Altered Phenotypes of Breast Epithelial × Breast Cancer Hybrids after ZEB1 Knock-Out
Abstract
:1. Introduction
2. Results
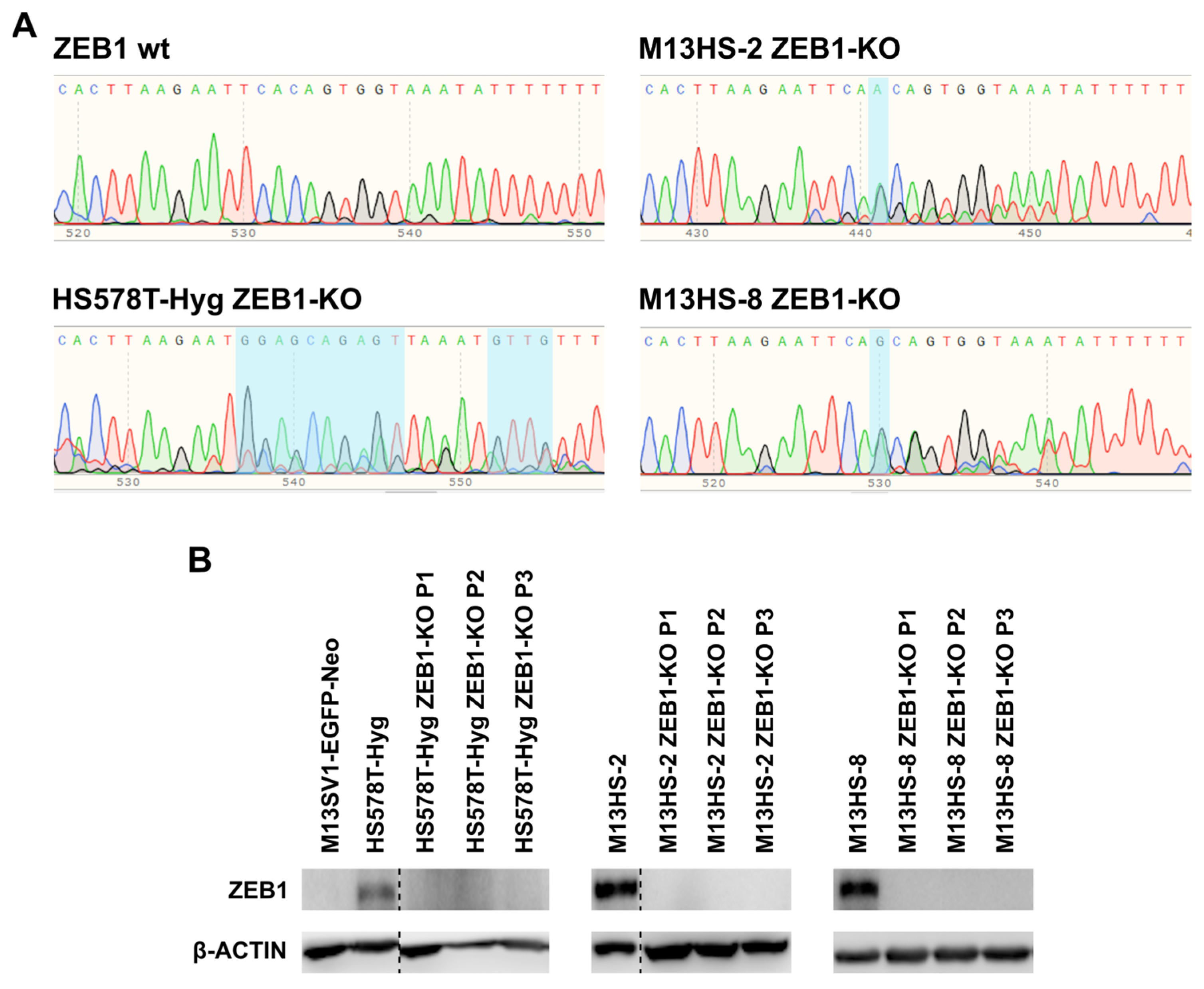
2.1. Succesful ZEB1-KO in HS578T-Hyg and M13HS-2 and -8 Tumor Hybrids
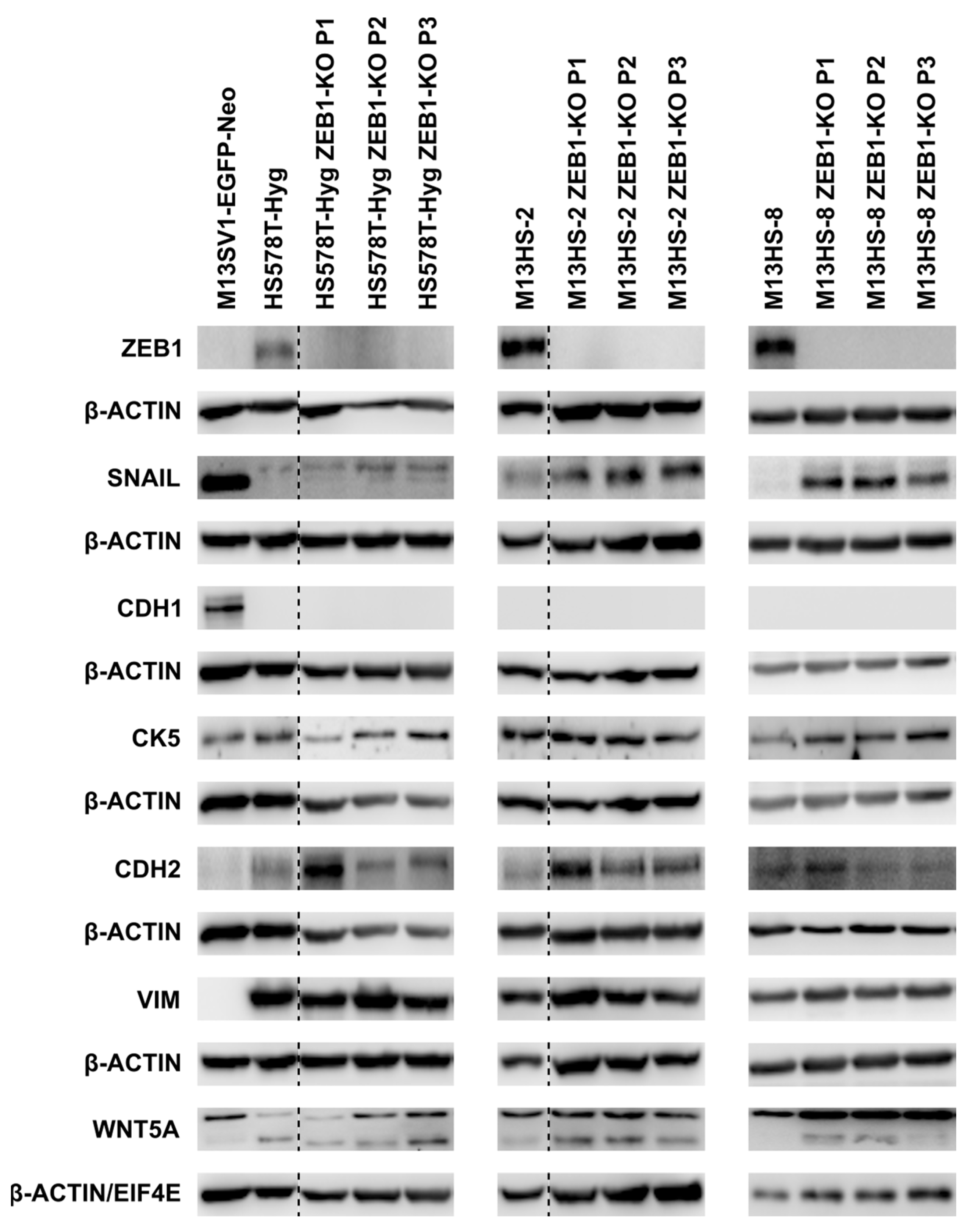
2.2. ZEB1-KO Is Not Correlated to a Markedly Altered E/M Gene Expression Profile
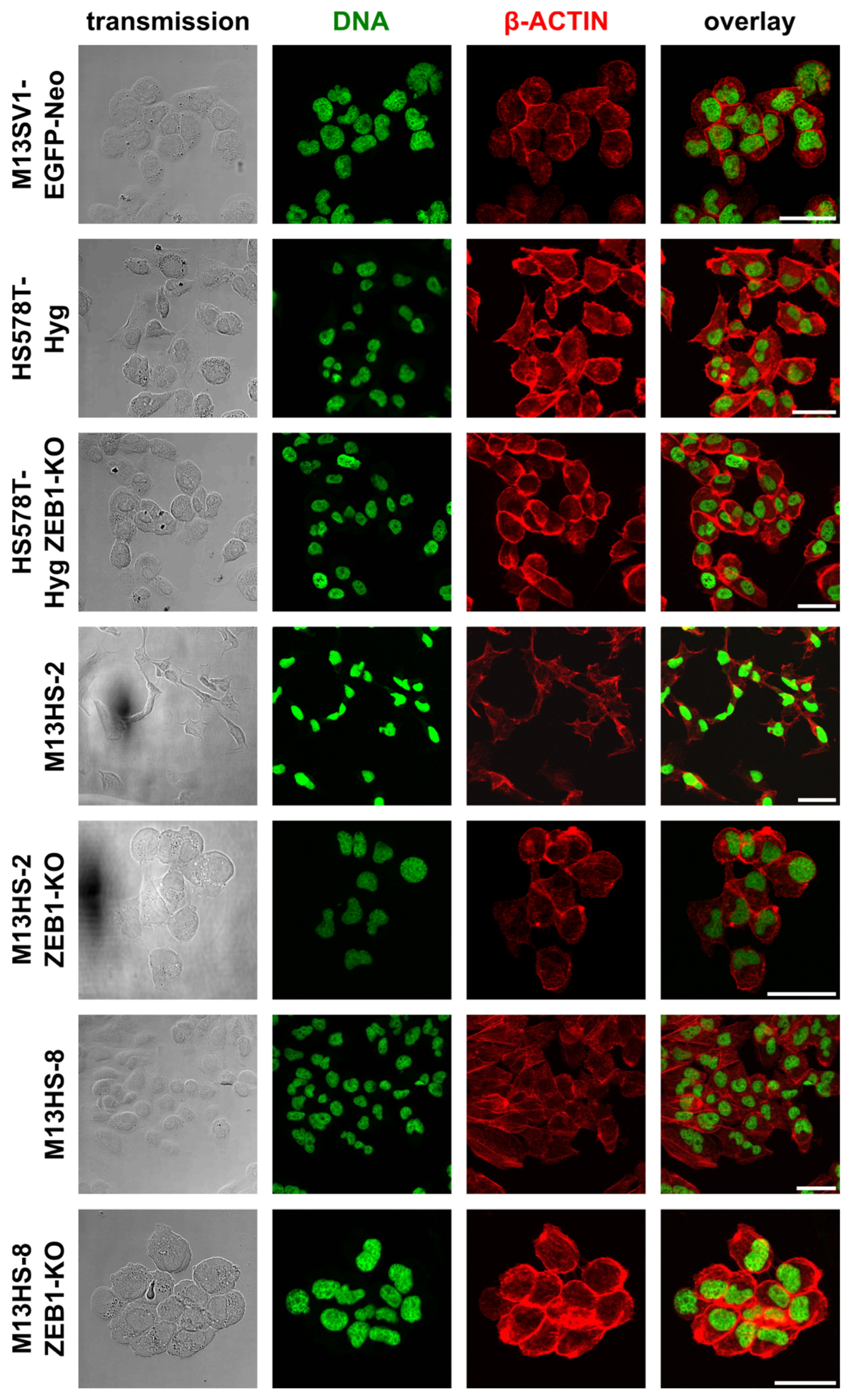
2.3. ZEB1-KO Cells Exhibit a More Round-Shaped Epithelial Morphology
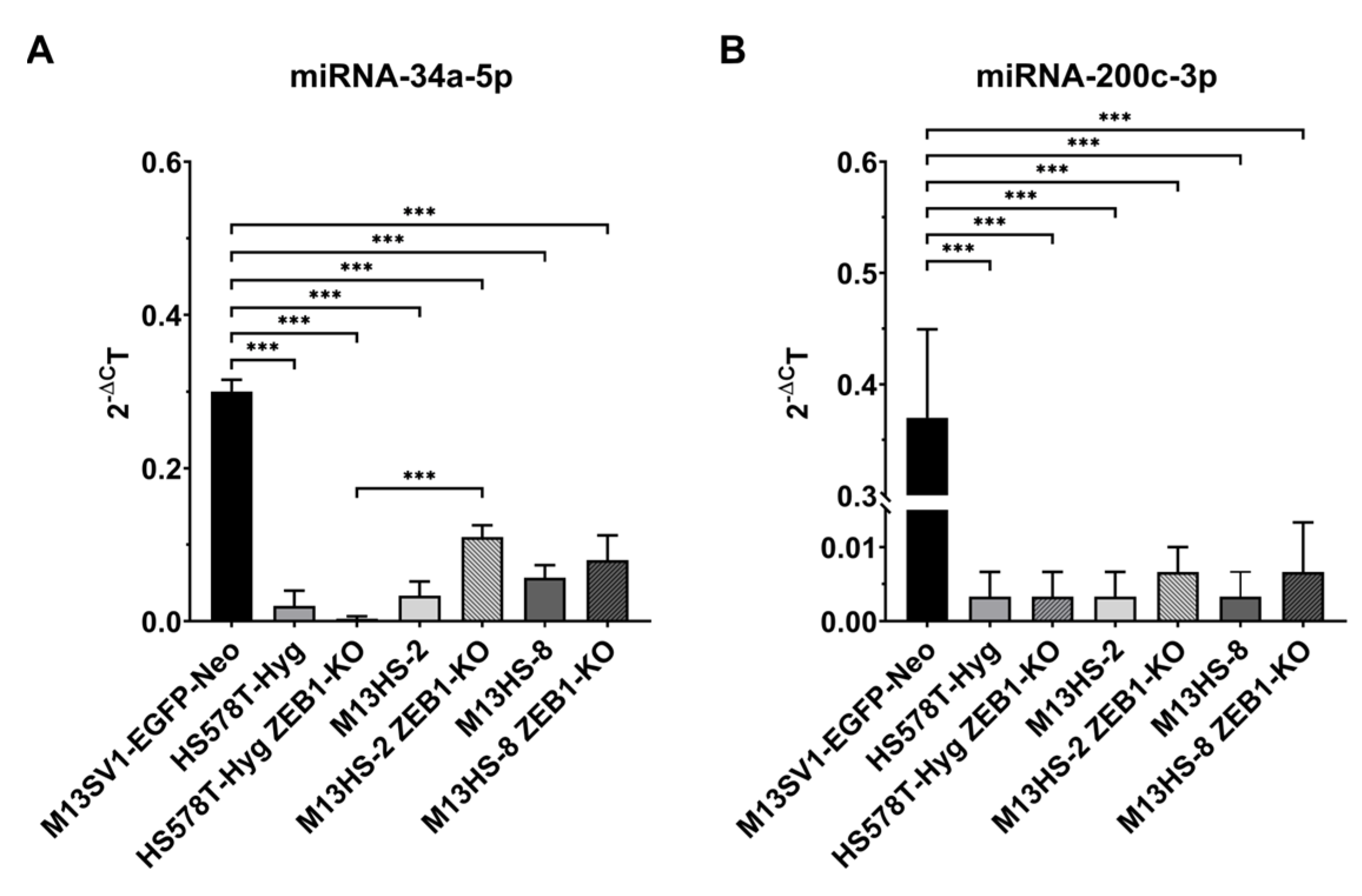
2.4. miRNA-34a-5p, but Not miRNA-200c-3p Levels Are Slightly Increased in ZEB1-KO Cells
2.5. ZEB2 Levels of ZEB1-KO Cells Were Comparable to Wildtype Cells
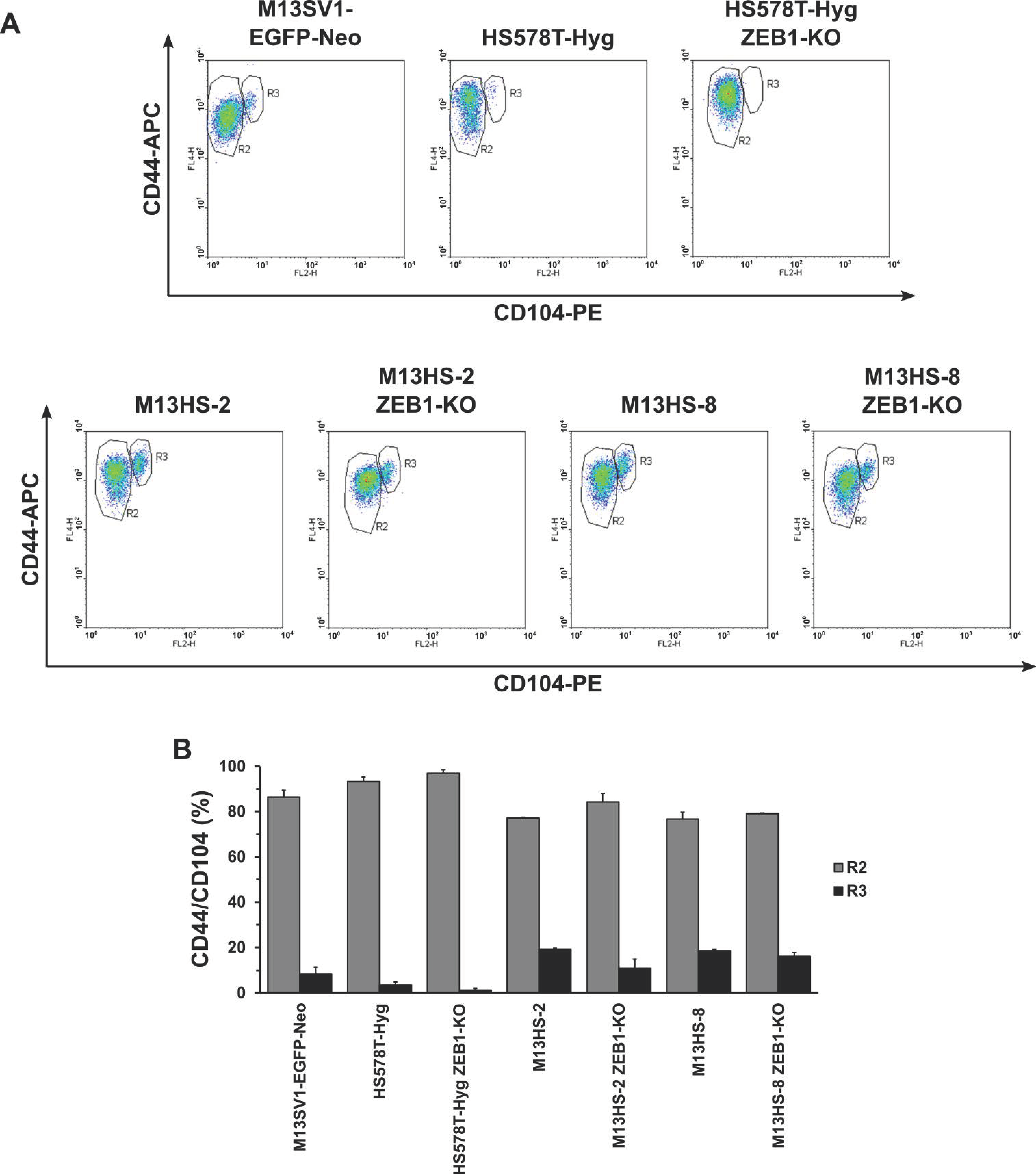
2.6. CD44/CD104 Expression Profile
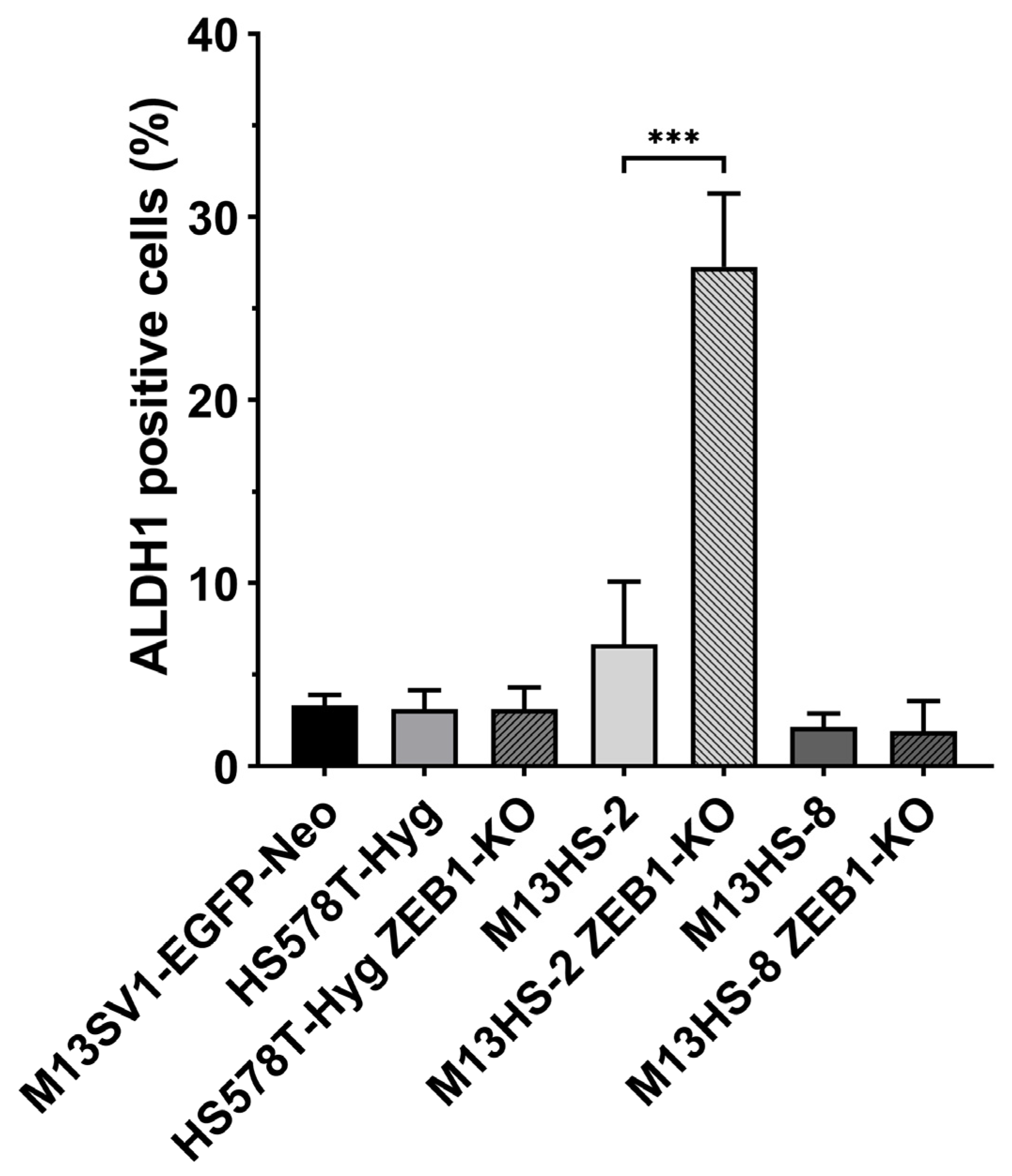
2.7. M13HS-2 ZEB1-KO Cells Exhibit a Markedly Enriched ALDH1 Positive Population
2.8. ZEB1-KO Cells Exhibit a Decreased Colony Formation Capacity
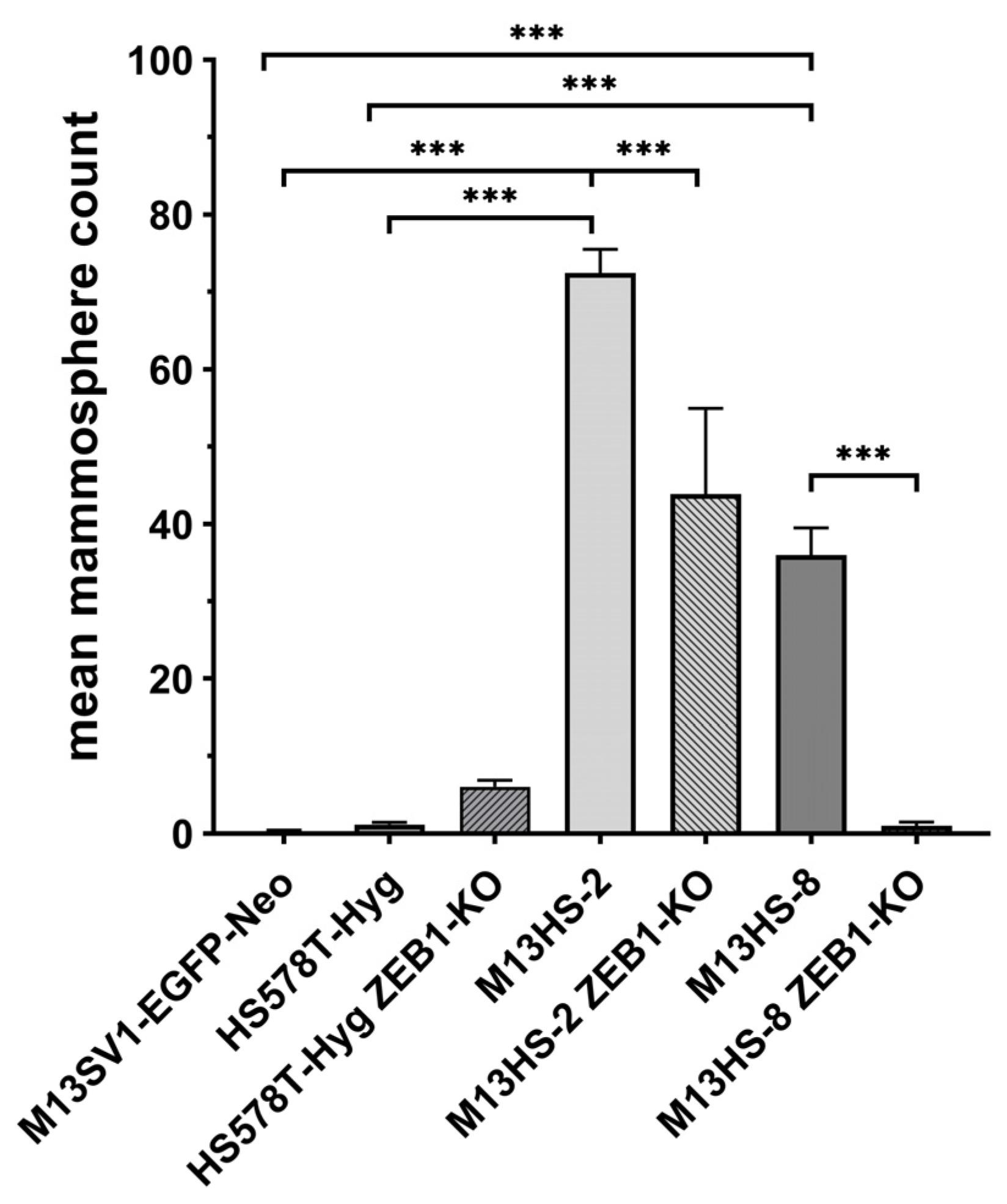
2.9. M13HS ZEB1-KO Cells Exhibit a Decreased Mammopshere Formation Capacity
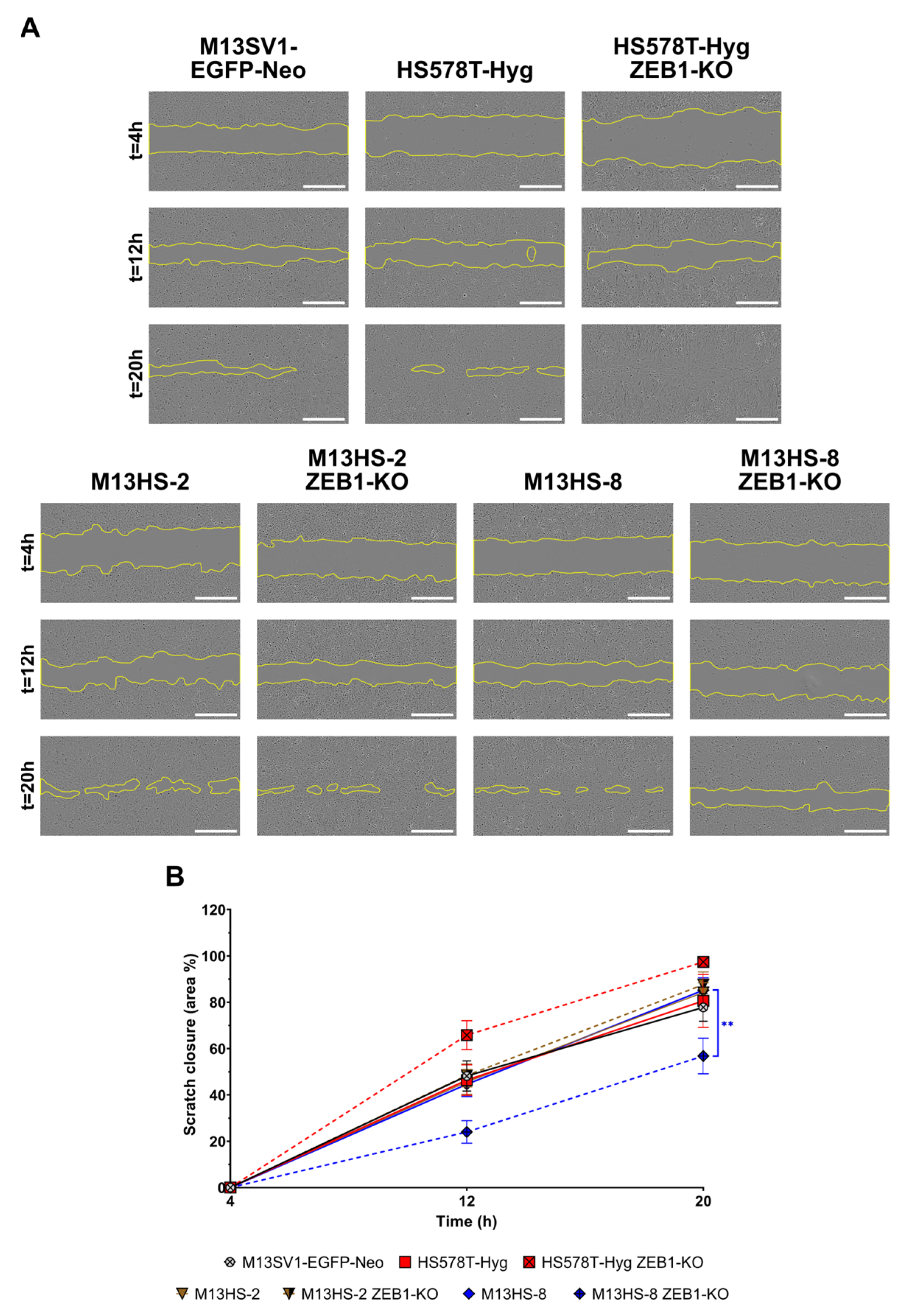
2.10. ZEB1-KO Cells Exhibit Different Migratory Properties
2.11. ZEB1-KO Cells Exhibit a Decrased Migratory Activity in Transwell/ Boyden Chamber Assays
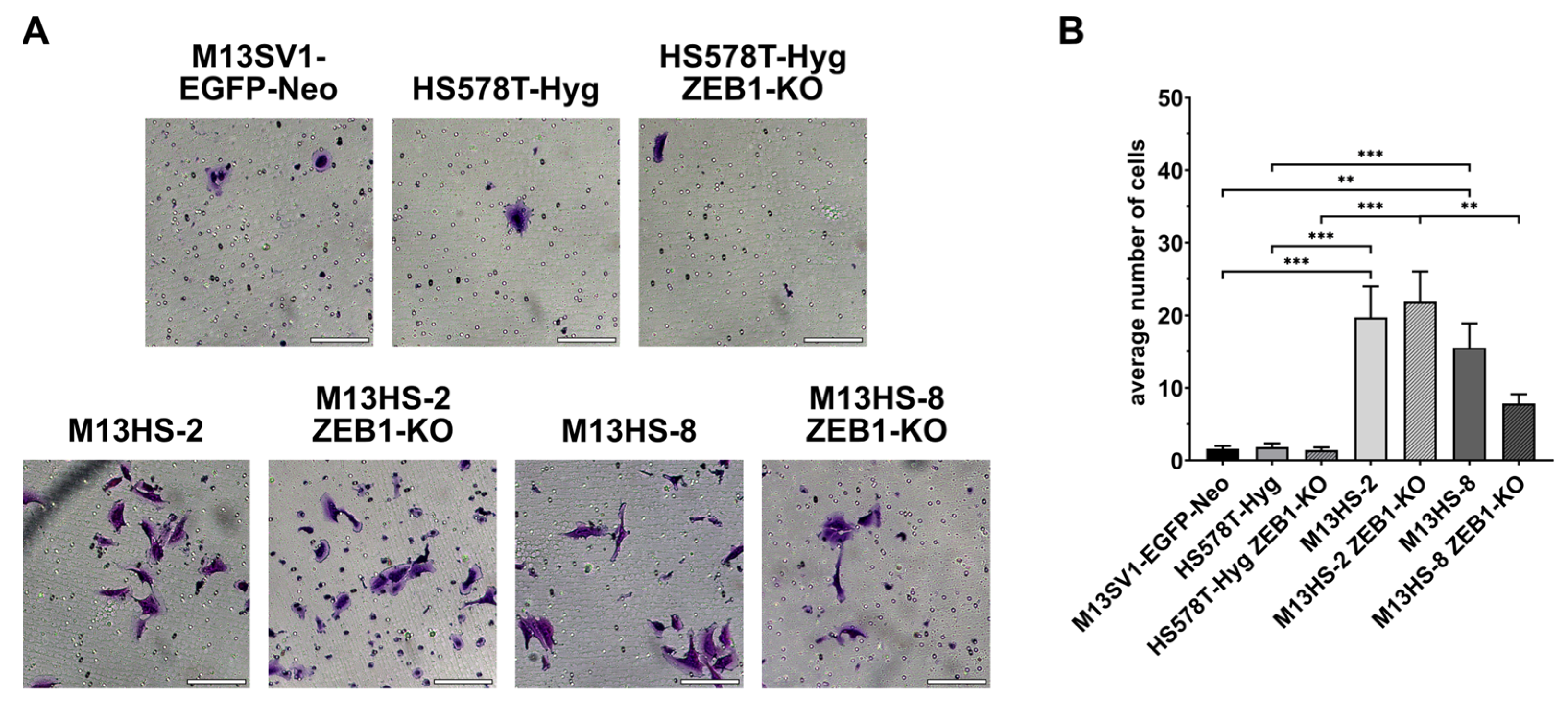
2.12. ZEB1-KO Cells Exhibit Different Invasion Capacities
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Generation of ZEB1-Knock-Out (KO) Cells
4.3. Colony Formation Assay
4.4. qPCR Analysis of miRNA Expression
4.5. Flow Cytometry
4.6. Western Blot Analysis
4.7. Mammosphere Formation Assay
4.8. Confocal Laser Scanning Microscopy
4.9. Scratch/Wound-Healing Assay
4.10. Transwell/Boydenchamber Assay and Invasion Assay
4.11. AldeRed Assay
4.12. Statistical Analysis
5. Conclusion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ALDH1 | aldehyde dehydrogenase 1 |
| BSA | bovine serum albumin |
| CK5 | CYTOKERATIN-5 |
| CSCs | cancer stem cells |
| DEAB | diethylaminobenzaldehyde |
| E | epithelial |
| CDH1 | E-CADHERIN |
| E/M | epithelial/mesenchymal |
| EMT | epithelial-to-mesenchymal transition |
| ESRP1 | epithelial splicing regulatory protein 1 |
| HAS2 | hyaluronic acid synthase 2 |
| KO | knock-out |
| M | mesenchymal |
| CDH2 | N-CADHERIN |
| PBS | phosphate-buffered saline |
| PVDF | polyvinyldifluoride |
| SDS-PAGE | sodium dodecylsulfate-polyacrylamide gel electrophoresis |
| TBS-T | Tris-buffered saline with 1% (v/v) Tween 20 |
| TF | transcription factor |
| TGF-β | transforming growth factor-β |
| VIM | VIMENTIN |
| ZEB1 | zinc finger E-box binding homeobox 1 |
References
- Lu, J.; Fei, F.; Wu, C.; Mei, J.; Xu, J.; Lu, P. ZEB1: Catalyst of immune escape during tumor metastasis. Biomed. Pharmacother. 2022, 153, 113490. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zheng, Y.L.; Jiang, C.; Fang, S.; Zeng, T.T.; Zhu, Y.H.; Li, Y.; Xie, D.; Guan, X.Y. HN1L-mediated transcriptional axis AP-2γ/METTL13/TCF3-ZEB1 drives tumor growth and metastasis in hepatocellular carcinoma. Cell Death Differ. 2019, 26, 2268–2283. [Google Scholar] [CrossRef] [PubMed]
- Drápela, S.; Bouchal, J.; Jolly, M.K.; Culig, Z.; Souček, K. ZEB1: A Critical Regulator of Cell Plasticity, DNA Damage Response, and Therapy Resistance. Front. Mol. Biosci. 2020, 7, 36. [Google Scholar] [CrossRef] [PubMed]
- Joseph, J.V.; Conroy, S.; Tomar, T.; Eggens-Meijer, E.; Bhat, K.; Copray, S.; Walenkamp, A.M.; Boddeke, E.; Balasubramanyian, V.; Wagemakers, M.; et al. TGF-β is an inducer of ZEB1-dependent mesenchymal transdifferentiation in glioblastoma that is associated with tumor invasion. Cell Death Dis. 2014, 5, e1443. [Google Scholar] [CrossRef] [PubMed]
- Lambert, A.W.; Weinberg, R.A. Linking EMT programmes to normal and neoplastic epithelial stem cells. Nat. Rev. Cancer 2021, 21, 325–338. [Google Scholar] [CrossRef] [PubMed]
- Kahlert, U.D.; Maciaczyk, D.; Doostkam, S.; Orr, B.A.; Simons, B.; Bogiel, T.; Reithmeier, T.; Prinz, M.; Schubert, J.; Niedermann, G.; et al. Activation of canonical WNT/β-catenin signaling enhances in vitro motility of glioblastoma cells by activation of ZEB1 and other activators of epithelial-to-mesenchymal transition. Cancer Lett. 2012, 325, 42–53. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Luo, Y.; Wang, Y.; Chen, Y.; Li, M.; Jiang, Y. Hepatocyte growth factor increases the invasive potential of PC-3 human prostate cancer cells via an ERK/MAPK and Zeb-1 signaling pathway. Oncol. Lett. 2016, 11, 753–759. [Google Scholar] [CrossRef]
- Burk, U.; Schubert, J.; Wellner, U.; Schmalhofer, O.; Vincan, E.; Spaderna, S.; Brabletz, T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008, 9, 582–589. [Google Scholar] [CrossRef]
- Sundararajan, V.; Gengenbacher, N.; Stemmler, M.P.; Kleemann, J.A.; Brabletz, T.; Brabletz, S. The ZEB1/miR-200c feedback loop regulates invasion via actin interacting proteins MYLK and TKS5. Oncotarget 2015, 6, 27083–27096. [Google Scholar] [CrossRef]
- Jolly, M.K.; Preca, B.T.; Tripathi, S.C.; Jia, D.; George, J.T.; Hanash, S.M.; Brabletz, T.; Stemmler, M.P.; Maurer, J.; Levine, H. Interconnected feedback loops among ESRP1, HAS2, and CD44 regulate epithelial-mesenchymal plasticity in cancer. APL Bioeng. 2018, 2, 031908. [Google Scholar] [CrossRef]
- Preca, B.T.; Bajdak, K.; Mock, K.; Sundararajan, V.; Pfannstiel, J.; Maurer, J.; Wellner, U.; Hopt, U.T.; Brummer, T.; Brabletz, S.; et al. A self-enforcing CD44s/ZEB1 feedback loop maintains EMT and stemness properties in cancer cells. Int. J. Cancer 2015, 137, 2566–2577. [Google Scholar] [CrossRef] [PubMed]
- Jolly, M.K.; Boareto, M.; Huang, B.; Jia, D.; Lu, M.; Ben-Jacob, E.; Onuchic, J.N.; Levine, H. Implications of the Hybrid Epithelial/Mesenchymal Phenotype in Metastasis. Front. Oncol. 2015, 5, 155. [Google Scholar] [CrossRef] [PubMed]
- Larsen, J.E.; Nathan, V.; Osborne, J.K.; Farrow, R.K.; Deb, D.; Sullivan, J.P.; Dospoy, P.D.; Augustyn, A.; Hight, S.K.; Sato, M.; et al. ZEB1 drives epithelial-to-mesenchymal transition in lung cancer. J. Clin. Investig. 2016, 126, 3219–3235. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Guo, F.; Zhu, X.; Guo, W.; Fu, T.; Wang, W. Death Domain-Associated Protein Promotes Colon Cancer Metastasis through Direct Interaction with ZEB1. J. Cancer 2020, 11, 750–758. [Google Scholar] [CrossRef]
- Zhang, D.; Liu, X.; Li, Y.; Sun, L.; Liu, S.S.; Ma, Y.; Zhang, H.; Wang, X.; Yu, Y. LINC01189-miR-586-ZEB1 feedback loop regulates breast cancer progression through Wnt/β-catenin signaling pathway. Mol. Ther. Nucleic Acids 2021, 25, 455–467. [Google Scholar] [CrossRef] [PubMed]
- Jagle, S.; Dertmann, A.; Schrempp, M.; Hecht, A. ZEB1 is neither sufficient nor required for epithelial-mesenchymal transition in LS174T colorectal cancer cells. Biochem. Biophys. Res. Commun. 2017, 482, 1226–1232. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Tillo, E.; Pedrosa, L.; Vila, I.; Chen, Y.; Gyorffy, B.; Sanchez-Moral, L.; Siles, L.; Lozano, J.J.; Esteve-Codina, A.; Darling, D.S.; et al. The EMT factor ZEB1 paradoxically inhibits EMT in BRAF-mutant carcinomas. JCI Insight 2023, 8, e164629. [Google Scholar] [CrossRef]
- Jolly, M.K.; Tripathi, S.C.; Jia, D.; Mooney, S.M.; Celiktas, M.; Hanash, S.M.; Mani, S.A.; Pienta, K.J.; Ben-Jacob, E.; Levine, H. Stability of the hybrid epithelial/mesenchymal phenotype. Oncotarget 2016, 7, 27067–27084. [Google Scholar] [CrossRef]
- Kroger, C.; Afeyan, A.; Mraz, J.; Eaton, E.N.; Reinhardt, F.; Khodor, Y.L.; Thiru, P.; Bierie, B.; Ye, X.; Burge, C.B.; et al. Acquisition of a hybrid E/M state is essential for tumorigenicity of basal breast cancer cells. Proc. Natl. Acad. Sci. USA 2019, 116, 7353–7362. [Google Scholar] [CrossRef]
- Chandra, A.; Jahangiri, A.; Chen, W.; Nguyen, A.T.; Yagnik, G.; Pereira, M.P.; Jain, S.; Garcia, J.H.; Shah, S.S.; Wadhwa, H.; et al. Clonal ZEB1-Driven Mesenchymal Transition Promotes Targetable Oncologic Antiangiogenic Therapy Resistance. Cancer Res. 2020, 80, 1498–1511. [Google Scholar] [CrossRef]
- De Craene, B.; Berx, G. Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Cancer 2013, 13, 97–110. [Google Scholar] [CrossRef] [PubMed]
- Bocci, F.; Levine, H.; Onuchic, J.N.; Jolly, M.K. Deciphering the Dynamics of Epithelial-Mesenchymal Transition and Cancer Stem Cells in Tumor Progression. Curr. Stem Cell Rep. 2019, 5, 11–21. [Google Scholar] [CrossRef]
- Bierie, B.; Pierce, S.E.; Kroeger, C.; Stover, D.G.; Pattabiraman, D.R.; Thiru, P.; Liu Donaher, J.; Reinhardt, F.; Chaffer, C.L.; Keckesova, Z.; et al. Integrin-beta4 identifies cancer stem cell-enriched populations of partially mesenchymal carcinoma cells. Proc. Natl. Acad. Sci. USA 2017, 114, E2337–E2346. [Google Scholar] [CrossRef]
- Grosse-Wilde, A.; Fouquier d’Herouel, A.; McIntosh, E.; Ertaylan, G.; Skupin, A.; Kuestner, R.E.; del Sol, A.; Walters, K.A.; Huang, S. Stemness of the hybrid Epithelial/Mesenchymal State in Breast Cancer and Its Association with Poor Survival. PLoS ONE 2015, 10, e0126522. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Jolly, M.K.; Levine, H.; Onuchic, J.N.; Ben-Jacob, E. MicroRNA-based regulation of epithelial-hybrid-mesenchymal fate determination. Proc. Natl. Acad. Sci. USA 2013, 110, 18144–18149. [Google Scholar] [CrossRef] [PubMed]
- Wellner, U.; Schubert, J.; Burk, U.C.; Schmalhofer, O.; Zhu, F.; Sonntag, A.; Waldvogel, B.; Vannier, C.; Darling, D.; zur Hausen, A.; et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat. Cell Biol. 2009, 11, 1487–1495. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Jiang, H.; Zhang, Z.; Zhang, G.; Wang, H.; Zhang, Q.; Sun, P.; Xiang, R.; Yang, S. ZEB1 confers stem cell-like properties in breast cancer by targeting neurogenin-3. Oncotarget 2017, 8, 54388–54401. [Google Scholar] [CrossRef]
- Krebs, A.M.; Mitschke, J.; Lasierra Losada, M.; Schmalhofer, O.; Boerries, M.; Busch, H.; Boettcher, M.; Mougiakakos, D.; Reichardt, W.; Bronsert, P.; et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat. Cell Biol. 2017, 19, 518–529. [Google Scholar] [CrossRef]
- Demin, S.; Berdieva, M.; Goodkov, A. Cell-cell fusions and cell-in-cell phenomena in healthy cells and cancer: Lessons from protists and invertebrates. Semin. Cancer Biol. 2021, 81, 96–105. [Google Scholar] [CrossRef]
- Duelli, D.; Lazebnik, Y. Cell fusion: A hidden enemy? Cancer Cell 2003, 3, 445–448. [Google Scholar] [CrossRef]
- Kloc, M.; Subuddhi, A.; Uosef, A.; Kubiak, J.Z.; Ghobrial, R.M. Monocyte-Macrophage Lineage Cell Fusion. Int. J. Mol. Sci. 2022, 23, 6553. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Kang, Y. Cell fusion as a hidden force in tumor progression. Cancer Res. 2009, 69, 8536–8539. [Google Scholar] [CrossRef] [PubMed]
- Petrany, M.J.; Millay, D.P. Cell Fusion: Merging Membranes and Making Muscle. Trends Cell Biol. 2019, 29, 964–973. [Google Scholar] [CrossRef] [PubMed]
- Willkomm, L.; Bloch, W. State of the art in cell-cell fusion. Methods Mol. Biol. 2015, 1313, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, P.S.; Baylies, M.K.; Fleissner, A.; Helming, L.; Inoue, N.; Podbilewicz, B.; Wang, H.; Wong, M. Genetic basis of cell-cell fusion mechanisms. Trends Genet. 2013, 29, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, J.M.; Podbilewicz, B. The hallmarks of cell-cell fusion. Development 2017, 144, 4481–4495. [Google Scholar] [CrossRef] [PubMed]
- Perez-Vargas, J.; Krey, T.; Valansi, C.; Avinoam, O.; Haouz, A.; Jamin, M.; Raveh-Barak, H.; Podbilewicz, B.; Rey, F.A. Structural basis of eukaryotic cell-cell fusion. Cell 2014, 157, 407–419. [Google Scholar] [CrossRef]
- Dittmar, T.; Hass, R. Extracellular Events Involved in Cancer Cell-Cell Fusion. Int. J. Mol. Sci. 2022, 23, 16071. [Google Scholar] [CrossRef]
- Dittmar, T.; Hass, R. Intrinsic signalling factors associated with cancer cell-cell fusion. Cell Commun. Signal. 2023, 21, 68. [Google Scholar] [CrossRef]
- Dittmar, T.; Weiler, J.; Luo, T.; Hass, R. Cell-Cell Fusion Mediated by Viruses and HERV-Derived Fusogens in Cancer Initiation and Progression. Cancers 2021, 13, 5363. [Google Scholar] [CrossRef]
- Jiang, E.; Yan, T.; Xu, Z.; Shang, Z. Tumor Microenvironment and Cell Fusion. BioMed Res. Int. 2019, 2019, 5013592. [Google Scholar] [CrossRef]
- Leroy, H.; Han, M.; Woottum, M.; Bracq, L.; Bouchet, J.; Xie, M.; Benichou, S. Virus-Mediated Cell-Cell Fusion. Int. J. Mol. Sci. 2020, 21, 9644. [Google Scholar] [CrossRef] [PubMed]
- Manjunath, Y.; Porciani, D.; Mitchem, J.B.; Suvilesh, K.N.; Avella, D.M.; Kimchi, E.T.; Staveley-O’Carroll, K.F.; Burke, D.H.; Li, G.; Kaifi, J.T. Tumor-Cell-Macrophage Fusion Cells as Liquid Biomarkers and Tumor Enhancers in Cancer. Int. J. Mol. Sci. 2020, 21, 1872. [Google Scholar] [CrossRef]
- Podbilewicz, B. Virus and cell fusion mechanisms. Annu. Rev. Cell Dev. Biol. 2014, 30, 111–139. [Google Scholar] [CrossRef] [PubMed]
- Searles, S.C.; Santosa, E.K.; Bui, J.D. Cell-cell fusion as a mechanism of DNA exchange in cancer. Oncotarget 2018, 9, 6156–6173. [Google Scholar] [CrossRef]
- Hass, R.; von der Ohe, J.; Ungefroren, H. Potential Role of MSC/Cancer Cell Fusion and EMT for Breast Cancer Stem Cell Formation. Cancers 2019, 11, 1432. [Google Scholar] [CrossRef] [PubMed]
- Uygur, B.; Leikina, E.; Melikov, K.; Villasmil, R.; Verma, S.K.; Vary, C.P.H.; Chernomordik, L.V. Interactions with Muscle Cells Boost Fusion, Stemness, and Drug Resistance of Prostate Cancer Cells. Mol. Cancer Res. 2019, 17, 806–820. [Google Scholar] [CrossRef]
- Tajima, Y.; Shibasaki, F.; Masai, H. Cell fusion upregulates PD-L1 expression for evasion from immunosurveillance. Cancer Gene Ther. 2023. [Google Scholar] [CrossRef]
- Minowa, T.; Hirohashi, Y.; Murata, K.; Sasaki, K.; Handa, T.; Nakatsugawa, M.; Mizue, Y.; Murai, A.; Kubo, T.; Kanaseki, T.; et al. Fusion with type 2 macrophages induces melanoma cell heterogeneity that potentiates immunological escape from cytotoxic T lymphocytes. J. Pathol. 2023, 260, 304–316. [Google Scholar] [CrossRef]
- Miroshnychenko, D.; Baratchart, E.; Ferrall-Fairbanks, M.C.; Velde, R.V.; Laurie, M.A.; Bui, M.M.; Tan, A.C.; Altrock, P.M.; Basanta, D.; Marusyk, A. Spontaneous cell fusions as a mechanism of parasexual recombination in tumour cell populations. Nat. Ecol. Evol. 2021, 5, 379–391. [Google Scholar] [CrossRef]
- Dittmar, T.; Schwitalla, S.; Seidel, J.; Haverkampf, S.; Reith, G.; Meyer-Staeckling, S.; Brandt, B.H.; Niggemann, B.; Zanker, K.S. Characterization of hybrid cells derived from spontaneous fusion events between breast epithelial cells exhibiting stem-like characteristics and breast cancer cells. Clin. Exp. Metastasis 2011, 28, 75–90. [Google Scholar] [CrossRef] [PubMed]
- Gauck, D.; Keil, S.; Niggemann, B.; Zanker, K.S.; Dittmar, T. Hybrid clone cells derived from human breast epithelial cells and human breast cancer cells exhibit properties of cancer stem/initiating cells. BMC Cancer 2017, 17, 515. [Google Scholar] [CrossRef]
- Berndt, B.; Haverkampf, S.; Reith, G.; Keil, S.; Niggemann, B.; Zanker, K.S.; Dittmar, T. Fusion of CCL21 non-migratory active breast epithelial and breast cancer cells give rise to CCL21 migratory active tumor hybrid cell lines. PLoS ONE 2013, 8, e63711. [Google Scholar] [CrossRef] [PubMed]
- Jolly, M.K.; Huang, B.; Lu, M.; Mani, S.A.; Levine, H.; Ben-Jacob, E. Towards elucidating the connection between epithelial-mesenchymal transitions and stemness. J. R. Soc. Interface 2014, 11, 20140962. [Google Scholar] [CrossRef] [PubMed]
- Graham, T.R.; Yacoub, R.; Taliaferro-Smith, L.; Osunkoya, A.O.; Odero-Marah, V.A.; Liu, T.; Kimbro, K.S.; Sharma, D.; O’Regan, R.M. Reciprocal regulation of ZEB1 and AR in triple negative breast cancer cells. Breast Cancer Res. Treat. 2010, 123, 139–147. [Google Scholar] [CrossRef]
- Sanchez-Tillo, E.; Lazaro, A.; Torrent, R.; Cuatrecasas, M.; Vaquero, E.C.; Castells, A.; Engel, P.; Postigo, A. ZEB1 represses E-cadherin and induces an EMT by recruiting the SWI/SNF chromatin-remodeling protein BRG1. Oncogene 2010, 29, 3490–3500. [Google Scholar] [CrossRef]
- DaSilva-Arnold, S.C.; Kuo, C.Y.; Davra, V.; Remache, Y.; Kim, P.C.W.; Fisher, J.P.; Zamudio, S.; Al-Khan, A.; Birge, R.B.; Illsley, N.P. ZEB2, a master regulator of the epithelial-mesenchymal transition, mediates trophoblast differentiation. Mol. Hum. Reprod. 2019, 25, 61–75. [Google Scholar] [CrossRef]
- Nam, E.H.; Lee, Y.; Zhao, X.F.; Park, Y.K.; Lee, J.W.; Kim, S. ZEB2-Sp1 cooperation induces invasion by upregulating cadherin-11 and integrin alpha5 expression. Carcinogenesis 2014, 35, 302–314. [Google Scholar] [CrossRef]
- Wang, G.; Guo, X.; Hong, W.; Liu, Q.; Wei, T.; Lu, C.; Gao, L.; Ye, D.; Zhou, Y.; Chen, J.; et al. Critical regulation of miR-200/ZEB2 pathway in Oct4/Sox2-induced mesenchymal-to-epithelial transition and induced pluripotent stem cell generation. Proc. Natl. Acad. Sci. USA 2013, 110, 2858–2863. [Google Scholar] [CrossRef]
- Chen, H.; Li, Z.; Zhang, L.; Zhang, L.; Zhang, Y.; Wang, Y.; Xu, M.; Zhong, Q. MicroRNA-200c Inhibits the Metastasis of Triple-Negative Breast Cancer by Targeting ZEB2, an Epithelial-Mesenchymal Transition Regulator. Ann. Clin. Lab. Sci. 2020, 50, 519–527. [Google Scholar]
- Ginestier, C.; Hur, M.H.; Charafe-Jauffret, E.; Monville, F.; Dutcher, J.; Brown, M.; Jacquemier, J.; Viens, P.; Kleer, C.G.; Liu, S.; et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 2007, 1, 555–567. [Google Scholar] [CrossRef] [PubMed]
- Fahlbusch, S.S.; Keil, S.; Epplen, J.T.; Zanker, K.S.; Dittmar, T. Comparison of hybrid clones derived from human breast epithelial cells and three different cancer cell lines regarding in vitro cancer stem/ initiating cell properties. BMC Cancer 2020, 20, 446. [Google Scholar] [CrossRef]
- Guaita, S.; Puig, I.; Franci, C.; Garrido, M.; Dominguez, D.; Batlle, E.; Sancho, E.; Dedhar, S.; De Herreros, A.G.; Baulida, J. Snail induction of epithelial to mesenchymal transition in tumor cells is accompanied by MUC1 repression and ZEB1 expression. J. Biol. Chem. 2002, 277, 39209–39216. [Google Scholar] [CrossRef]
- Medici, D.; Hay, E.D.; Olsen, B.R. Snail and Slug promote epithelial-mesenchymal transition through beta-catenin-T-cell factor-4-dependent expression of transforming growth factor-beta3. Mol. Biol. Cell 2008, 19, 4875–4887. [Google Scholar] [CrossRef]
- Li, X.; Li, Y.; Du, X.; Wang, X.; Guan, S.; Cao, Y.; Jin, F.; Li, F. HES1 promotes breast cancer stem cells by elevating Slug in triple-negative breast cancer. Int. J. Biol. Sci. 2021, 17, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Jiao, D.; Qiao, J.; Yang, S.; Yan, M.; Cui, S.; Liu, Z. Restin suppressed epithelial-mesenchymal transition and tumor metastasis in breast cancer cells through upregulating mir-200a/b expression via association with p73. Mol. Cancer 2015, 14, 102. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Jiao, W.Y. Down-Regulation of ZEB1 by miR-199a-3p Overexpression Restrains Tumor Stem-Like Properties and Mitochondrial Function of Non-Small Cell Lung Cancer. OncoTargets Ther. 2020, 13, 4607–4616. [Google Scholar] [CrossRef]
- Ferreira, L.A.M.; Bezerra, M.; Kawasaki-Oyama, R.S.; Fernandes, G.M.M.; Castanhole-Nunes, M.M.U.; Serafim Junior, V.; Castilho, R.M.; Pavarino, E.C.; Maniglia, J.V.; Goloni-Bertollo, E.M. Effect of ZEB1 Associated with microRNAs on Tumor Stem Cells in Head and Neck Cancer. Int. J. Mol. Sci. 2023, 24, 5916. [Google Scholar] [CrossRef]
- Vandamme, N.; Denecker, G.; Bruneel, K.; Blancke, G.; Akay, O.; Taminau, J.; De Coninck, J.; De Smedt, E.; Skrypek, N.; Van Loocke, W.; et al. The EMT Transcription Factor ZEB2 Promotes Proliferation of Primary and Metastatic Melanoma While Suppressing an Invasive, Mesenchymal-Like Phenotype. Cancer Res. 2020, 80, 2983–2995. [Google Scholar] [CrossRef]
- Zhao, F.; He, X.; Wang, Y.; Shi, F.; Wu, D.; Pan, M.; Li, M.; Wu, S.; Wang, X.; Dou, J. Decrease of ZEB1 expression inhibits the B16F10 cancer stem-like properties. Biosci. Trends 2015, 9, 325–334. [Google Scholar] [CrossRef]
- Li, L.Y.; Yang, J.F.; Rong, F.; Luo, Z.P.; Hu, S.; Fang, H.; Wu, Y.; Yao, R.; Kong, W.H.; Feng, X.W.; et al. ZEB1 serves an oncogenic role in the tumourigenesis of HCC by promoting cell proliferation, migration, and inhibiting apoptosis via Wnt/β-catenin signaling pathway. Acta Pharmacol. Sin. 2021, 42, 1676–1689. [Google Scholar] [CrossRef] [PubMed]
- Kitz, J.; Lefebvre, C.; Carlos, J.; Lowes, L.E.; Allan, A.L. Reduced ZEB1 expression in prostate cancer cells leads to an aggressive partial-EMT phenotype associated with altered global methylation patterns. Int. J. Mol. Sci. 2021, 22, 12840. [Google Scholar] [CrossRef] [PubMed]
- Pawelek, J.; Chakraborty, A.; Lazova, R.; Yilmaz, Y.; Cooper, D.; Brash, D.; Handerson, T. Co-opting macrophage traits in cancer progression: A consequence of tumor cell fusion? Contrib. Microbiol. 2006, 13, 138–155. [Google Scholar]
- Dittmar, T. Generation of Cancer Stem/Initiating Cells by Cell-Cell Fusion. Int. J. Mol. Sci. 2022, 23, 4514. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.H.; Lee, J.Y.; Yu, S.; Cho, Y.; Hur, S.; Nam, K.T.; Kim, M.H. RAE1 mediated ZEB1 expression promotes epithelial-mesenchymal transition in breast cancer. Sci. Rep. 2019, 9, 2977. [Google Scholar] [CrossRef]
- Mohammadi Ghahhari, N.; Sznurkowska, M.K.; Hulo, N.; Bernasconi, L.; Aceto, N.; Picard, D. Cooperative interaction between ERalpha and the EMT-inducer ZEB1 reprograms breast cancer cells for bone metastasis. Nat. Commun. 2022, 13, 2104. [Google Scholar] [CrossRef]
- Chang, C.C.; Sun, W.; Cruz, A.; Saitoh, M.; Tai, M.H.; Trosko, J.E. A human breast epithelial cell type with stem cell characteristics as target cells for carcinogenesis. Radiat. Res. 2001, 155, 201–207. [Google Scholar] [CrossRef]
- Dittmar, T.; Nagler, C.; Schwitalla, S.; Reith, G.; Niggemann, B.; Zanker, K.S. Recurrence cancer stem cells--made by cell fusion? Med. Hypotheses 2009, 73, 542–547. [Google Scholar] [CrossRef]



| Antibody | Clone; Catalog Number | Manufacturer |
|---|---|---|
| anti-β-ACTIN (mouse monoclonal) | clone AC-15; A5441 | Merck KGaA, Darmstadt, Germany |
| E-CADHERIN (CDH1) (rabbit monoclonal) | clone 24E10; 3195S | Cell Signaling Technology Europe B.V., Frankfurt am Main, Germany |
| eIF4E (rabbit polyclonal) | 9742S | Cell Signaling Technology Europe B.V., Frankfurt am Main, Germany |
| CYTOKERATIN-5 (rabbit monoclonal) | clone D4U8Q; 25807S | Cell Signaling Technology Europe B.V., Frankfurt am Main, Germany |
| N-CADHERIN (CDH2) (mouse monoclonal) | clone 13A9; 14215S | Cell Signaling Technology Europe B.V., Frankfurt am Main, Germany |
| SNAIL (rabbit monoclonal) | clone C15D3; 3879S | Cell Signaling Technology Europe B.V., Frankfurt am Main, Germany |
| VIMENTIN (rabbit polyclonal) | 3932S | Cell Signaling Technology Europe B.V., Frankfurt am Main, Germany |
| WNT5A (rabbit monoclonal) | clone G.307.7; MA5-14946 | Thermo Fisher Scientific, Wesel, Germany |
| ZEB1 (rabbit monoclonal) | clone D808D3; 3396S | Cell Signaling Technology Europe B.V., Frankfurt am Main, Germany |
| ZEB2 (rabbit polyclonal) | 14026-1-AP | Proteintech Germany GmbH Planegg-Martinsried, Germany |
| anti-mouse IgG, HRP linked (horse polyclonal) | 7076S | Cell Signaling Technology Europe B.V., Frankfurt am Main, Germany |
| anti-rabbit IgG, HRP linked (horse polyclonal) | 7074S | Cell Signaling Technology Europe B.V., Frankfurt am Main, Germany |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Merckens, A.; Sieler, M.; Keil, S.; Dittmar, T. Altered Phenotypes of Breast Epithelial × Breast Cancer Hybrids after ZEB1 Knock-Out. Int. J. Mol. Sci. 2023, 24, 17310. https://doi.org/10.3390/ijms242417310
Merckens A, Sieler M, Keil S, Dittmar T. Altered Phenotypes of Breast Epithelial × Breast Cancer Hybrids after ZEB1 Knock-Out. International Journal of Molecular Sciences. 2023; 24(24):17310. https://doi.org/10.3390/ijms242417310
Chicago/Turabian StyleMerckens, Alexander, Mareike Sieler, Silvia Keil, and Thomas Dittmar. 2023. "Altered Phenotypes of Breast Epithelial × Breast Cancer Hybrids after ZEB1 Knock-Out" International Journal of Molecular Sciences 24, no. 24: 17310. https://doi.org/10.3390/ijms242417310





