Functional Characterization of Potato UBC13-UEV1s Genes Required for Ubiquitin Lys63 Chain to Polyubiquitination
Abstract
:1. Introduction
2. Results
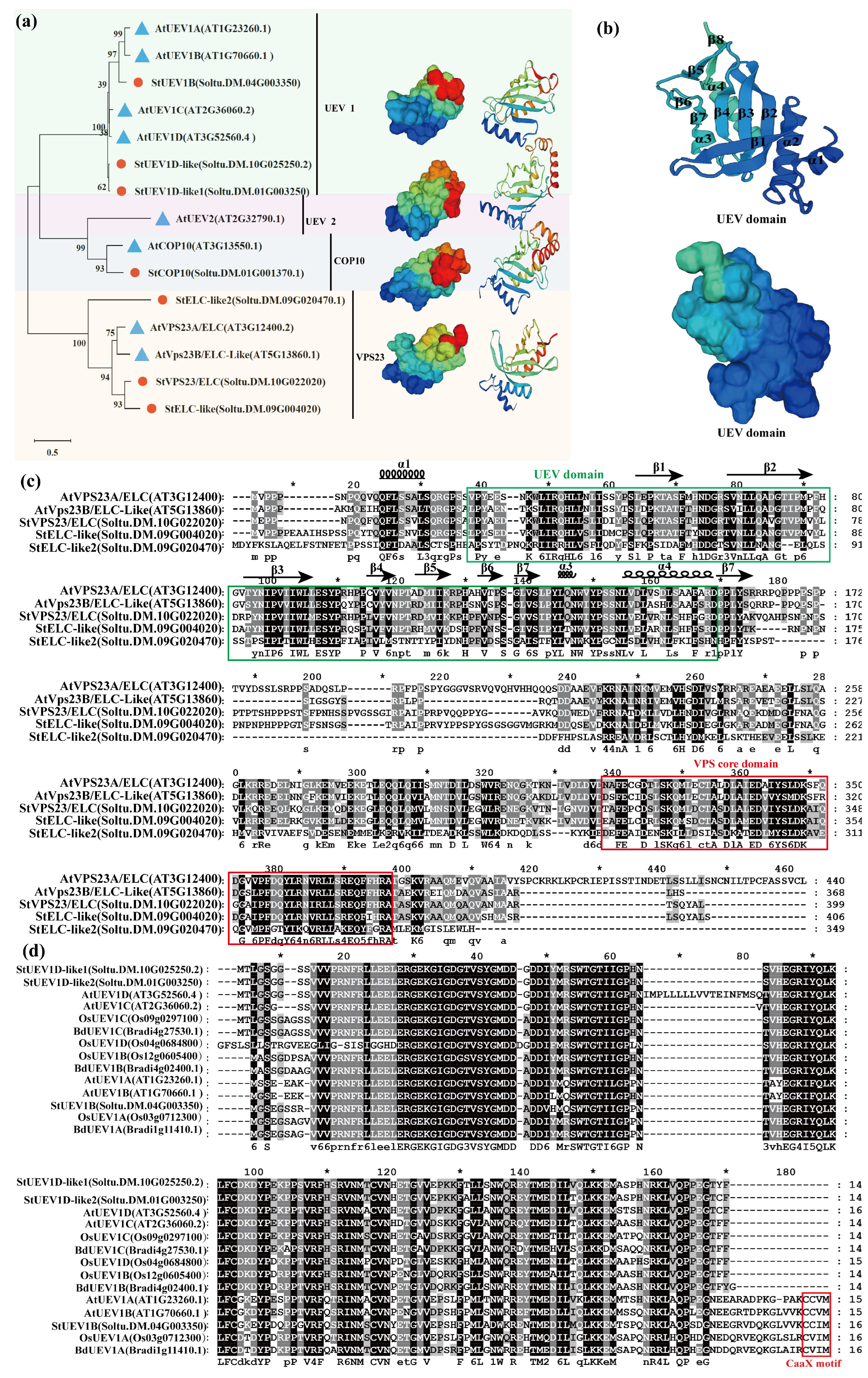
2.1. Identification and Characterization of Potato UEVs
2.2. The Cis-Regulatory Elements Analysis
2.3. Conserved Motif and Gene Structure Analyses
2.4. Identification and Characterization of Potato StUBC13
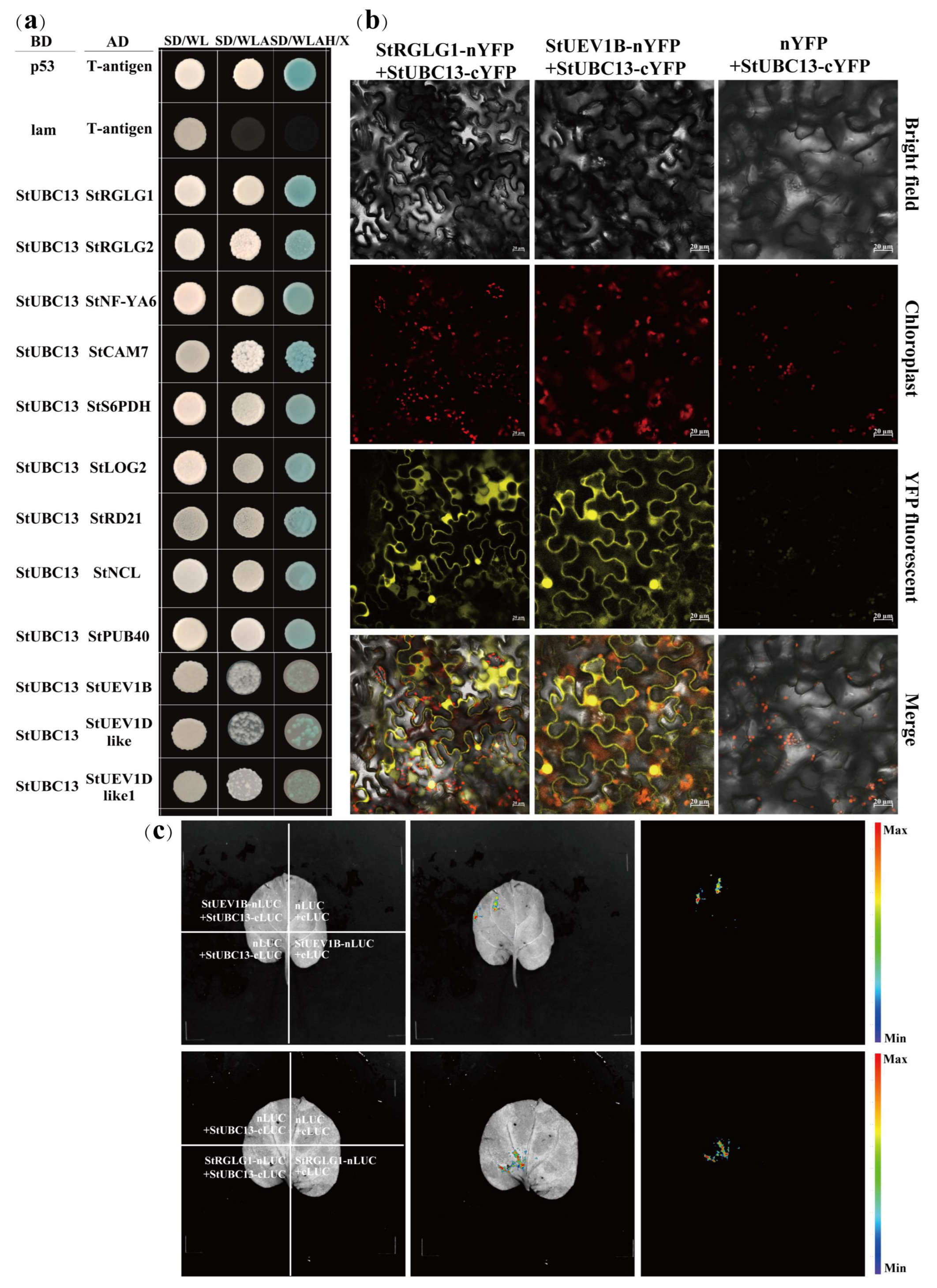
2.5. StUBC13 Interacted with StUEV1s and E3 Ligase Enzyme StRGLG1
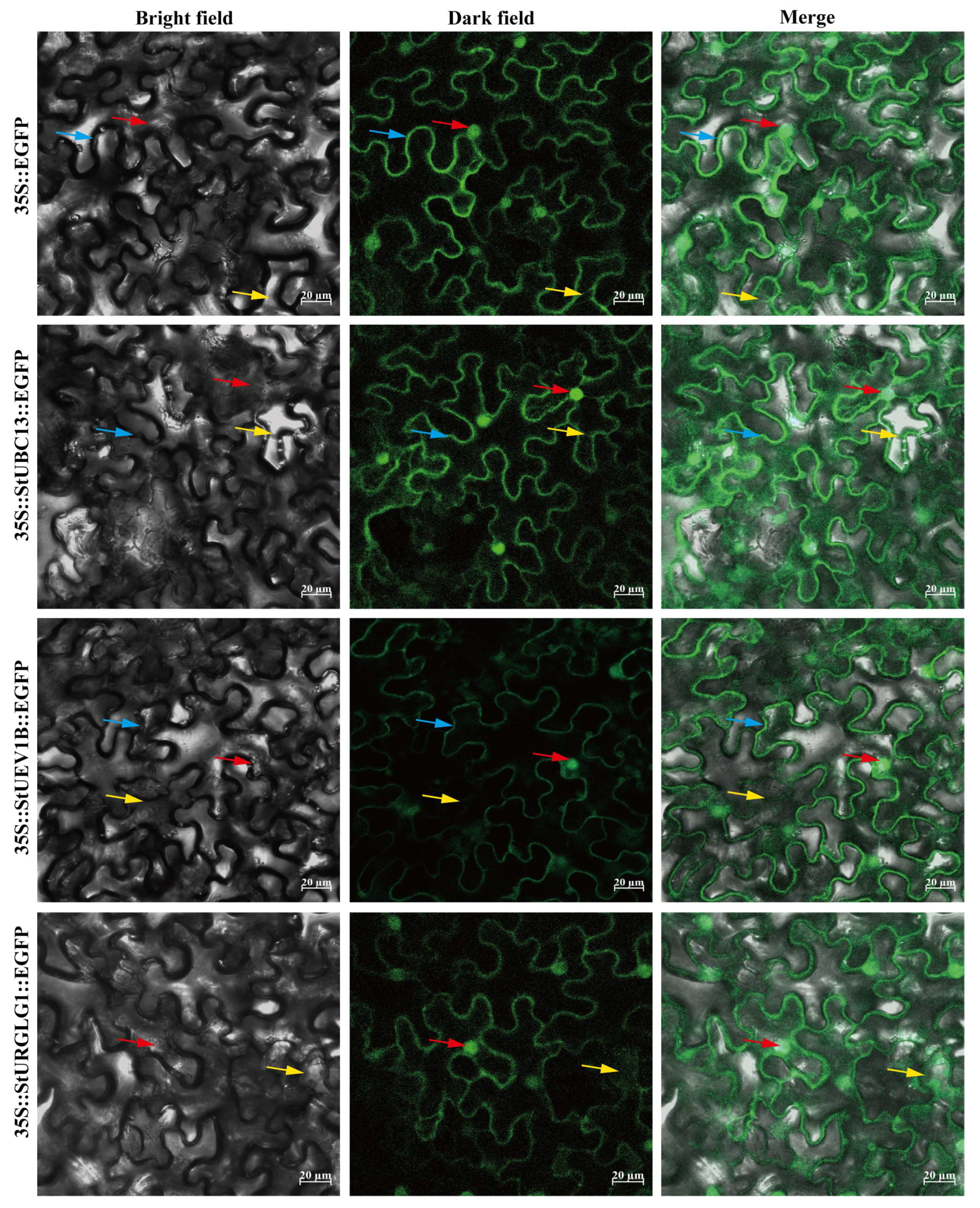
2.6. Subcellular Localization of UBC13 and Its Partners
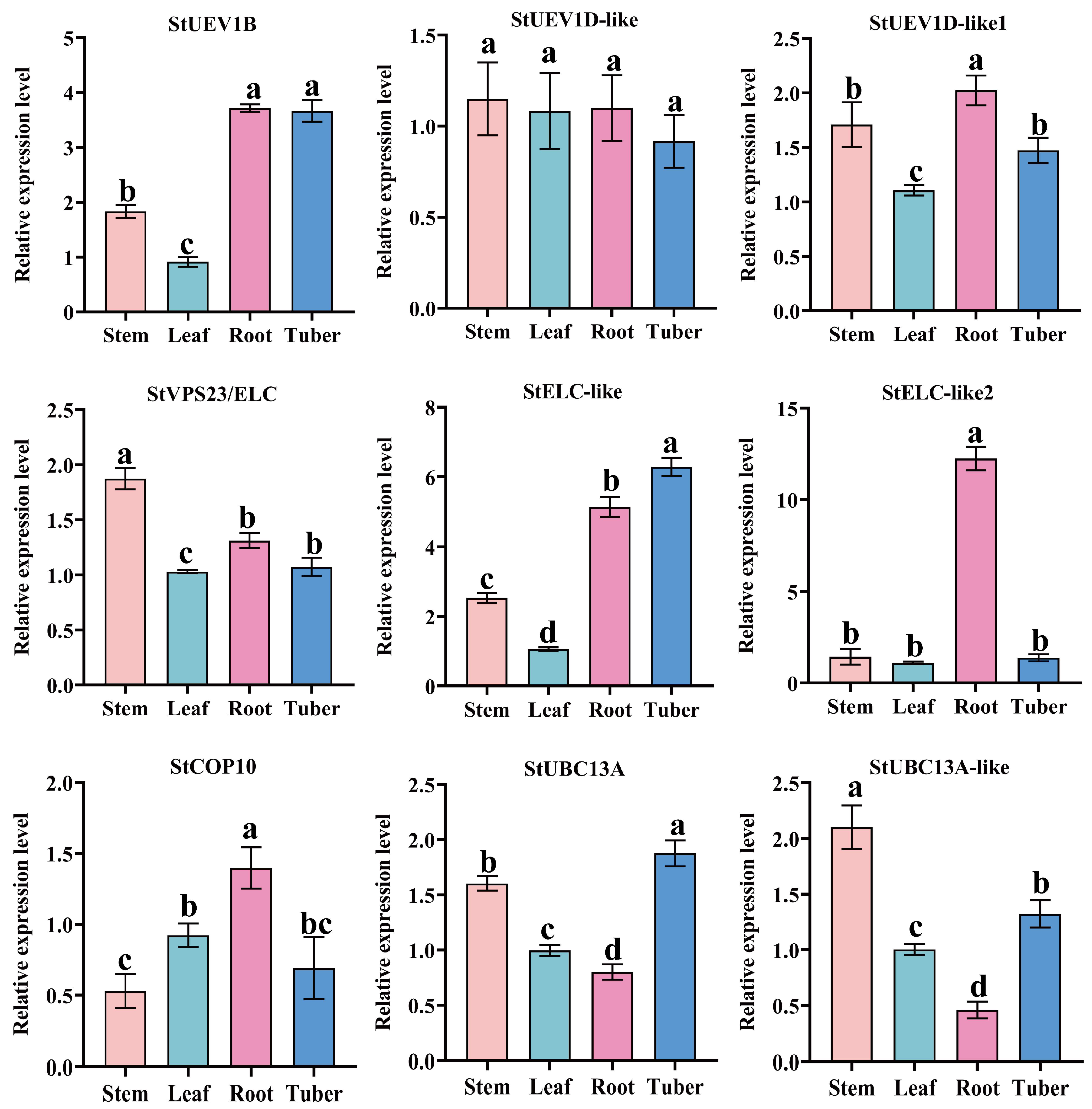
2.7. Expression Patterns of StUEVs and StUBC13
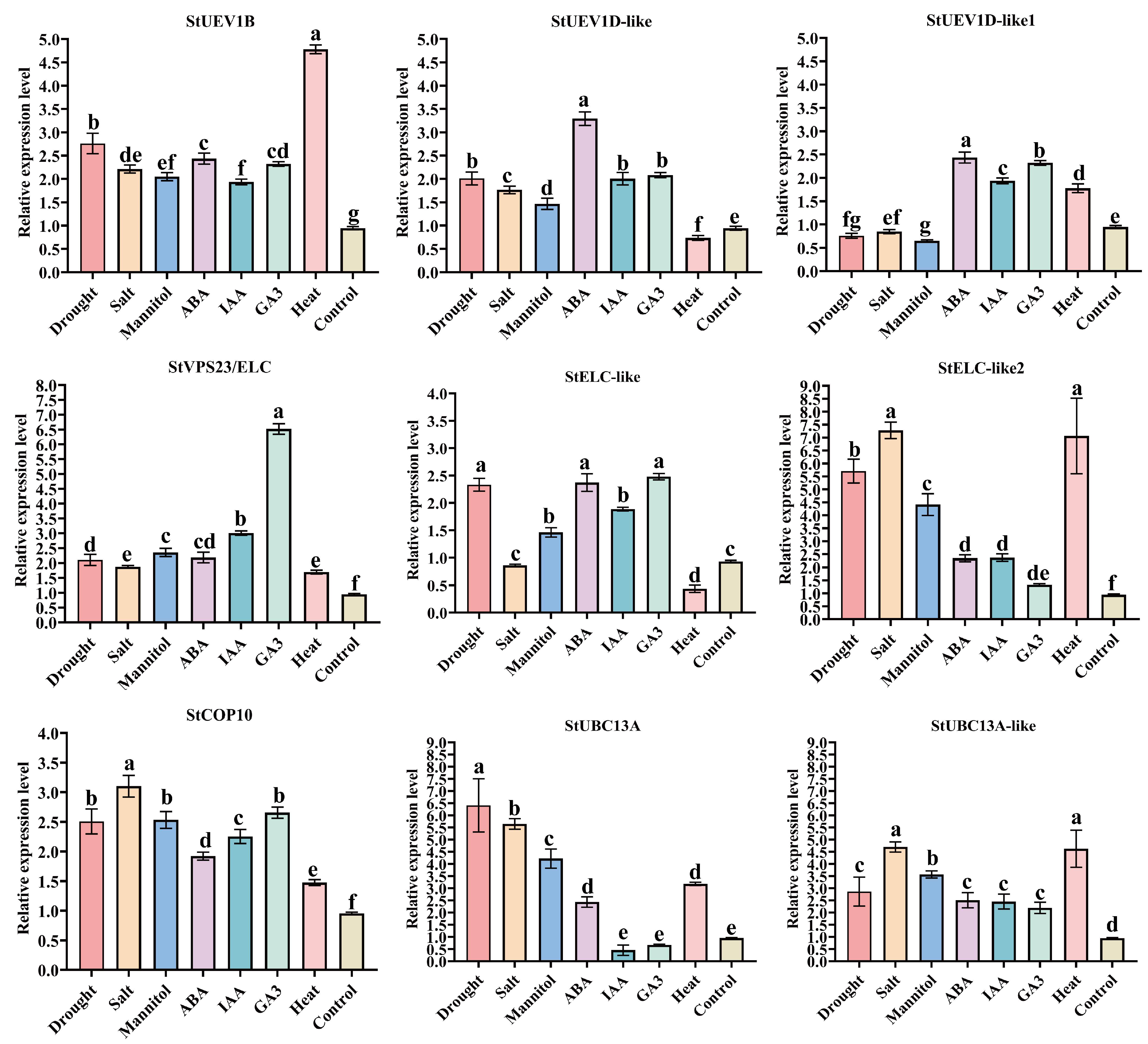
2.8. The qRT-PCR Analysis of StUEVs and StUBC13 in Potato
3. Discussion
4. Materials and Methods
4.1. Identification and Phylogenetic Analysis of StUBC13s and StUEVs
4.2. Multiple Sequence Alignment, Gene Structure and Motif, and Cis-Acting Elements Analysis
4.3. Yeast Two-Hybrid Assay
4.4. Gene Ontology (GO) and KEGG Analysis
4.5. Subcellular Localization and BiFC, SLC Analysis
4.6. Expression Profiles of StUBC13s and StUEV1s
4.7. Plant Materials and Growth Condition
4.8. RNA Extraction and qRT-PCR Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xu, F.Q.; Xue, H.W. The ubiquitin-proteasome system in plant responses to environments. Plant Cell Environ. 2019, 42, 2931–2944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komander, D.; Rape, M. The Ubiquitin Code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wickliffe, K.E.; Williamson, A.; Meyer, H.-J.; Kelly, A.; Rape, M. K11-linked ubiquitin chains as novel regulators of cell division. Trends Cell Biol. 2011, 21, 656–663. [Google Scholar] [CrossRef] [Green Version]
- Cheng, M.-C.; Kuo, W.-C.; Wang, Y.-M.; Chen, H.-Y.; Lin, T.-P. UBC18 mediates ERF1 degradation under light–dark cycles. New Phytol. 2017, 213, 1156–1167. [Google Scholar] [CrossRef]
- Saeki, Y.; Kudo, T.; Sone, T.; Kikuchi, Y.; Yokosawa, H.; Toh-e, A.; Tanaka, K. Lysine 63-linked polyubiquitin chain may serve as a targeting signal for the 26S proteasome. EMBO J. 2009, 28, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Romero-Barrios, N.; Vert, G. Proteasome-independent functions of lysine-63 polyubiquitination in plants. New Phytol. 2018, 217, 995–1011. [Google Scholar] [CrossRef] [Green Version]
- Wen, R.; Wang, S.; Xiang, D.; Venglat, P.; Shi, X.; Zang, Y.; Datla, R.; Xiao, W.; Wang, H. UBC13, an E2 enzyme for Lys63-linked ubiquitination, functions in root development by affecting auxin signaling and Aux/IAA protein stability. Plant J. 2014, 80, 424–436. [Google Scholar] [CrossRef]
- Romero-Barrios, N.; Monachello, D.; Dolde, U.; Wong, A.; San Clemente, H.; Cayrel, A.; Johnson, A.; Lurin, C.; Vert, G. advanced cataloging of Lysine-63 polyubiquitin networks by genomic, interactome, and sensor-based proteomic analyses. Plant Cell 2020, 32, 123–138. [Google Scholar] [CrossRef]
- Guo, H.; Wang, L.; Hu, R.; He, Y.; Xiao, W. Molecular cloning and functional characterization of Physcomitrella patens UBC13-UEV1 genes required for Lys63-linked polyubiquitination. Plant Sci. 2020, 297, 110518. [Google Scholar] [CrossRef]
- Wen, R.; Torres-Acosta, J.A.; Pastushok, L.; Lai, X.; Pelzer, L.; Wang, H.; Xiao, W. Arabidopsis UEV1D promotes Lysine-63–linked polyubiquitination and is involved in DNA damage response. Plant Cell 2008, 20, 213–227. [Google Scholar] [CrossRef]
- Wang, Q.; Zang, Y.; Zhou, X.; Xiao, W. Characterization of four rice UEV1 genes required for Lys63-linked polyubiquitination and distinct functions. BMC Plant Biol. 2017, 17, 126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Liu, M.; Zang, Y.; Xiao, W. The C-terminal extension of Arabidopsis Uev1A/B with putative prenylation site plays critical roles in protein interaction, subcellular distribution and membrane association. Plant Sci. 2020, 291, 110324. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liao, W.; Nie, B.; Zhang, J.; Xu, W. OsUEV1B, an Ubc enzyme variant protein, is required for phosphate homeostasis in rice. Plant J. 2021, 106, 706–719. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Tang, X.; Qi, X.; Fu, X.; Ghimire, S.; Ma, R.; Li, S.; Zhang, N.; Si, H. The ubiquitin conjugating enzyme: An important ubiquitin transfer platform in ubiquitin-proteasome system. In Int. J. Mol. Sci. 2020, 21, 2894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Wen, P.; Lau, O.-S.; Deng, X.-W. Characterization of the ubiquitin E2 enzyme variant gene family in Arabidopsis. J. Integr. Plant Biol. 2007, 49, 120–126. [Google Scholar]
- Bachmair, A.; Novatchkova, M.; Potuschak, T.; Eisenhaber, F. Ubiquitylation in plants: A post-genomic look at a post-translational modification. Trends Plant Sci. 2001, 6, 463–470. [Google Scholar] [CrossRef]
- Zang, Y.; Wang, Q.; Xue, C.; Li, M.; Wen, R.; Xiao, W. Rice UBC13, a candidate housekeeping gene, is required for K63-linked polyubiquitination and tolerance to DNA damage. Rice 2012, 5, 24. [Google Scholar] [CrossRef] [Green Version]
- Wen, R.; Newton, L.; Li, G.; Wang, H.; Xiao, W. Arabidopsis thaliana UBC13: Implication of error-free DNA damage tolerance and Lys63-linked polyubiquitylation in plants. Plant Mol. Biol. 2006, 61, 241–253. [Google Scholar] [CrossRef]
- Liu, W.; Tang, X.; Zhu, X.; Qi, X.; Zhang, N.; Si, H. Genome-wide identification and expression analysis of the E2 gene family in potato. Mol. Biol. Rep. 2019, 46, 777–791. [Google Scholar] [CrossRef]
- Guo, H.; Wen, R.; Liu, Z.; Datla, R.; Xiao, W. Molecular cloning and functional characterization of two Brachypodium distachyon UBC13 genes whose products promote K63-linked polyubiquitination. Front. Plant Sci. 2016, 6, 1222. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.; Wen, R.; Wang, Q.; Datla, R.; Xiao, W. Three Brachypodium distachyon Uev1s promote Ubc13-mediated Lys63-linked polyubiquitination and confer different functions. Front. Plant Sci. 2016, 7, 1551. [Google Scholar] [CrossRef] [PubMed]
- Belda-Palazon, B.; Julian, J.; Coego, A.; Wu, Q.; Zhang, X.; Batistic, O.; Alquraishi, S.A.; Kudla, J.; An, C.; Rodriguez, P.L. ABA inhibits myristoylation and induces shuttling of the RGLG1 E3 ligase to promote nuclear degradation of PP2CA. Plant J. 2019, 98, 813–825. [Google Scholar] [CrossRef] [PubMed]
- Sekiguchi, N.; Sasaki, K.; Oshima, Y.; Mitsuda, N. Ectopic expression of AtNF-YA6-VP16 in petals results in a novel petal phenotype in Torenia fournieri. Planta 2022, 255, 105. [Google Scholar] [CrossRef] [PubMed]
- Abbas, N.; Maurya, J.P.; Senapati, D.; Gangappa, S.N.; Chattopadhyay, S. Arabidopsis CAM7 and HY5 physically interact and directly bind to the HY5 promoter to regulate its expression and thereby promote photomorphogenesis. Plant Cell 2014, 26, 1036–1052. [Google Scholar] [CrossRef] [Green Version]
- Shen, C.; Wang, J.; Shi, X.; Kang, Y.; Xie, C.; Peng, L.; Dong, C.; Shen, Q.; Xu, Y. Transcriptome analysis of differentially expressed genes induced by low and high potassium levels provides insight into fruit sugar metabolism of pear. Front. Plant Sci. 2017, 8, 938. [Google Scholar] [CrossRef] [Green Version]
- Pratelli, R.; Guerra, D.D.; Yu, S.; Wogulis, M.; Kraft, E.; Frommer, W.B.; Callis, J.; Pilot, G. The Ubiquitin E3 ligase LOSS OF GDU2 is required for GLUTAMINE DUMPER1-induced amino acid secretion in Arabidopsis. Plant Phytol. 2012, 158, 1628–1642. [Google Scholar] [CrossRef] [Green Version]
- Lampl, N.; Alkan, N.; Davydov, O.; Fluhr, R. Set-point control of RD21 protease activity by AtSerpin1 controls cell death in Arabidopsis. Plant J. 2013, 74, 498–510. [Google Scholar] [CrossRef]
- Jia, W.; Yao, Z.; Zhao, J.; Guan, Q.; Gao, L. New perspectives of physiological and pathological functions of nucleolin (NCL). Life Sci. 2017, 186, 1–10. [Google Scholar] [CrossRef]
- Kim, E.-J.; Lee, S.-H.; Park, C.-H.; Kim, S.-H.; Hsu, C.-C.; Xu, S.; Wang, Z.-Y.; Kim, S.-K.; Kim, T.-W. Plant U-Box40 mediates degradation of the brassinosteroid-responsive transcription factor BZR1 in Arabidopsis roots. Plant Cell 2019, 31, 791–808. [Google Scholar] [CrossRef]
- Pan, I.-C.; Schmidt, W. Functional implications of K63-linked ubiquitination in the iron deficiency response of Arabidopsis roots. Front. Plant Sci. 2014, 4, 542. [Google Scholar] [CrossRef] [Green Version]
- Turek, I.; Tischer, N.; Lassig, R.; Trujillo, M. Multi-tiered pairing selectivity between E2 ubiquitin–conjugating enzymes and E3 ligases. J. Biol. Chem. 2018, 293, 16324–16336. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wen, R.; Wang, J.; Xiang, D.; Wang, Q.; Zang, Y.; Wang, Z.; Huang, S.; Li, X.; Datla, R.; et al. Arabidopsis UBC13 differentially regulates two programmed cell death pathways in responses to pathogen and low-temperature stress. New Phytol. 2019, 221, 919–934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Schmidt, W. A lysine-63-linked ubiquitin chain-forming conjugase, UBC13, promotes the developmental responses to iron deficiency in Arabidopsis roots. Front. Plant Sci. 2010, 62, 330–343. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Wang, S.; Dong, D.; Zhou, R.; Wang, H. Arabidopsis ubiquitin-conjugating enzymes UBC7, UBC13, and UBC14 are required in plant responses to multiple stress conditions. Plants 2020, 9, 723. [Google Scholar] [CrossRef]
- Mural, R.V.; Liu, Y.; Rosebrock, T.R.; Brady, J.J.; Hamera, S.; Connor, R.A.; Martin, G.B.; Zeng, L. The tomato Fni3 Lysine-63–specific ubiquitin-conjugating enzyme and Suv ubiquitin E2 variant positively regulate plant immunity. Plant Cell 2013, 25, 3615–3631. [Google Scholar] [CrossRef] [Green Version]
- Diambra, L.A. Genome sequence and analysis of the tuber crop potato. Nature 2011, 475, 189–195. [Google Scholar]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: Protein domains identifier. Nucleic Acids Res. 2005, 33 (Suppl. S2), W116–W120. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Nicholas, K.B. GeneDoc: Analysis and visualization of genetic variation. Embnew. News 1997, 4, 14. [Google Scholar]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37 (Suppl. S2), W202–W208. [Google Scholar] [CrossRef]
- Fields, S.; Song, O.-K. A novel genetic system to detect protein–protein interactions. Nature 1989, 340, 245–246. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R.J.M. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]



| Gene Name | Accession Number | Protein/AA1 | Chrom2 | MW (Da)3 | pI4 | Instability Index | GRAVY5 |
|---|---|---|---|---|---|---|---|
| StUEV1B | Soltu.DM.04G003350 | 161 AA | chr4:3,714,877–3,722,749 | 19,476.32 | 9.25 | 34.47 | −0.617 |
| StUEV1D-like | Soltu.DM.01G003250 | 147AA | chr1:3,436,712–3,441,067 | 16,560.81 | 6.20 | 29.72 | −0.601 |
| StUEV1D-like1 | Soltu.DM.10G025250 | 147AA | chr10:56,826,223–56,830,996 | 16,648.90 | 6.20 | 30.24 | −0.601 |
| StVPS23/ ELC | Soltu.DM.10G022020 | 400AA | chr10:54,196,275–54,199,731 | 44,902.08 | 6.07 | 47.44 | −0.434 |
| StELC-like | Soltu.DM.09G004020 | 407AA | chr9:3,420,331–3,426,759 | 45,550.78 | 6.17 | 48.10 | −0.451 |
| StELC-like2 | Soltu.DM.09G020470 | 350AA | chr9:55,278,138–55,279,565 | 40,160.35 | 4.97 | 50.05 | −0.336 |
| StCOP10 | Soltu.DM.01G001370 | 181AA | chr1:1,515,501–1,519,690 | 19,476.32 | 9.25 | 34.36 | −0.222 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, W.; Tang, X.; Fu, X.; Zhang, H.; Zhu, C.; Zhang, N.; Si, H. Functional Characterization of Potato UBC13-UEV1s Genes Required for Ubiquitin Lys63 Chain to Polyubiquitination. Int. J. Mol. Sci. 2023, 24, 2412. https://doi.org/10.3390/ijms24032412
Liu W, Tang X, Fu X, Zhang H, Zhu C, Zhang N, Si H. Functional Characterization of Potato UBC13-UEV1s Genes Required for Ubiquitin Lys63 Chain to Polyubiquitination. International Journal of Molecular Sciences. 2023; 24(3):2412. https://doi.org/10.3390/ijms24032412
Chicago/Turabian StyleLiu, Weigang, Xun Tang, Xue Fu, Huanhuan Zhang, Cunlan Zhu, Ning Zhang, and Huaijun Si. 2023. "Functional Characterization of Potato UBC13-UEV1s Genes Required for Ubiquitin Lys63 Chain to Polyubiquitination" International Journal of Molecular Sciences 24, no. 3: 2412. https://doi.org/10.3390/ijms24032412





