google Scholar] [CrossRef]
- Herzog, M.; Dorne, A.M.; Grellet, F. GASA, a gibberellin-regulated gene family from Arabidopsis thaliana related to the tomato GAST1 gene. Plant Mol. Biol. 1995, 27, 743–752. [Google Scholar] [CrossRef]
- Wang, H.; Wei, T.; Wang, X.; Zhang, L.; Yang, M.; Chen, L.; Song, W.; Wang, C.; Chen, C. Transcriptome Analyses from Mutant Salvia miltiorrhiza Reveals Important Roles for SmGASA4 during Plant Development. Int. J. Mol. Sci. 2018, 19, 2088. [Google Scholar] [CrossRef]
- Ko, C.B.; Woo, Y.M.; Lee, M.C.; Kim, C.S. Enhanced tolerance to heatstress in transgenic plants expressing the GASA4 gene. Plant Physiol. Biochem. 2007, 45, 722–728. [Google Scholar] [CrossRef]
- Alonso-Ramírez, A.; Rodríguez, D.; Reyes, D.; Jiménez, J.A.; Nicolás, G.; López-Climent, M.; Gómez-Cadenas, A.; Nicolás, C. Evidence for a Role of Gibberellins in Salicylic Acid-Modulated Early Plant Responses to Abiotic Stress in Arabidopsis Seeds. Plant Physiol. 2009, 150, 1335–1344. [Google Scholar] [CrossRef] [PubMed]
- Li, K.L.; Bai, X.; Li, Y.; Cai, H.; Ji, W.; Tang, L.L.; Wen, Y.D.; Zhu, Y.M. GsGASA1 mediated root growth inhibition in response to chronic cold stress is marked by the accumulation of DELLAs. J. Plant Physiol. 2011, 168, 2153–2160. [Google Scholar] [CrossRef]
- Kovalskaya, N.; Hammond, R.W. Expression and functional characterization of the plant antimicrobial snakin-1 and defensin recombinant proteins. Protein Expr. Purif. 2009, 63, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Almasia, N.I.; Bazzini, A.A.; Hopp, H.E.; Vazquez-Rovere, C. Overexpression of snakin-1 gene enhances resistance to Rhizoctonia solani and Erwinia carotovora in transgenic potato plants. Mol. Plant Pathol. 2008, 9, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Berrocal-Lobo, M.; Segura, A.; Moreno, M.; López, G.; Garcıa-Olmedo, F.; Molina, A. Snakin-2, an antimicrobial peptide from potato whose gene is locally induced by wounding and responds to pathogen infection. Plant Physiol. 2002, 128, 951–961. [Google Scholar] [CrossRef]
- Wu, T.; Cheng, C.; Zhong, Y.; Lv, Y.; Ma, Y.; Zhong, G. Molecular characterization of the gibberellin-stimulated transcript of GASA4 in Citrus. Plant Growth Regul. 2020, 91, 89–99. [Google Scholar] [CrossRef]
- He, H.; Yang, X.; Xun, H.; Lou, X.; Li, S.; Zhang, Z.; Jiang, L.; Dong, Y.; Wang, S.; Pang, J. Over-expression of GmSN1 enhances virus resistance in Arabidopsis and soybean. Plant Cell Rep. 2017, 36, 1441–1455. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, X. Expression pattern of GASA, downstream genes of DELLA, in Arabidopsis. Chin. Sci. Bull. 2008, 53, 3839–3846. [Google Scholar] [CrossRef]
- Balaji, V.; Smart, C.D. Over-expression of snakin-2 and extensin-like protein genes restricts pathogen invasiveness and enhances tolerance to Clavibacter michiganensis subsp. michiganensis in transgenic tomato (Solanum lycopersicum). Transgenic Res. 2012, 21, 23–37. [Google Scholar] [CrossRef]
- Rubinovich, L.; Weiss, D. The Arabidopsis cysteine-rich protein GASA4 promotes GA responses and exhibits redox activity in bacteria and in planta. Plant J. 2010, 64, 1018–1027. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Kang, S.G.; Hah, C.; Jang, J. Molecular and cellular characterization of GA-Stimulated Transcripts GASA4 and GASA6 in Arabidopsis thaliana. Plant Sci. 2016, 246, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zhong, C.; Xu, H.; Ye, S.; Wang, S.; Li, L.; Zhang, S.; Wang, X. AtGASA6 Serves as an Integrator of Gibberellin-, Abscisic Acid- and Glucose-Signaling during Seed Germination in Arabidopsis. Plant Physiol. 2015, 169, 858–2015. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, Z.; Xu, Y.; Joo, S.H.; Kim, S.K.; Xue, Z.; Xu, Z.; Wang, Z.; Chong, K. OsGSR is involved in crosstalk between gibberellins and brassinosteroids in rice. Plant J. 2009, 57, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Liu, W.; Li, X.; Li, H. Overexpression of GmGASA32 promoted soybean height by interacting with GmCDC25. Plant Signal. Behav. 2021, 16, 1855017. [Google Scholar] [CrossRef]
- Raventos, D.; Meier, C.; Mattsson, O.; Jensen, A.B.; Mundy, J. Fusion genetic analysis of gibberellin signaling mutants. Plant J. 2001, 22, 427–438. [Google Scholar] [CrossRef]
- Shi, L.; Gast, R.T.; Gopalraj, M.; Olszewski, N.E. Characterization of a shoot-specific, GA3-and ABA-regulated gene from tomato. Plant J. 1992, 2, 153–159. [Google Scholar] [CrossRef]
- Fan, S.; Zhang, D.; Zhang, L.; Gao, C.; Xin, M.; Tahir, M.M.; Li, Y.; Ma, J.; Han, M. Comprehensive analysis of GASA family members in the Malus domestica genome: Identification, characterization, and their expressions in response to apple flower induction. BMC Genom. 2017, 18, 827. [Google Scholar] [CrossRef]
- An, B.; Wang, Q.; Zhang, X.; Zhang, B.; Luo, H.; He, C. Comprehensive transcriptional and functional analyses of HbGASA genes reveal their roles in fungal pathogen resistance in Hevea brasiliensis. Tree Genet. Genomes 2018, 14, 41. [Google Scholar] [CrossRef]
- Lobo, M.G.; Siddiq, M. Overview of pineapple production, postharvest physiology, processing and nutrition. In Handbook of Pineapple Technology: Production, Postharvest Science, Processing and Nutrition, 1st ed.; Lobo, M.G., Paull, R., Eds.; John Wiley & Sons: Chichester, UK, 2017; Volume 1, pp. 1–15. [Google Scholar]
- Baruwa, O.I. Profitability and constraints of pineapple production in Osun State, Nigeria. J. Hortic. Res. 2013, 21, 59–64. [Google Scholar] [CrossRef]
- d’Eeckenbrugge, G.C.; Leal, F. History, distribution and world production. In The Pineapple: Botany, Production and Uses, 2nd ed.; Sanewski, G.M., Bartholomew, D.P., Paull, R.E., Eds.; CABI Publishing: Wallingford, UK, 2018; Volume 1, pp. 1–10. [Google Scholar]
- Lynch, M.; Conery, J. The Evolutionary Fate and Consequences of Duplicate Genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [PubMed]
- Moore, R.C.; Purugganan, M.D. The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 15682–15687. [Google Scholar] [CrossRef] [PubMed]
- Maere, S.; Bodt, S.D.; Raes, J.; Casneuf, T.; Montagu, M.V.; Kuiper, M.; de Peer, Y.V. Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. USA 2005, 102, 5454–5459. [Google Scholar] [CrossRef]
- Mao, Q.; Chen, C.; Xie, T.; Luan, A.; Liu, C.; He, Y. Comprehensive tissue-specific transcriptome profiling of pineapple (Ananas comosus) and building an eFP-browser for further study. PeerJ 2018, 6, e6028. [Google Scholar] [CrossRef]
- Wang, L.; Li, Y.; Jin, X.; Liu, L.; Dai, X.; Liu, Y.; Zhao, L.; Zheng, P.; Wang, X.; Liu, Y.; et al. Floral transcriptomes reveal gene networks in pineapple floral growth and fruit development. Commun. Biol. 2020, 3, 500. [Google Scholar] [CrossRef] [PubMed]
- Yusuf, N.H.M.; Ong, W.D.; Redwan, R.M.; Latip, M.A.; Kumar, S.V. Discovery of precursor and mature microRNAs and their putative gene targets using high-throughput sequencing in pineapple (Ananas comosus var. comosus). Gene 2015, 571, 71–80. [Google Scholar] [CrossRef]
- Meslin, C.; Mugnier, S.; Callebaut, I.; Laurin, M.; Pascal, G.; Poupon, A.; Goudet, G.; Monget, P. Evolution of genes involved in gamete interaction: Evidence for positive selection, duplications and losses in vertebrates. PLoS ONE 2012, 7, e44548. [Google Scholar] [CrossRef]
- Zhang, M.; Cheng, W.; Wang, J.; Cheng, T.; Zhang, Q. Genome-Wide Identification, Evolution, and Expression Analysis of GASA Gene Family in Prunus mume. Int. J. Mol. Sci. 2022, 23, 10923. [Google Scholar] [CrossRef]
- Sun, S.; Wang, H.; Yu, H.; Zhong, C.; Zhang, X.; Peng, J.; Wang, X. GASA14 regulates leaf expansion and abiotic stress resistance by modulating reactive oxygen species accumulation. J. Exp. Bot. 2013, 64, 1637–1647. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, A.; Kumar, P.; Sarkar, A.K. Giberellic Acid-Stimulated Transcript Proteins Evolved through Successive Conjugation of Novel Motifs and Their Subfunctionalization. Plant Physiol. 2019, 180, 998–1012. [Google Scholar] [CrossRef] [PubMed]
- Zan, T.; Zhang, L.; Xie, T.; Li, L. Genome-Wide Identification and Analysis of the Growth-Regulating Factor (GRF) Gene Family and GRF-Interacting Factor Family in Triticum aestivum L. Biochem. Genet. 2020, 58, 705–724. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon–intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef] [PubMed]
- Paterson, A.H.; Freeling, M.; Tang, H.; Wang, X. Insights from the comparison of plant genome sequences. Annu Rev. Plant Biol. 2010, 61, 349–372. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chen, J.; Liang, C.; Liu, F.; Hou, X.; Zou, X. Genome-Wide Identification and Characterization of the bHLH Transcription Factor Family in Pepper (Capsicum annuum L.). Front. Genet. 2020, 11, 570156. [Google Scholar] [CrossRef]
- Chow, C.; Lee, T.; Hung, Y.; Li, G.; Tseng, K.; Liu, Y.; Kuo, P.; Zheng, H.; Chang, W. PlantPAN3.0: A new and updated resource for reconstructing transcriptional regulatory networks from ChIP-seq experiments in plants. Nucleic Acids Res. 2019, 47, D1155–D1163. [Google Scholar] [CrossRef]
- Peng, J.; Lai, L.; Wang, X. Temporal and spatial expression analysis of PRGL in Gerbera hybrida. Mol. Biol. Rep. 2010, 37, 3311–3317. [Google Scholar] [CrossRef]
- Muhammad, I.; Li, W.Q.; Jing, X.Q.; Zhou, M.R.; Shalmani, A.; Ali, M.; Wei, X.Y.; Sharif, R.; Liu, W.T.; Chen, K.M. A Systematic In Silico Prediction of Gibberellic Acid Stimulated GASA Family Members: A Novel Small Peptide Contributes to Floral Architecture and Transcriptomic Changes Induced by External Stimuli in Rice. J. Plant Physiol. 2019, 234–235, 117–132. [Google Scholar] [CrossRef]
- Kaikai, Q.; Changkai, M.; Jiaoyan, L.; Chaojun, Z.; Qifeng, M.; Shuli, F. Identification, characterization, and expression profiles of the GASA genes in cotton. J. Cotton Res. 2021, 4, 7. [Google Scholar]
- Ming, R.; VanBuren, R.; Wai, C.M.; Tang, H.; Schatz, M.C.; Bowers, J.E.; Lyons, E.; Wang, M.-L.; Chen, J.; Biggers, E.; et al. The pineapple genome and the evolution of CAM photosynthesis. Nat. Genet. 2015, 47, 1435–1442. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “One for All, All for One” Bioinformatics Platform for Biological Big-data Mining. Mol Plant 2023, 22, S1674–S2052. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef] [PubMed]






| Gene Name | Gene ID | PL (aa) | MW (kDa) | pI | GRAVY | AI | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| AcGASA1 | Aco012370 | 90 | 10.08 | 9.07 | −0.149 | 64.00 | Cell membrane, Golgi, nucleus |
| AcGASA2 | Aco011225 | 94 | 10.17 | 8.48 | 0.007 | 72.66 | Golgi |
| AcGASA3 | Aco011224 | 63 | 6.96 | 9.16 | −0.805 | 17.14 | Golgi, nucleus |
| AcGASA4 | Aco001116 | 111 | 12.35 | 8.84 | −0.307 | 56.31 | Golgi |
| AcGASA5 | Aco001067 | 95 | 10.3 | 8.98 | 0.014 | 58.53 | Cell membrane, Golgi, nucleus |
| AcGASA6 | Aco000980 | 63 | 6.67 | 8.96 | −0.398 | 29.52 | Golgi, nucleus |
| AcGASA7 | Aco000979 | 116 | 12.37 | 8.59 | 0.011 | 69.14 | Cell wall |
| AcGASA8 | Aco002373 | 155 | 17.39 | 9.25 | −0.691 | 45.42 | Nucleus |
| AcGASA9 | Aco004893 | 95 | 10.72 | 9.20 | −0.229 | 74.00 | Cell membrane, Golgi, nucleus |
| AcGASA10 | Aco008536 | 97 | 10.69 | 8.98 | −0.070 | 71.44 | Cell membrane, cell wall, Golgi |
| AcGASA11 | Aco000098 | 88 | 9.85 | 8.88 | −0.365 | 54.43 | Golgi |
| AcGASA12 | Aco003244 | 114 | 12.51 | 9.23 | −0.104 | 65.96 | Golgi |
| AcGASA13 | Aco015815 | 93 | 10.54 | 9.17 | −0.932 | 38.92 | Golgi, nucleus |
| AcGASA14 | Aco015632 | 106 | 11.52 | 8.65 | −0.010 | 73.77 | nucleus |
| AcGASA15 | Aco010336 | 67 | 7.12 | 8.79 | −0.482 | 35.07 | Golgi, nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, M.; Liu, C.; Zhang, W.; Wu, J.; Zhong, Z.; Yi, W.; Liu, H.; Leng, Y.; Sun, W.; Luan, A.; et al. Genome-Wide Identification and Characterization of Gibberellic Acid-Stimulated Arabidopsis Gene Family in Pineapple (Ananas comosus). Int. J. Mol. Sci. 2023, 24, 17063. https://doi.org/10.3390/ijms242317063
Yang M, Liu C, Zhang W, Wu J, Zhong Z, Yi W, Liu H, Leng Y, Sun W, Luan A, et al. Genome-Wide Identification and Characterization of Gibberellic Acid-Stimulated Arabidopsis Gene Family in Pineapple (Ananas comosus). International Journal of Molecular Sciences. 2023; 24(23):17063. https://doi.org/10.3390/ijms242317063
Chicago/Turabian StyleYang, Mingzhe, Chaoyang Liu, Wei Zhang, Jing Wu, Ziqin Zhong, Wen Yi, Hui Liu, Yan Leng, Weisheng Sun, Aiping Luan, and et al. 2023. "Genome-Wide Identification and Characterization of Gibberellic Acid-Stimulated Arabidopsis Gene Family in Pineapple (Ananas comosus)" International Journal of Molecular Sciences 24, no. 23: 17063. https://doi.org/10.3390/ijms242317063





