The PPP1R15 Family of eIF2-alpha Phosphatase Targeting Subunits (GADD34 and CReP)
Abstract
:1. Introduction
2. The Role of eIF2α Phosphatases in Cellular Stress Responses
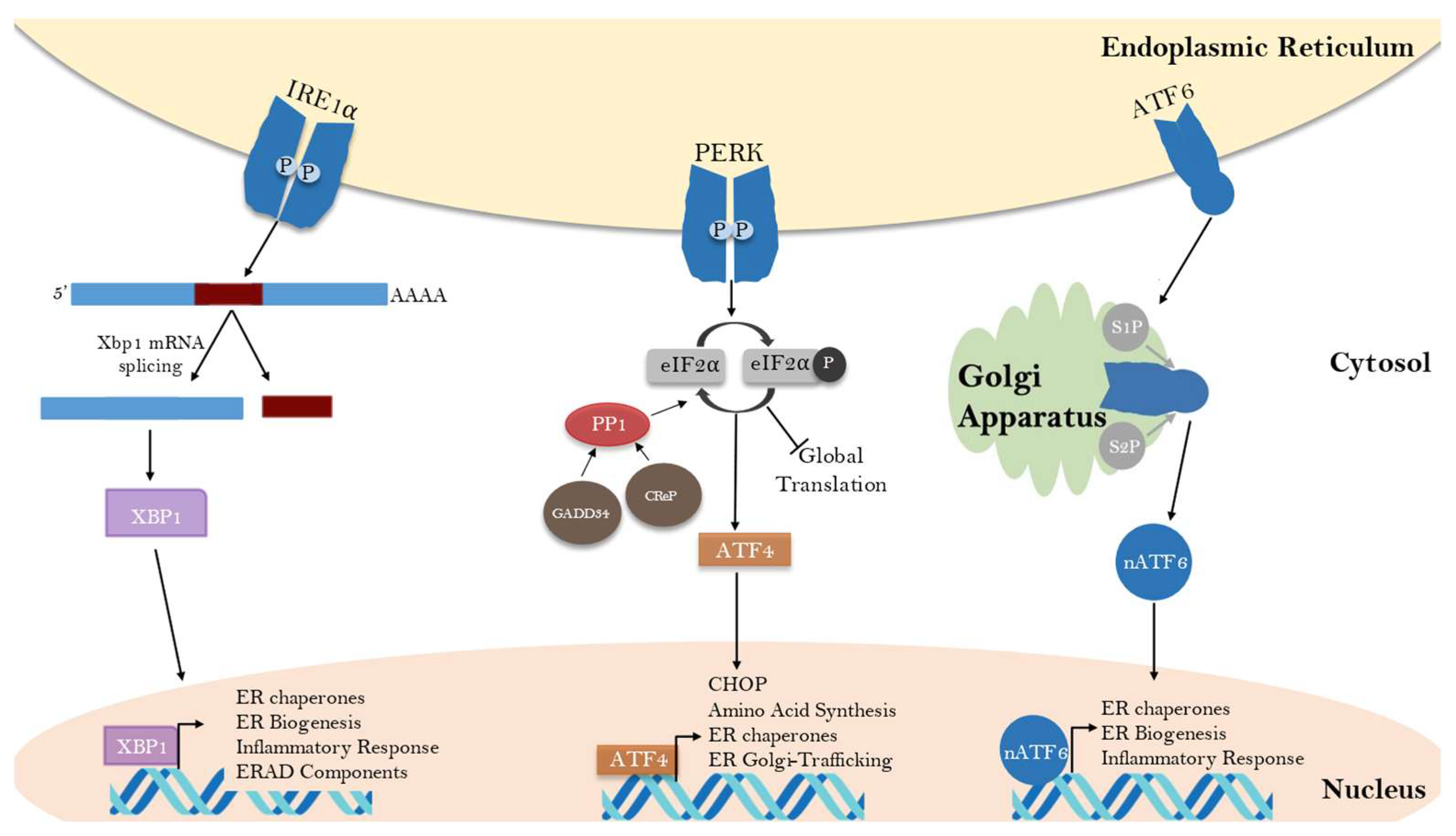
2.1. The Unfolded Protein Response
2.2. The Genetics of eIF2α Phosphatases
2.3. eIF2α Phosphatases in Health and Disease
2.4. Pharmacological Modulation of eIF2α Phosphorylation
3. Molecular Properties and Binding Partners of GADD34 and CReP
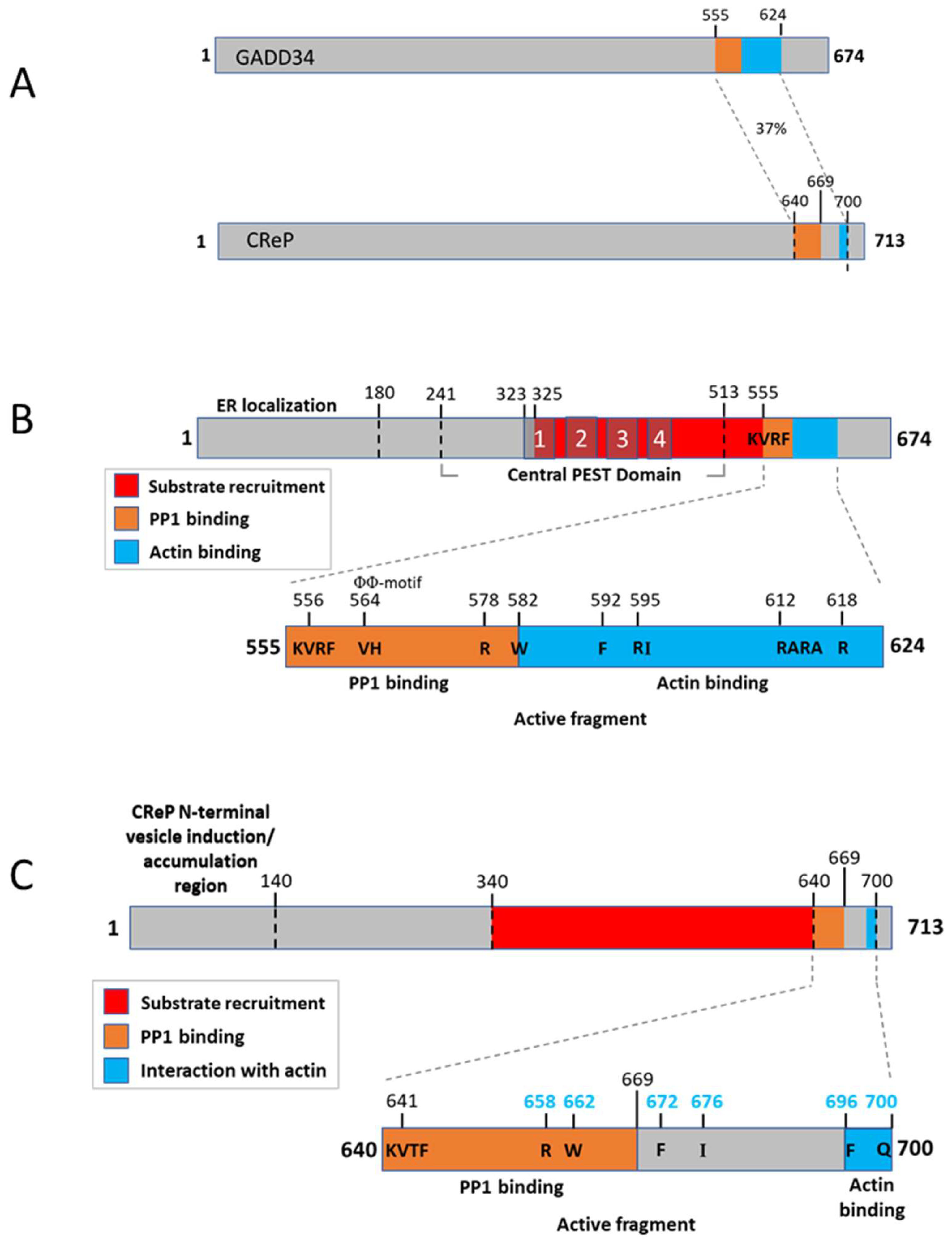
3.1. Biochemistry of eIF2α Phosphatases
3.2. Transcriptional and Translational Regulation of GADD34 and CReP
3.3. GADD34 and CReP Binding Partners and Posttranslational Regulation
4. Diverse Cellular Functions of GADD34 and CReP
5. Future Directions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Costa-Mattioli, M.; Walter, P. The integrated stress response: From mechanism to disease. Science 2020, 368, eaat5314. [Google Scholar] [CrossRef] [PubMed]
- Wek, R.C.; Anthony, T.G.; Staschke, K.A. Surviving and Adapting to Stress: Translational Control and the Integrated Stress Response. Antioxid. Redox Signal. 2023, 39, 351–373. [Google Scholar] [CrossRef]
- Bollen, M.; Peti, W.; Ragusa, M.J.; Beullens, M. The extended PP1 toolkit: Designed to create specificity. Trends Biochem. Sci. 2010, 35, 450–458. [Google Scholar] [CrossRef] [PubMed]
- Virshup, D.M.; Shenolikar, S. From promiscuity to precision: Protein phosphatases get a makeover. Mol. Cell 2009, 33, 537–545. [Google Scholar] [CrossRef] [PubMed]
- Ceulemans, H.; Bollen, M. Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol. Rev. 2004, 84, 1–39. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P.T.W. Protein phosphatase 1—targeted in many directions. J. Cell Sci. 2002, 115, 241–256. [Google Scholar] [CrossRef]
- Brautigan, D.L.; Shenolikar, S. Protein Serine/Threonine Phosphatases: Keys to Unlocking Regulators and Substrates. Annu. Rev. Biochem. 2018, 87, 921–964. [Google Scholar] [CrossRef]
- Eto, M. Regulation of cellular protein phosphatase-1 (PP1) by phosphorylation of the CPI-17 family, C-kinase-activated PP1 inhibitors. J. Biol. Chem. 2009, 284, 35273–35277. [Google Scholar] [CrossRef]
- Casamayor, A.; Arino, J. Controlling Ser/Thr protein phosphatase PP1 activity and function through interaction with regulatory subunits. Adv. Protein Chem. Struct. Biol. 2020, 122, 231–288. [Google Scholar] [CrossRef]
- Bertolotti, A. The split protein phosphatase system. Biochem. J. 2018, 475, 3707–3723. [Google Scholar] [CrossRef]
- Eto, M.; Karginov, A.; Brautigan, D.L. A novel phosphoprotein inhibitor of protein type-1 phosphatase holoenzymes. Biochemistry 1999, 38, 16952–16957. [Google Scholar] [CrossRef]
- Eto, M.; Elliott, E.; Prickett, T.D.; Brautigan, D.L. Inhibitor-2 regulates protein phosphatase-1 complexed with NimA-related kinase to induce centrosome separation. J. Biol. Chem. 2002, 277, 44013–44020. [Google Scholar] [CrossRef]
- Eto, M.; Brautigan, D.L. Endogenous inhibitor proteins that connect Ser/Thr kinases and phosphatases in cell signaling. IUBMB Life 2012, 64, 732–739. [Google Scholar] [CrossRef]
- Terry-Lorenzo, R.T.; Elliot, E.; Weiser, D.C.; Prickett, T.D.; Brautigan, D.L.; Shenolikar, S. Neurabins recruit protein phosphatase-1 and inhibitor-2 to the actin cytoskeleton. J. Biol. Chem. 2002, 277, 46535–46543. [Google Scholar] [CrossRef]
- Connor, J.H.; Weiser, D.C.; Li, S.; Hallenbeck, J.M.; Shenolikar, S. Growth arrest and DNA damage-inducible protein GADD34 assembles a novel signaling complex containing protein phosphatase 1 and inhibitor 1. Mol. Cell Biol. 2001, 21, 6841–6850. [Google Scholar] [CrossRef] [PubMed]
- Jayashankar, V.; Nguyen, M.J.; Carr, B.W.; Zheng, D.C.; Rosales, J.B.; Rosales, J.B.; Weiser, D.C. Protein phosphatase 1 beta paralogs encode the zebrafish myosin phosphatase catalytic subunit. PLoS ONE 2013, 8, e75766. [Google Scholar] [CrossRef] [PubMed]
- Varmuza, S.; Jurisicova, A.; Okano, K.; Hudson, J.; Boekelheide, K.; Shipp, E.B. Spermiogenesis is impaired in mice bearing a targeted mutation in the protein phosphatase 1cgamma gene. Dev. Biol. 1999, 205, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Kirchner, J.; Gross, S.; Bennett, D.; Alphey, L. Essential, overlapping and redundant roles of the Drosophila protein phosphatase 1 alpha and 1 beta genes. Genetics 2007, 176, 273–281. [Google Scholar] [CrossRef]
- Gibbons, J.A.; Kozubowski, L.; Tatchell, K.; Shenolikar, S. Expression of human protein phosphatase-1 in Saccharomyces cerevisiae highlights the role of phosphatase isoforms in regulating eukaryotic functions. J. Biol. Chem. 2007, 282, 21838–21847. [Google Scholar] [CrossRef]
- Korrodi-Gregorio, L.; Esteves, S.L.C.; Fardilha, M. Protein phosphatase 1 catalytic isoforms: Specificity toward interacting proteins. Transl. Res. 2014, 164, 366–391. [Google Scholar] [CrossRef]
- Ceulemans, H.; Stalmans, W.; Bollen, M. Regulator-driven functional diversification of protein phosphatase-1 in eukaryotic evolution. Bioessays 2002, 24, 371–381. [Google Scholar] [CrossRef]
- Novoa, I.; Zeng, H.; Harding, H.P.; Ron, D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J. Cell Biol. 2001, 153, 1011–1022. [Google Scholar] [CrossRef]
- Jousse, C.; Oyadomari, S.; Novoa, I.; Lu, P.; Zhang, Y.; Harding, H.P.; Ron, D. Inhibition of a constitutive translation initiation factor 2alpha phosphatase, CReP, promotes survival of stressed cells. J. Cell Biol. 2003, 163, 767–775. [Google Scholar] [CrossRef] [PubMed]
- Brush, M.H.; Weiser, D.C.; Shenolikar, S. Growth arrest and DNA damage-inducible protein GADD34 targets protein phosphatase 1 alpha to the endoplasmic reticulum and promotes dephosphorylation of the alpha subunit of eukaryotic translation initiation factor 2. Mol. Cell Biol. 2003, 23, 1292–1303. [Google Scholar] [CrossRef]
- Hinnebusch, A.G. The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem. 2014, 83, 779–812. [Google Scholar] [CrossRef]
- Baird, T.D.; Wek, R.C. Eukaryotic initiation factor 2 phosphorylation and translational control in metabolism. Adv. Nutr. 2012, 3, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Wek, R.C. Role of eIF2alpha Kinases in Translational Control and Adaptation to Cellular Stress. Cold Spring Harb. Perspect. Biol. 2018, 10, a032870. [Google Scholar] [CrossRef] [PubMed]
- Reid, D.W.; Tay, A.S.L.; Sundaram, J.R.; Lee, I.C.J.; Chen, Q.; George, S.E.; Nicchitta, C.V.; Shenolikar, S. Complementary Roles of GADD34- and CReP-Containing Eukaryotic Initiation Factor 2alpha Phosphatases during the Unfolded Protein Response. Mol. Cell Biol. 2016, 36, 1868–1880. [Google Scholar] [CrossRef] [PubMed]
- Baird, T.D.; Palam, L.R.; Fusakio, M.E.; Willy, J.A.; Davis, C.M.; McClintick, J.N.; Anthony, T.G.; Wek, R.C. Selective mRNA translation during eIF2 phosphorylation induces expression of IBTKalpha. Mol. Biol. Cell 2014, 25, 1686–1697. [Google Scholar] [CrossRef] [PubMed]
- Dey, S.; Baird, T.D.; Zhou, D.; Palam, L.R.; Spandau, D.F.; Wek, R.C. Both transcriptional regulation and translational control of ATF4 are central to the integrated stress response. J. Biol. Chem. 2010, 285, 33165–33174. [Google Scholar] [CrossRef]
- Harding, H.P.; Novoa, I.; Zhang, Y.; Zeng, H.; Wek, R.; Schapira, M.; Ron, D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 2000, 6, 1099–1108. [Google Scholar] [CrossRef]
- Komar, A.A.; Merrick, W.C. A Retrospective on eIF2A-and Not the Alpha Subunit of eIF2. Int. J. Mol. Sci. 2020, 21, 2054. [Google Scholar] [CrossRef]
- Golovko, A.; Kojukhov, A.; Guan, B.; Morpurgo, B.; Merrick, W.C.; Mazumder, B.; Hatzoglou, M.; Komar, A.A. The eIF2A knockout mouse. Cell Cycle 2016, 15, 3115–3120. [Google Scholar] [CrossRef]
- Starck, S.R.; Tsai, J.C.; Chen, K.; Shodiya, M.; Wang, L.; Yahiro, K.; Martins-Green, M.; Shastri, N.; Walter, P. Translation from the 5′ untranslated region shapes the integrated stress response. Science 2016, 351, aad3867. [Google Scholar] [CrossRef]
- Sendoel, A.; Dunn, J.G.; Rodriguez, E.H.; Naik, S.; Gomez, N.C.; Hurwitz, B.; Levorse, J.; Dill, B.D.; Schramek, D.; Molina, H.; et al. Translation from unconventional 5’ start sites drives tumour initiation. Nature 2017, 541, 494–499. [Google Scholar] [CrossRef]
- Chen, L.; He, J.; Zhou, J.; Xiao, Z.; Ding, N.; Duan, Y.; Li, W.; Sun, L. EIF2A promotes cell survival during paclitaxel treatment in vitro and in vivo. J. Cell Mol. Med. 2019, 23, 6060–6071. [Google Scholar] [CrossRef]
- Sonobe, Y.; Ghadge, G.; Masaki, K.; Sendoel, A.; Fuchs, E.; Roos, R.P. Translation of dipeptide repeat proteins from the C9ORF72 expanded repeat is associated with cellular stress. Neurobiol. Dis. 2018, 116, 155–165. [Google Scholar] [CrossRef]
- Balendra, R.; Isaacs, A.M. C9orf72-mediated ALS and FTD: Multiple pathways to disease. Nat. Rev. Neurol. 2018, 14, 544–558. [Google Scholar] [CrossRef]
- Rao, R.V.; Poksay, K.S.; Castro-Obregon, S.; Schilling, B.; Row, R.H.; del Rio, G.; Gibson, B.W.; Ellerby, H.M.; Bredesen, D.E. Molecular components of a cell death pathway activated by endoplasmic reticulum stress. J. Biol. Chem. 2004, 279, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Kaufman, R.J. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature 2016, 529, 326–335. [Google Scholar] [CrossRef] [PubMed]
- Wiseman, R.L.; Mesgarzadeh, J.S.; Hendershot, L.M. Reshaping endoplasmic reticulum quality control through the unfolded protein response. Mol. Cell 2022, 82, 1477–1491. [Google Scholar] [CrossRef]
- Dill, K.A.; Chan, H.S. From Levinthal to pathways to funnels. Nat. Struct. Biol. 1997, 4, 10–19. [Google Scholar] [CrossRef]
- Gething, M.J.; Sambrook, J. Protein folding in the cell. Nature 1992, 355, 33–45. [Google Scholar] [CrossRef]
- Braakman, I.; Bulleid, N.J. Protein folding and modification in the mammalian endoplasmic reticulum. Annu. Rev. Biochem. 2011, 80, 71–99. [Google Scholar] [CrossRef]
- Ron, D.; Walter, P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007, 8, 519–529. [Google Scholar] [CrossRef]
- Aridor, M.; Balch, W.E. Integration of endoplasmic reticulum signaling in health and disease. Nat. Med. 1999, 5, 745–751. [Google Scholar] [CrossRef]
- Tirasophon, W.; Welihinda, A.A.; Kaufman, R.J. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes. Dev. 1998, 12, 1812–1824. [Google Scholar] [CrossRef]
- Liu, C.Y.; Schroder, M.; Kaufman, R.J. Ligand-independent dimerization activates the stress response kinases IRE1 and PERK in the lumen of the endoplasmic reticulum. J. Biol. Chem. 2000, 275, 24881–24885. [Google Scholar] [CrossRef]
- Sidrauski, C.; Walter, P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 1997, 90, 1031–1039. [Google Scholar] [CrossRef]
- Travers, K.J.; Patil, C.K.; Wodicka, L.; Lockhart, D.J.; Weissman, J.S.; Walter, P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 2000, 101, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Kopito, R.R. ER quality control: The cytoplasmic connection. Cell 1997, 88, 427–430. [Google Scholar] [CrossRef]
- Hollien, J.; Lin, J.H.; Li, H.; Stevens, N.; Walter, P.; Weissman, J.S. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J. Cell Biol. 2009, 186, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Haze, K.; Yoshida, H.; Yanagi, H.; Yura, T.; Mori, K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell 1999, 10, 3787–3799. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Shen, J.; Prywes, R. The luminal domain of ATF6 senses endoplasmic reticulum (ER) stress and causes translocation of ATF6 from the ER to the Golgi. J. Biol. Chem. 2002, 277, 13045–13052. [Google Scholar] [CrossRef]
- Yoshida, H.; Matsui, T.; Yamamoto, A.; Okada, T.; Mori, K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 2001, 107, 881–891. [Google Scholar] [CrossRef] [PubMed]
- Walter, F.; O’Brien, A.; Concannon, C.G.; Dussmann, H.; Prehn, J.H.M. ER stress signaling has an activating transcription factor 6alpha (ATF6)-dependent “off-switch”. J. Biol. Chem. 2018, 293, 18270–18284. [Google Scholar] [CrossRef]
- Andreev, D.E.; O’Connor, P.B.F.; Fahey, C.; Kenny, E.M.; Terenin, I.M.; Dmitriev, S.E.; Cormican, P.; Morris, D.W.; Shatsky, I.N.; Baranov, P.V. Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression. Elife 2015, 4, e03971. [Google Scholar] [CrossRef] [PubMed]
- Harding, H.P.; Zhang, Y.; Ron, D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 1999, 397, 271–274. [Google Scholar] [CrossRef]
- Pavitt, G.D.; Ron, D. New insights into translational regulation in the endoplasmic reticulum unfolded protein response. Cold Spring Harb. Perspect. Biol. 2012, 4, a012278. [Google Scholar] [CrossRef]
- Brostrom, C.O.; Brostrom, M.A. Regulation of translational initiation during cellular responses to stress. Prog. Nucleic Acid. Res. Mol. Biol. 1998, 58, 79–125. [Google Scholar] [CrossRef]
- Lu, P.D.; Jousse, C.; Marciniak, S.J.; Zhang, Y.; Novoa, I.; Scheuner, D.; Kaufman, R.J.; Ron, D.; Harding, H.P. Cytoprotection by pre-emptive conditional phosphorylation of translation initiation factor 2. EMBO J. 2004, 23, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Reid, D.W.; Chen, Q.; Tay, A.S.; Shenolikar, S.; Nicchitta, C.V. The unfolded protein response triggers selective mRNA release from the endoplasmic reticulum. Cell 2014, 158, 1362–1374. [Google Scholar] [CrossRef] [PubMed]
- Neill, G.; Masson, G.R. A stay of execution: ATF4 regulation and potential outcomes for the integrated stress response. Front. Mol. Neurosci. 2023, 16, 1112253. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Brewer, J.W.; Diehl, J.A.; Hendershot, L.M. Two distinct stress signaling pathways converge upon the CHOP promoter during the mammalian unfolded protein response. J. Mol. Biol. 2002, 318, 1351–1365. [Google Scholar] [CrossRef]
- Fawcett, T.W.; Martindale, J.L.; Guyton, K.Z.; Hai, T.; Holbrook, N.J. Complexes containing activating transcription factor (ATF)/cAMP-responsive-element-binding protein (CREB) interact with the CCAAT/enhancer-binding protein (C/EBP)-ATF composite site to regulate Gadd153 expression during the stress response. Biochem. J. 1999, 339 Pt 1, 135–141. [Google Scholar] [CrossRef]
- Marciniak, S.J.; Yun, C.Y.; Oyadomari, S.; Novoa, I.; Zhang, Y.; Jungreis, R.; Nagata, K.; Harding, H.P.; Ron, D. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes. Dev. 2004, 18, 3066–3077. [Google Scholar] [CrossRef]
- Young, S.K.; Palam, L.R.; Wu, C.; Sachs, M.S.; Wek, R.C. Ribosome Elongation Stall Directs Gene-specific Translation in the Integrated Stress Response. J. Biol. Chem. 2016, 291, 6546–6558. [Google Scholar] [CrossRef]
- Palam, L.R.; Baird, T.D.; Wek, R.C. Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. J. Biol. Chem. 2011, 286, 10939–10949. [Google Scholar] [CrossRef] [PubMed]
- Preissler, S.; Ron, D. Early Events in the Endoplasmic Reticulum Unfolded Protein Response. Cold Spring Harb. Perspect. Biol. 2019, 11, a033894. [Google Scholar] [CrossRef] [PubMed]
- Marton, M.; Banhegyi, G.; Gyongyosi, N.; Kalman, E.E.; Pettko-Szandtner, A.; Kaldi, K.; Kapuy, O. A systems biological analysis of the ATF4-GADD34-CHOP regulatory triangle upon endoplasmic reticulum stress. FEBS Open Bio 2022, 12, 2065–2082. [Google Scholar] [CrossRef]
- Marton, M.; Kurucz, A.; Lizak, B.; Margittai, E.; Banhegyi, G.; Kapuy, O. A Systems Biological View of Life-and-Death Decision with Respect to Endoplasmic Reticulum Stress-The Role of PERK Pathway. Int. J. Mol. Sci. 2017, 18, 58. [Google Scholar] [CrossRef]
- Kapuy, O.; Marton, M.; Banhegyi, G.; Vinod, P.K. Multiple system-level feedback loops control life-and-death decisions in endoplasmic reticulum stress. FEBS Lett. 2020, 594, 1112–1123. [Google Scholar] [CrossRef]
- Pyati, U.J.; Gjini, E.; Carbonneau, S.; Lee, J.; Guo, F.; Jette, C.A.; Kelsell, D.P.; Look, A.T. p63 mediates an apoptotic response to pharmacological and disease-related ER stress in the developing epidermis. Dev. Cell 2011, 21, 492–505. [Google Scholar] [CrossRef]
- Ubeda, M.; Wang, X.Z.; Zinszner, H.; Wu, I.; Habener, J.F.; Ron, D. Stress-induced binding of the transcriptional factor CHOP to a novel DNA control element. Mol. Cell Biol. 1996, 16, 1479–1489. [Google Scholar] [CrossRef]
- Ron, D.; Habener, J.F. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes Dev. 1992, 6, 439–453. [Google Scholar] [CrossRef]
- Matsumoto, M.; Minami, M.; Takeda, K.; Sakao, Y.; Akira, S. Ectopic expression of CHOP (GADD153) induces apoptosis in M1 myeloblastic leukemia cells. FEBS Lett. 1996, 395, 143–147. [Google Scholar] [CrossRef]
- Maytin, E.V.; Ubeda, M.; Lin, J.C.; Habener, J.F. Stress-inducible transcription factor CHOP/gadd153 induces apoptosis in mammalian cells via p38 kinase-dependent and -independent mechanisms. Exp. Cell Res. 2001, 267, 193–204. [Google Scholar] [CrossRef]
- Oyadomari, S.; Araki, E.; Mori, M. Endoplasmic reticulum stress-mediated apoptosis in pancreatic beta-cells. Apoptosis 2002, 7, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Novoa, I.; Zhang, Y.; Zeng, H.; Jungreis, R.; Harding, H.P.; Ron, D. Stress-induced gene expression requires programmed recovery from translational repression. EMBO J. 2003, 22, 1180–1187. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Back, S.H.; Hur, J.; Lin, Y.; Gildersleeve, R.; Shan, J.; Yuan, C.L.; Krokowski, D.; Wang, S.; Hatzoglou, M.; et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol. 2013, 15, 481–490. [Google Scholar] [CrossRef] [PubMed]
- McCullough, K.D.; Martindale, J.L.; Klotz, L.O.; Aw, T.Y.; Holbrook, N.J. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Mol. Cell Biol. 2001, 21, 1249–1259. [Google Scholar] [CrossRef] [PubMed]
- Rojas, M.; Gingras, A.; Dever, T.E. Protein phosphatase PP1/GLC7 interaction domain in yeast eIF2gamma bypasses targeting subunit requirement for eIF2alpha dephosphorylation. Proc. Natl. Acad. Sci. USA 2014, 111, 1344. [Google Scholar] [CrossRef] [PubMed]
- Wakula, P.; Beullens, M.; Ceulemans, H.; Stalmans, W.; Bollen, M. Degeneracy and function of the ubiquitous RVXF motif that mediates binding to protein phosphatase-1. J. Biol. Chem. 2003, 278, 18817–18823. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, J.A.; Weiser, D.C.; Shenolikar, S. Importance of a surface hydrophobic pocket on protein phosphatase-1 catalytic subunit in recognizing cellular regulators. J. Biol. Chem. 2005, 280, 15903–15911. [Google Scholar] [CrossRef] [PubMed]
- Egloff, M.P.; Johnson, D.F.; Moorhead, G.; Cohen, P.T.; Cohen, P.; Barford, D. Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J. 1997, 16, 1876–1887. [Google Scholar] [CrossRef]
- Malzer, E.; Dominicus, C.S.; Chambers, J.E.; Dickens, J.A.; Mookerjee, S.; Marciniak, S.J. The integrated stress response regulates BMP signalling through effects on translation. BMC Biol. 2018, 16, 34. [Google Scholar] [CrossRef]
- Malzer, E.; Szajewska-Skuta, M.; Dalton, L.E.; Thomas, S.E.; Hu, N.; Skaer, H.; Lomas, D.A.; Crowther, D.C.; Marciniak, S.J. Coordinate regulation of eIF2alpha phosphorylation by PPP1R15 and GCN2 is required during Drosophila development. J. Cell Sci. 2013, 126, 1406–1415. [Google Scholar] [CrossRef]
- Harding, H.P.; Zhang, Y.; Scheuner, D.; Chen, J.; Kaufman, R.J.; Ron, D. Ppp1r15 gene knockout reveals an essential role for translation initiation factor 2 alpha (eIF2alpha) dephosphorylation in mammalian development. Proc. Natl. Acad. Sci. USA 2009, 106, 1832–1837. [Google Scholar] [CrossRef]
- Kojima, E.; Takeuchi, A.; Haneda, M.; Yagi, A.; Hasegawa, T.; Yamaki, K.; Takeda, K.; Akira, S.; Shimokata, K.; Isobe, K. The function of GADD34 is a recovery from a shutoff of protein synthesis induced by ER stress: Elucidation by GADD34-deficient mice. FASEB J. 2003, 17, 1573–1575. [Google Scholar] [CrossRef]
- Nishio, N.; Isobe, K. GADD34-deficient mice develop obesity, nonalcoholic fatty liver disease, hepatic carcinoma and insulin resistance. Sci. Rep. 2015, 5, 13519. [Google Scholar] [CrossRef]
- Nishio, N.; Hasegawa, T.; Tatsuno, I.; Isaka, M.; Isobe, K. Female GADD34 mice develop age-related inflammation and hepatocellular carcinoma. Geriatr. Gerontol. Int. 2017, 17, 2593–2601. [Google Scholar] [CrossRef] [PubMed]
- Kernohan, K.D.; Tetreault, M.; Liwak-Muir, U.; Geraghty, M.T.; Qin, W.; Venkateswaran, S.; Davila, J.; Care4Rare Canada Consortium; Holcik, M.; Majewski, J.; et al. Homozygous mutation in the eukaryotic translation initiation factor 2alpha phosphatase gene, PPP1R15B, is associated with severe microcephaly, short stature and intellectual disability. Hum. Mol. Genet. 2015, 24, 6293–6300. [Google Scholar] [CrossRef] [PubMed]
- Abdulkarim, B.; Nicolino, M.; Igoillo-Esteve, M.; Daures, M.; Romero, S.; Philippi, A.; Senee, V.; Lopes, M.; Cunha, D.A.; Harding, H.P.; et al. A Missense Mutation in PPP1R15B Causes a Syndrome Including Diabetes, Short Stature, and Microcephaly. Diabetes 2015, 64, 3951–3962. [Google Scholar] [CrossRef]
- Zhang, P.; McGrath, B.; Li, S.; Frank, A.; Zambito, F.; Reinert, J.; Gannon, M.; Ma, K.; McNaughton, K.; Cavener, D.R. The PERK eukaryotic initiation factor 2 alpha kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Mol. Cell Biol. 2002, 22, 3864–3874. [Google Scholar] [CrossRef] [PubMed]
- Delepine, M.; Nicolino, M.; Barrett, T.; Golamaully, M.; Lathrop, G.M.; Julier, C. EIF2AK3, encoding translation initiation factor 2-alpha kinase 3, is mutated in patients with Wolcott-Rallison syndrome. Nat. Genet. 2000, 25, 406–409. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Kaufman, R.J. The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nat. Rev. Cancer 2014, 14, 581–597. [Google Scholar] [CrossRef] [PubMed]
- Eisele, Y.S.; Monteiro, C.; Fearns, C.; Encalada, S.E.; Wiseman, R.L.; Powers, E.T.; Kelly, J.W. Targeting protein aggregation for the treatment of degenerative diseases. Nat. Rev. Drug Discov. 2015, 14, 759–780. [Google Scholar] [CrossRef]
- Baleriola, J.; Walker, C.A.; Jean, Y.Y.; Crary, J.F.; Troy, C.M.; Nagy, P.L.; Hengst, U. Axonally synthesized ATF4 transmits a neurodegenerative signal across brain regions. Cell 2014, 158, 1159–1172. [Google Scholar] [CrossRef]
- Credle, J.J.; Forcelli, P.A.; Delannoy, M.; Oaks, A.W.; Permaul, E.; Berry, D.L.; Duka, V.; Wills, J.; Sidhu, A. alpha-Synuclein-mediated inhibition of ATF6 processing into COPII vesicles disrupts UPR signaling in Parkinson’s disease. Neurobiol. Dis. 2015, 76, 112–125. [Google Scholar] [CrossRef]
- Wang, S.; Kaufman, R.J. The impact of the unfolded protein response on human disease. J. Cell Biol. 2012, 197, 857–867. [Google Scholar] [CrossRef]
- Oyadomari, S.; Harding, H.P.; Zhang, Y.; Oyadomari, M.; Ron, D. Dephosphorylation of translation initiation factor 2alpha enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metab. 2008, 7, 520–532. [Google Scholar] [CrossRef]
- Sequeira, S.J.; Ranganathan, A.C.; Adam, A.P.; Iglesias, B.V.; Farias, E.F.; Aguirre-Ghiso, J.A. Inhibition of proliferation by PERK regulates mammary acinar morphogenesis and tumor formation. PLoS ONE 2007, 2, e615. [Google Scholar] [CrossRef]
- Sequeira, S.J.; Wen, H.C.; Avivar-Valderas, A.; Farias, E.F.; Aguirre-Ghiso, J.A. Inhibition of eIF2alpha dephosphorylation inhibits ErbB2-induced deregulation of mammary acinar morphogenesis. BMC Cell Biol. 2009, 10, 64. [Google Scholar] [CrossRef] [PubMed]
- Bobrovnikova-Marjon, E.; Grigoriadou, C.; Pytel, D.; Zhang, F.; Ye, J.; Koumenis, C.; Cavener, D.; Diehl, J.A. PERK promotes cancer cell proliferation and tumor growth by limiting oxidative DNA damage. Oncogene 2010, 29, 3881–3895. [Google Scholar] [CrossRef] [PubMed]
- Bi, M.; Naczki, C.; Koritzinsky, M.; Fels, D.; Blais, J.; Hu, N.; Harding, H.; Novoa, I.; Varia, M.; Raleigh, J.; et al. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J. 2005, 24, 3470–3481. [Google Scholar] [CrossRef] [PubMed]
- Rouschop, K.M.; Dubois, L.J.; Keulers, T.G.; van den Beucken, T.; Lambin, P.; Bussink, J.; van der Kogel, A.J.; Koritzinsky, M.; Wouters, B.G. PERK/eIF2alpha signaling protects therapy resistant hypoxic cells through induction of glutathione synthesis and protection against ROS. Proc. Natl. Acad. Sci. USA 2013, 110, 4622–4627. [Google Scholar] [CrossRef] [PubMed]
- Thevenot, P.T.; Sierra, R.A.; Raber, P.L.; Al-Khami, A.A.; Trillo-Tinoco, J.; Zarreii, P.; Ochoa, A.C.; Cui, Y.; Del Valle, L.; Rodriguez, P.C. The stress-response sensor chop regulates the function and accumulation of myeloid-derived suppressor cells in tumors. Immunity 2014, 41, 389–401. [Google Scholar] [CrossRef]
- Hetz, C.; Axten, J.M.; Patterson, J.B. Pharmacological targeting of the unfolded protein response for disease intervention. Nat. Chem. Biol. 2019, 15, 764–775. [Google Scholar] [CrossRef]
- Ghemrawi, R.; Khair, M. Endoplasmic Reticulum Stress and Unfolded Protein Response in Neurodegenerative Diseases. Int. J. Mol. Sci. 2020, 21, 6127. [Google Scholar] [CrossRef]
- Rozpedek-Kaminska, W.; Siwecka, N.; Wawrzynkiewicz, A.; Wojtczak, R.; Pytel, D.; Diehl, J.A.; Majsterek, I. The PERK-Dependent Molecular Mechanisms as a Novel Therapeutic Target for Neurodegenerative Diseases. Int. J. Mol. Sci. 2020, 21, 2108. [Google Scholar] [CrossRef]
- Marciniak, S.J.; Chambers, J.E.; Ron, D. Pharmacological targeting of endoplasmic reticulum stress in disease. Nat. Rev. Drug Discov. 2022, 21, 115–140. [Google Scholar] [CrossRef] [PubMed]
- Swingle, M.R.; Honkanen, R.E. Inhibitors of Serine/Threonine Protein Phosphatases: Biochemical and Structural Studies Provide Insight for Further Development. Curr. Med. Chem. 2019, 26, 2634–2660. [Google Scholar] [CrossRef]
- Weiser, D.C.; Shenolikar, S. Use of protein phosphatase inhibitors. Curr. Protoc. Mol. Biol. 2003, 62, 10. [Google Scholar] [CrossRef]
- Vagnarelli, P.; Alessi, D.R. PP1 Phosphatase Complexes: Undruggable No Longer. Cell 2018, 174, 1049–1051. [Google Scholar] [CrossRef] [PubMed]
- Grandjean, J.M.D.; Wiseman, R.L. Small molecule strategies to harness the unfolded protein response: Where do we go from here? J. Biol. Chem. 2020, 295, 15692–15711. [Google Scholar] [CrossRef] [PubMed]
- Boyce, M.; Bryant, K.F.; Jousse, C.; Long, K.; Harding, H.P.; Scheuner, D.; Kaufman, R.J.; Ma, D.; Coen, D.M.; Ron, D.; et al. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science 2005, 307, 935–939. [Google Scholar] [CrossRef] [PubMed]
- Saxena, S.; Cabuy, E.; Caroni, P. A role for motoneuron subtype-selective ER stress in disease manifestations of FALS mice. Nat. Neurosci. 2009, 12, 627–636. [Google Scholar] [CrossRef]
- Colla, E.; Coune, P.; Liu, Y.; Pletnikova, O.; Troncoso, J.C.; Iwatsubo, T.; Schneider, B.L.; Lee, M.K. Endoplasmic reticulum stress is important for the manifestations of alpha-synucleinopathy in vivo. J. Neurosci. 2012, 32, 3306–3320. [Google Scholar] [CrossRef]
- Moreno, J.A.; Radford, H.; Peretti, D.; Steinert, J.R.; Verity, N.; Martin, M.G.; Halliday, M.; Morgan, J.; Dinsdale, D.; Ortori, C.A.; et al. Sustained translational repression by eIF2alpha-P mediates prion neurodegeneration. Nature 2012, 485, 507–511. [Google Scholar] [CrossRef]
- O’Connor, T.; Sadleir, K.R.; Maus, E.; Velliquette, R.A.; Zhao, J.; Cole, S.L.; Eimer, W.A.; Hitt, B.; Bembinster, L.A.; Lammich, S.; et al. Phosphorylation of the translation initiation factor eIF2alpha increases BACE1 levels and promotes amyloidogenesis. Neuron 2008, 60, 988–1009. [Google Scholar] [CrossRef]
- Lee, D.Y.; Lee, K.; Lee, H.J.; Kim, D.H.; Noh, Y.H.; Yu, K.; Jung, H.; Lee, S.H.; Lee, J.Y.; Youn, Y.C.; et al. Activation of PERK signaling attenuates Abeta-mediated ER stress. PLoS ONE 2010, 5, e10489. [Google Scholar] [CrossRef]
- Robert, F.; Kapp, L.D.; Khan, S.N.; Acker, M.G.; Kolitz, S.; Kazemi, S.; Kaufman, R.J.; Merrick, W.C.; Koromilas, A.E.; Lorsch, J.R.; et al. Initiation of protein synthesis by hepatitis C virus is refractory to reduced eIF2.GTP.Met-tRNA(i)(Met) ternary complex availability. Mol. Biol. Cell 2006, 17, 4632–4644. [Google Scholar] [CrossRef] [PubMed]
- Tsaytler, P.; Harding, H.P.; Ron, D.; Bertolotti, A. Selective inhibition of a regulatory subunit of protein phosphatase 1 restores proteostasis. Science 2011, 332, 91–94. [Google Scholar] [CrossRef]
- Wiseman, R.L.; Kelly, J.W. Cell biology. Phosphatase inhibition delays translational recovery. Science 2011, 332, 44–45. [Google Scholar] [CrossRef]
- Das, I.; Krzyzosiak, A.; Schneider, K.; Wrabetz, L.; D’Antonio, M.; Barry, N.; Sigurdardottir, A.; Bertolotti, A. Preventing proteostasis diseases by selective inhibition of a phosphatase regulatory subunit. Science 2015, 348, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Krzyzosiak, A.; Sigurdardottir, A.; Luh, L.; Carrara, M.; Das, I.; Schneider, K.; Bertolotti, A. Target-Based Discovery of an Inhibitor of the Regulatory Phosphatase PPP1R15B. Cell 2018, 174, 1216–1228.e19. [Google Scholar] [CrossRef] [PubMed]
- Sundaram, J.R.; Wu, Y.; Lee, I.C.; George, S.E.; Hota, M.; Ghosh, S.; Kesavapany, S.; Ahmed, M.; Tan, E.; Shenolikar, S. PromISR-6, a Guanabenz Analogue, Improves Cellular Survival in an Experimental Model of Huntington’s Disease. ACS Chem. Neurosci. 2019, 10, 3575–3589. [Google Scholar] [CrossRef]
- Crespillo-Casado, A.; Chambers, J.E.; Fischer, P.M.; Marciniak, S.J.; Ron, D. PPP1R15A-mediated dephosphorylation of eIF2alpha is unaffected by Sephin1 or Guanabenz. Elife 2017, 6, e26109. [Google Scholar] [CrossRef]
- Carrara, M.; Sigurdardottir, A.; Bertolotti, A. Decoding the selectivity of eIF2alpha holophosphatases and PPP1R15A inhibitors. Nat. Struct. Mol. Biol. 2017, 24, 708–716. [Google Scholar] [CrossRef]
- Shacham, T.; Patel, C.; Lederkremer, G.Z. PERK Pathway and Neurodegenerative Disease: To Inhibit or to Activate? Biomolecules 2021, 11, 354. [Google Scholar] [CrossRef]
- Szaruga, M.; Janssen, D.A.; de Miguel, C.; Hodgson, G.; Fatalska, A.; Pitera, A.P.; Andreeva, A.; Bertolotti, A. Activation of the integrated stress response by inhibitors of its kinases. Nat. Commun. 2023, 14, 5535–5538. [Google Scholar] [CrossRef]
- Axten, J.M.; Medina, J.R.; Feng, Y.; Shu, A.; Romeril, S.P.; Grant, S.W.; Li, W.H.H.; Heerding, D.A.; Minthorn, E.; Mencken, T.; et al. Discovery of 7-methyl-5-(1-[3-(trifluoromethyl)phenyl]acetyl-2,3-dihydro-1H-indol-5-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a potent and selective first-in-class inhibitor of protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK). J. Med. Chem. 2012, 55, 7193–7207. [Google Scholar] [CrossRef]
- Moreno, J.A.; Halliday, M.; Molloy, C.; Radford, H.; Verity, N.; Axten, J.M.; Ortori, C.A.; Willis, A.E.; Fischer, P.M.; Barrett, D.A.; et al. Oral treatment targeting the unfolded protein response prevents neurodegeneration and clinical disease in prion-infected mice. Sci. Transl. Med. 2013, 5, 206ra138. [Google Scholar] [CrossRef] [PubMed]
- Mercado, G.; Castillo, V.; Soto, P.; Lopez, N.; Axten, J.M.; Sardi, S.P.; Hoozemans, J.J.M.; Hetz, C. Targeting PERK signaling with the small molecule GSK2606414 prevents neurodegeneration in a model of Parkinson’s disease. Neurobiol. Dis. 2018, 112, 136–148. [Google Scholar] [CrossRef] [PubMed]
- Grande, V.; Ornaghi, F.; Comerio, L.; Restelli, E.; Masone, A.; Corbelli, A.; Tolomeo, D.; Capone, V.; Axten, J.M.; Laping, N.J.; et al. PERK inhibition delays neurodegeneration and improves motor function in a mouse model of Marinesco-Sjogren syndrome. Hum. Mol. Genet. 2018, 27, 2477–2489. [Google Scholar] [CrossRef] [PubMed]
- Radford, H.; Moreno, J.A.; Verity, N.; Halliday, M.; Mallucci, G.R. PERK inhibition prevents tau-mediated neurodegeneration in a mouse model of frontotemporal dementia. Acta Neuropathol. 2015, 130, 633–642. [Google Scholar] [CrossRef] [PubMed]
- Rojas-Rivera, D.; Delvaeye, T.; Roelandt, R.; Nerinckx, W.; Augustyns, K.; Vandenabeele, P.; Bertrand, M.J.M. When PERK inhibitors turn out to be new potent RIPK1 inhibitors: Critical issues on the specificity and use of GSK2606414 and GSK2656157. Cell Death Differ. 2017, 24, 1100–1110. [Google Scholar] [CrossRef] [PubMed]
- Sidrauski, C.; McGeachy, A.M.; Ingolia, N.T.; Walter, P. The small molecule ISRIB reverses the effects of eIF2alpha phosphorylation on translation and stress granule assembly. Elife 2015, 4, e05033. [Google Scholar] [CrossRef] [PubMed]
- Sidrauski, C.; Acosta-Alvear, D.; Khoutorsky, A.; Vedantham, P.; Hearn, B.R.; Li, H.; Gamache, K.; Gallagher, C.M.; Ang, K.K.; Wilson, C.; et al. Pharmacological brake-release of mRNA translation enhances cognitive memory. Elife 2013, 2, e00498. [Google Scholar] [CrossRef]
- Halliday, M.; Radford, H.; Sekine, Y.; Moreno, J.; Verity, N.; le Quesne, J.; Ortori, C.A.; Barrett, D.A.; Fromont, C.; Fischer, P.M.; et al. Partial restoration of protein synthesis rates by the small molecule ISRIB prevents neurodegeneration without pancreatic toxicity. Cell Death Dis. 2015, 6, e1672. [Google Scholar] [CrossRef]
- Hosoi, T.; Kakimoto, M.; Tanaka, K.; Nomura, J.; Ozawa, K. Unique pharmacological property of ISRIB in inhibition of Abeta-induced neuronal cell death. J. Pharmacol. Sci. 2016, 131, 292–295. [Google Scholar] [CrossRef]
- Choy, M.S.; Yusoff, P.; Lee, I.C.; Newton, J.C.; Goh, C.W.; Page, R.; Shenolikar, S.; Peti, W. Structural and Functional Analysis of the GADD34:PP1 eIF2alpha Phosphatase. Cell Rep. 2015, 11, 1885–1891. [Google Scholar] [CrossRef]
- Zhou, W.; Brush, M.H.; Choy, M.S.; Shenolikar, S. Association with endoplasmic reticulum promotes proteasomal degradation of GADD34 protein. J. Biol. Chem. 2011, 286, 21687–21696. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Gross, M.; Roizman, B. The gamma134.5 protein of herpes simplex virus 1 has the structural and functional attributes of a protein phosphatase 1 regulatory subunit and is present in a high molecular weight complex with the enzyme in infected cells. J. Biol. Chem. 1998, 273, 20737–20743. [Google Scholar] [CrossRef] [PubMed]
- Cheng, G.; Gross, M.; Brett, M.E.; He, B. AlaArg motif in the carboxyl terminus of the gamma(1)34.5 protein of herpes simplex virus type 1 is required for the formation of a high-molecular-weight complex that dephosphorylates eIF-2alpha. J. Virol. 2001, 75, 3666–3674. [Google Scholar] [CrossRef] [PubMed]
- Peti, W.; Nairn, A.C.; Page, R. Structural basis for protein phosphatase 1 regulation and specificity. FEBS J. 2013, 280, 596–611. [Google Scholar] [CrossRef]
- Rojas, M.; Vasconcelos, G.; Dever, T.E. An eIF2alpha-binding motif in protein phosphatase 1 subunit GADD34 and its viral orthologs is required to promote dephosphorylation of eIF2alpha. Proc. Natl. Acad. Sci. USA 2015, 112, 3466. [Google Scholar] [CrossRef]
- Chen, R.; Rato, C.; Yan, Y.; Crespillo-Casado, A.; Clarke, H.J.; Harding, H.P.; Marciniak, S.J.; Read, R.J.; Ron, D. G-actin provides substrate-specificity to eukaryotic initiation factor 2alpha holophosphatases. Elife 2015, 4, e04871. [Google Scholar] [CrossRef]
- Yan, Y.; Harding, H.P.; Ron, D. Higher-order phosphatase-substrate contacts terminate the integrated stress response. Nat. Struct. Mol. Biol. 2021, 28, 835–846. [Google Scholar] [CrossRef]
- Chambers, J.E.; Dalton, L.E.; Clarke, H.J.; Malzer, E.; Dominicus, C.S.; Patel, V.; Moorhead, G.; Ron, D.; Marciniak, S.J. Actin dynamics tune the integrated stress response by regulating eukaryotic initiation factor 2alpha dephosphorylation. Elife 2015, 4, e04872. [Google Scholar] [CrossRef]
- Kloft, N.; Neukirch, C.; von Hoven, G.; Bobkiewicz, W.; Weis, S.; Boller, K.; Husmann, M. A subunit of eukaryotic translation initiation factor 2alpha-phosphatase (CreP/PPP1R15B) regulates membrane traffic. J. Biol. Chem. 2012, 287, 35299–35317. [Google Scholar] [CrossRef]
- Hodgson, G.; Andreeva, A.; Bertolotti, A. Substrate recognition determinants of human eIF2alpha phosphatases. Open Biol. 2021, 11, 210205. [Google Scholar] [CrossRef]
- Kastan, J.P.; Dobrikova, E.Y.; Bryant, J.D.; Gromeier, M. CReP mediates selective translation initiation at the endoplasmic reticulum. Sci. Adv. 2020, 6, eaba0745. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Q.; Lord, K.A.; Alamo, I.J.; Hollander, M.C.; Carrier, F.; Ron, D.; Kohn, K.W.; Hoffman, B.; Liebermann, D.A.; Fornace, A.J.J. The gadd and MyD genes define a novel set of mammalian genes encoding acidic proteins that synergistically suppress cell growth. Mol. Cell Biol. 1994, 14, 2361–2371. [Google Scholar] [CrossRef] [PubMed]
- Hollander, M.C.; Zhan, Q.; Bae, I.; Fornace, A.J.J. Mammalian GADD34, an apoptosis- and DNA damage-inducible gene. J. Biol. Chem. 1997, 272, 13731–13737. [Google Scholar] [CrossRef]
- Amundson, S.A.; Zhan, Q.; Penn, L.Z.; Fornace, A.J.J. Myc suppresses induction of the growth arrest genes gadd34, gadd45, and gadd153 by DNA-damaging agents. Oncogene 1998, 17, 2149–2154. [Google Scholar] [CrossRef]
- Smith, M.L.; Fornace, A.J.J. Mammalian DNA damage-inducible genes associated with growth arrest and apoptosis. Mutat. Res. 1996, 340, 109–124. [Google Scholar] [CrossRef] [PubMed]
- Hollander, M.C.; Sheikh, M.S.; Yu, K.; Zhan, Q.; Iglesias, M.; Woodworth, C.; Fornace, A.J.J. Activation of Gadd34 by diverse apoptotic signals and suppression of its growth inhibitory effects by apoptotic inhibitors. Int. J. Cancer 2001, 96, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Brush, M.H.; Shenolikar, S. Control of cellular GADD34 levels by the 26S proteasome. Mol. Cell Biol. 2008, 28, 6989–7000. [Google Scholar] [CrossRef]
- Goh, C.W.; Lee, I.C.; Sundaram, J.R.; George, S.E.; Yusoff, P.; Brush, M.H.; Sze, N.S.K.; Shenolikar, S. Chronic oxidative stress promotes GADD34-mediated phosphorylation of the TAR DNA-binding protein TDP-43, a modification linked to neurodegeneration. J. Biol. Chem. 2018, 293, 163–176. [Google Scholar] [CrossRef]
- Vattem, K.M.; Wek, R.C. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. USA 2004, 101, 11269–11274. [Google Scholar] [CrossRef] [PubMed]
- Young, S.K.; Willy, J.A.; Wu, C.; Sachs, M.S.; Wek, R.C. Ribosome Reinitiation Directs Gene-specific Translation and Regulates the Integrated Stress Response. J. Biol. Chem. 2015, 290, 28257–28271. [Google Scholar] [CrossRef] [PubMed]
- Hulsmann, J.; Kravic, B.; Weith, M.; Gstaiger, M.; Aebersold, R.; Collins, B.C.; Meyer, H. AP-SWATH Reveals Direct Involvement of VCP/p97 in Integrated Stress Response Signaling through Facilitating CReP/PPP1R15B Degradation. Mol. Cell Proteom. 2018, 17, 1295–1307. [Google Scholar] [CrossRef] [PubMed]
- Loveless, T.B.; Topacio, B.R.; Vashisht, A.A.; Galaang, S.; Ulrich, K.M.; Young, B.D.; Wohlschlegel, J.A.; Toczyski, D.P. DNA Damage Regulates Translation through beta-TRCP Targeting of CReP. PLoS Genet. 2015, 11, e1005292. [Google Scholar] [CrossRef] [PubMed]
- So, J.; Cho, S.; Min, S.; Kimball, S.R.; Lee, A. IRE1alpha-Dependent Decay of CReP/Ppp1r15b mRNA Increases Eukaryotic Initiation Factor 2alpha Phosphorylation and Suppresses Protein Synthesis. Mol. Cell Biol. 2015, 35, 2761–2770. [Google Scholar] [CrossRef]
- Khan, R.; Kadamkode, V.; Kesharwani, D.; Purkayastha, S.; Banerjee, G.; Datta, M. Circulatory miR-98-5p levels are deregulated during diabetes and it inhibits proliferation and promotes apoptosis by targeting PPP1R15B in keratinocytes. RNA Biol. 2020, 17, 188–201. [Google Scholar] [CrossRef]
- Khan, R.; Verma, A.K.; Datta, M. mir-98-5p regulates gluconeogenesis and lipogenesis by targeting PPP1R15B in hepatocytes. J. Cell Commun. Signal 2023, 17, 881–895. [Google Scholar] [CrossRef]
- Zhou, W.; Jeyaraman, K.; Yusoff, P.; Shenolikar, S. Phosphorylation at tyrosine 262 promotes GADD34 protein turnover. J. Biol. Chem. 2013, 288, 33146–33155. [Google Scholar] [CrossRef]
- Lee, I.C.; Ho, X.Y.; George, S.E.; Goh, C.W.; Sundaram, J.R.; Pang, K.K.L.; Luo, W.; Yusoff, P.; Sze, N.S.K.; Shenolikar, S. Oxidative stress promotes SIRT1 recruitment to the GADD34/PP1alpha complex to activate its deacetylase function. Cell Death Differ. 2018, 25, 255–267. [Google Scholar] [CrossRef]
- Weiser, D.C.; Sikes, S.; Li, S.; Shenolikar, S. The inhibitor-1 C terminus facilitates hormonal regulation of cellular protein phosphatase-1: Functional implications for inhibitor-1 isoforms. J. Biol. Chem. 2004, 279, 48904–48914. [Google Scholar] [CrossRef]
- Hung, W.J.; Roberson, R.S.; Taft, J.; Wu, D.Y. Human BAG-1 proteins bind to the cellular stress response protein GADD34 and interfere with GADD34 functions. Mol. Cell Biol. 2003, 23, 3477–3486. [Google Scholar] [CrossRef]
- Adler, H.T.; Chinery, R.; Wu, D.Y.; Kussick, S.J.; Payne, J.M.; Fornace, A.J.J.; Tkachuk, D.C. Leukemic HRX fusion proteins inhibit GADD34-induced apoptosis and associate with the GADD34 and hSNF5/INI1 proteins. Mol. Cell Biol. 1999, 19, 7050–7060. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.Y.; Tkachuck, D.C.; Roberson, R.S.; Schubach, W.H. The human SNF5/INI1 protein facilitates the function of the growth arrest and DNA damage-inducible protein (GADD34) and modulates GADD34-bound protein phosphatase-1 activity. J. Biol. Chem. 2002, 277, 27706–27715. [Google Scholar] [CrossRef] [PubMed]
- Grishin, A.V.; Azhipa, O.; Semenov, I.; Corey, S.J. Interaction between growth arrest-DNA damage protein 34 and Src kinase Lyn negatively regulates genotoxic apoptosis. Proc. Natl. Acad. Sci. USA 2001, 98, 10172–10177. [Google Scholar] [CrossRef] [PubMed]
- Prola, A.; Pires Da Silva, J.; Guilbert, A.; Lecru, L.; Piquereau, J.; Ribeiro, M.; Mateo, P.; Gressette, M.; Fortin, D.; Boursier, C.; et al. SIRT1 protects the heart from ER stress-induced cell death through eIF2alpha deacetylation. Cell Death Differ. 2017, 24, 343–356. [Google Scholar] [CrossRef]
- Hasegawa, T.; Yagi, A.; Isobe, K. Interaction between GADD34 and kinesin superfamily, KIF3A. Biochem. Biophys. Res. Commun. 2000, 267, 593–596. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, T.; Xiao, H.; Hamajima, F.; Isobe, K. Interaction between DNA-damage protein GADD34 and a new member of the Hsp40 family of heat shock proteins that is induced by a DNA-damaging reagent. Biochem. J. 2000, 352 Pt 3, 795–800. [Google Scholar] [CrossRef]
- Hasegawa, T.; Isobe, K. Evidence for the interaction between Translin and GADD34 in mammalian cells. Biochim. Biophys. Acta 1999, 1428, 161–168. [Google Scholar] [CrossRef]
- Hasegawa, T.; Isobe, K. Characterization of a murine gene encoding an acidic-basic dipeptide repeat that interacts with GADD34. J. Cell Biochem. 2000, 77, 596–603. [Google Scholar] [CrossRef]
- Hasegawa, T.; Xiao, H.; Isobe, K. Cloning of a GADD34-like gene that interacts with the zinc-finger transcription factor which binds to the p21(WAF) promoter. Biochem. Biophys. Res. Commun. 1999, 256, 249–254. [Google Scholar] [CrossRef]
- Dedigama-Arachchige, P.M.; Acharige, N.P.N.; Pflum, M.K.H. Identification of PP1-Gadd34 substrates involved in the unfolded protein response using K-BIPS, a method for phosphatase substrate identification. Mol. Omics 2018, 14, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Sun, C.; He, B.; Xiong, W.; Shi, X.; Yao, D.; Cao, X. GADD34-PP1c recruited by Smad7 dephosphorylates TGFbeta type I receptor. J. Cell Biol. 2004, 164, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, H.; Wang, C.; Zhang, J.; Man, J.; Gao, Y.; Zhang, P.; Li, W.; Zhao, J.; Pan, X.; et al. Deactivation of the kinase IKK by CUEDC2 through recruitment of the phosphatase PP1. Nat. Immunol. 2008, 9, 533–541. [Google Scholar] [CrossRef] [PubMed]
- Zarogoulidis, P.; Lampaki, S.; Turner, J.F.; Huang, H.; Kakolyris, S.; Syrigos, K.; Zarogoulidis, K. mTOR pathway: A current, up-to-date mini-review (Review). Oncol. Lett. 2014, 8, 2367–2370. [Google Scholar] [CrossRef] [PubMed]
- Holczer, M.; Banhegyi, G.; Kapuy, O. GADD34 Keeps the mTOR Pathway Inactivated in Endoplasmic Reticulum Stress Related Autophagy. PLoS ONE 2016, 11, e0168359. [Google Scholar] [CrossRef]
- Watanabe, R.; Tambe, Y.; Inoue, H.; Isono, T.; Haneda, M.; Isobe, K.; Kobayashi, T.; Hino, O.; Okabe, H.; Chano, T. GADD34 inhibits mammalian target of rapamycin signaling via tuberous sclerosis complex and controls cell survival under bioenergetic stress. Int. J. Mol. Med. 2007, 19, 475–483. [Google Scholar] [CrossRef]
- Uddin, M.N.; Ito, S.; Nishio, N.; Suganya, T.; Isobe, K. Gadd34 induces autophagy through the suppression of the mTOR pathway during starvation. Biochem. Biophys. Res. Commun. 2011, 407, 692–698. [Google Scholar] [CrossRef]
- Akhurst, R.J.; Hata, A. Targeting the TGFbeta signalling pathway in disease. Nat. Rev. Drug Discov. 2012, 11, 790–811. [Google Scholar] [CrossRef]
- Derynck, R.; Turley, S.J.; Akhurst, R.J. TGFbeta biology in cancer progression and immunotherapy. Nat. Rev. Clin. Oncol. 2021, 18, 9–34. [Google Scholar] [CrossRef]
- Moustakas, A.; Heldin, C. The regulation of TGFbeta signal transduction. Development 2009, 136, 3699–3714. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hicks, D.; Giresh, K.; Wrischnik, L.A.; Weiser, D.C. The PPP1R15 Family of eIF2-alpha Phosphatase Targeting Subunits (GADD34 and CReP). Int. J. Mol. Sci. 2023, 24, 17321. https://doi.org/10.3390/ijms242417321
Hicks D, Giresh K, Wrischnik LA, Weiser DC. The PPP1R15 Family of eIF2-alpha Phosphatase Targeting Subunits (GADD34 and CReP). International Journal of Molecular Sciences. 2023; 24(24):17321. https://doi.org/10.3390/ijms242417321
Chicago/Turabian StyleHicks, Danielle, Krithika Giresh, Lisa A. Wrischnik, and Douglas C. Weiser. 2023. "The PPP1R15 Family of eIF2-alpha Phosphatase Targeting Subunits (GADD34 and CReP)" International Journal of Molecular Sciences 24, no. 24: 17321. https://doi.org/10.3390/ijms242417321




