Novel Insight into the Role of Squalene Epoxidase (SQLE) Gene in Determining Milk Production Traits in Buffalo
Abstract
:1. Introduction
2. Results
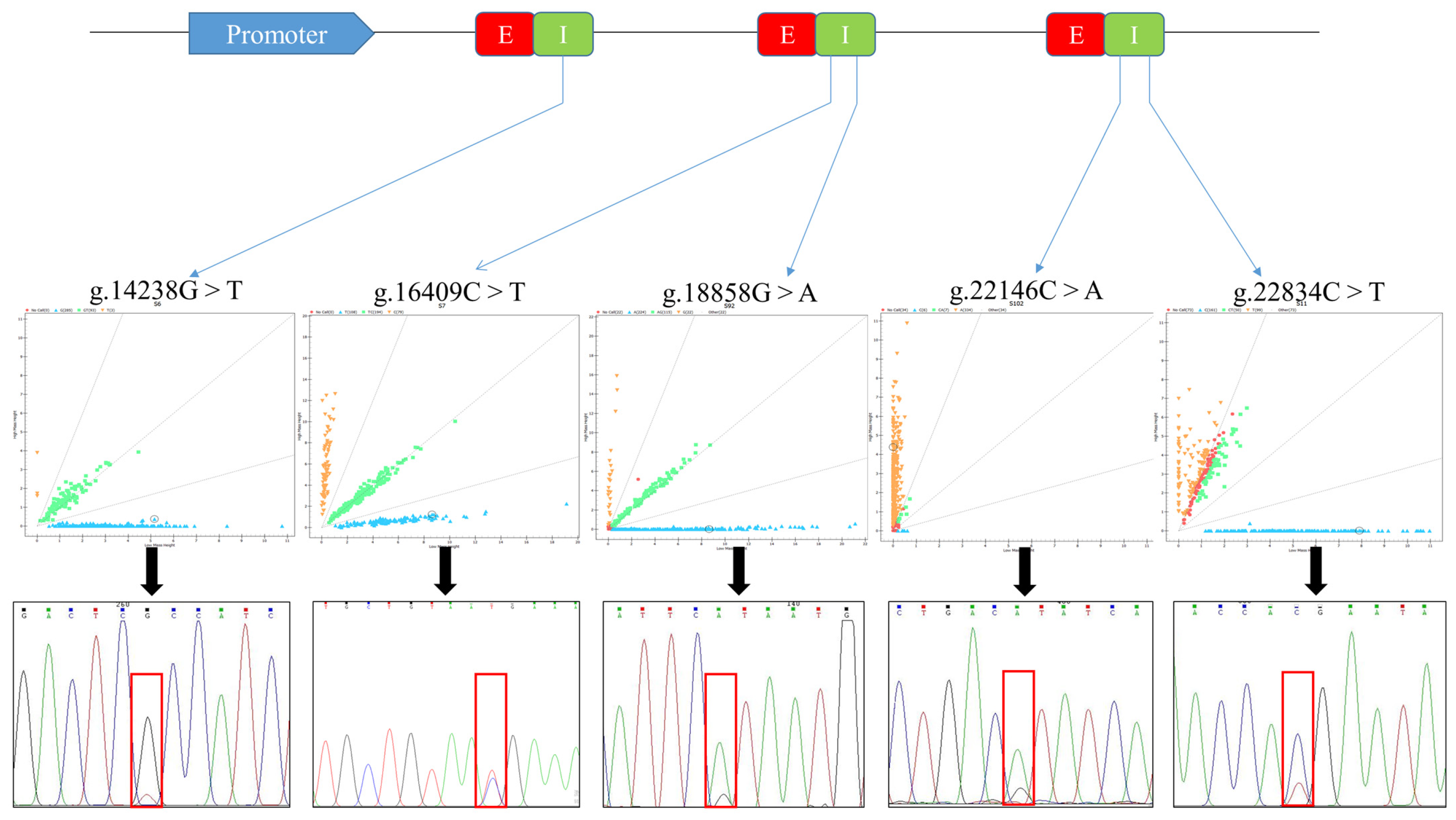
2.1. SNP Polymorphic Loci Identification and Genotyping of SQLE Gene
2.2. Association of SQLE Genotype with Milk Production Traits
2.3. Linkage Disequilibrium and Haplotypes Analysis
2.4. Identification of BuMECs, Interference Efficiency, and Expression Profile of SQLE
2.5. SQLE Regulates the BuMECs Proliferation, Cell Cycle, and Apoptosis
2.6. SQLE Regulates BuMECs Lipogenesis and Casein Synthesis
3. Discussion
4. Materials and Methods
4.1. Samples Collection
4.2. SNP Identification, Genotyping, and Linkage Disequilibrium Analysis
4.3. Association Analysis
4.4. Cell Culture and Transfection
4.5. RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
4.6. Western Blotting and Cellular Immunofluorescence
4.7. Cell Counts and Proliferation Assay
4.8. Cell Cycle and Apoptosis Assay
4.9. Enzyme-Linked Immunosorbent Assay Were Used to Detect Concentrations of α-, β-, and κ-Casein
4.10. Triglyceride Content Detection and Oil Red O Staining
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Arora, S.; Sindhu, J.S.; Khetra, Y. Buffalo milk. In Reference Module in Food Science; Elsevier: Amsterdam, The Netherlands, 2022. [Google Scholar]
- Nasr, M.A.F.; Awad, A.; El Araby, I.E. Associations of Leptin and Pituitary-Specific Transcription Factor Genes’ Polymorphisms with Reproduction and Production Traits in Dairy Buffalo. Reprod. Domest. Anim. 2016, 51, 597–603. [Google Scholar] [CrossRef]
- Hegde, N.G. Buffalo Husbandry for Sustainable Development of Small Farmers in India and other Developing Countries. Asian J. Res. Anim. Vet. Sci. 2019, 3, 1–20. [Google Scholar]
- Bhutto, B.; Arijo, A.; Phullan, M.S.; Rind, R. Prevalence of Fascioliasis in Buffaloes under Different Agro-Climatic Areas of Sindh Province of Pakistan. Int. J. Agric. Biol. 2012, 14, 241–245. [Google Scholar]
- Barłowska, J.; Szwajkowska, M.; Litwińczuk, Z.; Król, J. Nutritional Value and Technological Suitability of Milk from Various Animal Species Used for Dairy Production. Compr. Rev. Food Sci. Food Saf. 2011, 10, 291–302. [Google Scholar] [CrossRef]
- Jana, A.H.; Mandal, P.K. Manufacturing and quality of mozzarella cheese: A review. Int. J. Dairy Sci. 2011, 6, 199–226. [Google Scholar] [CrossRef]
- Pegolo, S.; Stocco, G.; Mele, M.; Schiavon, S.; Bittante, G.; Cecchinato, A. Factors affecting variations in the detailed fatty acid profile of Mediterranean buffalo milk determined by 2-dimensional gas chromatography. J. Dairy Sci. 2017, 100, 2564–2576. [Google Scholar] [CrossRef] [Green Version]
- De los Campos, G.; Hickey, J.M.; Pong-Wong, R.; Daetwyler, H.D.; Calus, M.P.L. Whole-Genome Regression and Prediction Methods Applied to Plant and Animal Breeding. Genetics 2013, 193, 327–345. [Google Scholar] [CrossRef] [Green Version]
- Goddard, M.E.; Hayes, B.J.; Meuwissen, T.H.E. Using the genomic relationship matrix to predict the accuracy of genomic selection. J. Anim. Breed. Genet. 2011, 128, 409–421. [Google Scholar] [CrossRef]
- Wang, R.J.; Gao, X.F.; Yang, J.; Kong, X.R. Genome-Wide Association Study to Identify Favorable SNP Allelic Variations and Candidate Genes That Control the Timing of Spring Bud Flush of Tea (Camellia sinensis) Using SLAF-seq. J. Agric. Food Chem. 2019, 67, 10380–10391. [Google Scholar] [CrossRef]
- Du, C.; Deng, T.; Zhou, Y.; Ye, T.; Zhou, Z.; Zhang, S.; Shao, B.; Wei, P.; Sun, H.; Khan, F.A.; et al. Systematic analyses for candidate genes of milk production traits in water buffalo (Bubalus bubalis). Anim. Genet. 2019, 50, 207–216. [Google Scholar] [CrossRef]
- Deng, T.; Pang, C.; Ma, X.; Duan, A.; Liang, S.; Lu, X.; Liang, X. Buffalo SREBP1: Molecular cloning, expression and association analysis with milk production traits. Anim. Genet. 2017, 48, 720–721. [Google Scholar] [CrossRef]
- Freitas, A.C.; de Camargo, G.M.F.; Aspilcueta-Borquis, R.R.; Stafuzza, N.B.; Venturini, G.C.; Tanamati, F.; Hurtado-Lugo, N.A.; Barros, C.C.; Tonhati, H. Polymorphism in the A2M gene associated with high-quality milk in Murrah buffaloes (Bubalus bubalis). Genet. Mol. Res. 2016, 15, gmr.15027637. [Google Scholar] [CrossRef]
- Houten, S.M.; Schneiders, M.S.; Wanders, R.; Waterham, H.R. Regulation of Isoprenoid/Cholesterol Biosynthesis in Cells from Mevalonate Kinase-deficient Patients. J. Biol. Chem. 2003, 278, 5736–5743. [Google Scholar] [CrossRef] [Green Version]
- Gill, S.; Stevenson, J.; Kristiana, I.; Brown, A.J. Cholesterol-Dependent Degradation of Squalene Monooxygenase, a Control Point in Cholesterol Synthesis beyond HMG-CoA Reductase. Cell Metab. 2011, 13, 260–273. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, S.; Bloch, K. Studies on Squalene Expoxidase of Rat Liver. J. Biol. Chem. 1970, 245, 1670–1674. [Google Scholar] [CrossRef] [PubMed]
- Ha, J.; Kwon, S.; Hwang, J.H.; Park, D.H.; Kim, T.W.; Kang, D.G.; Yu, G.E.; Park, H.C.; An, S.M.; Kim, C.W. Squalene epoxidase plays a critical role in determining pig meat quality by regulating adipogenesis, myogenesis, and ROS scavengers. Sci. Rep. 2017, 7, 16740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stopsack, K.H.; Gerke, T.A.; Sinnott, J.A.; Penney, K.L.; Tyekucheva, S.; Sesso, H.D.; Andersson, S.-O.; Andren, O.; Cerhan, J.R.; Giovannucci, E.L.; et al. Cholesterol Metabolism and Prostate Cancer Lethality. Cancer Res. 2016, 76, 4785–4790. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.; Kou, J.; Liu, Z.; Li, W.; Du, W. MYC Enhances Cholesterol Biosynthesis and Supports Cell Proliferation through SQLE. Front. Cell Dev. Biol. 2021, 9, 655889. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Pei, J.; Wu, X.; Kalwar, Q.; Yan, P.; Guo, X. Effect of Gender to Fat Deposition in Yaks Based on Transcriptomic and Metabolomics Analysis. Front. Cell Dev. Biol. 2021, 9, 653188. [Google Scholar] [CrossRef] [PubMed]
- Capuco, A.V.; Wood, D.L.; Baldwin, R.; McLeod, K.; Paape, M.J. Mammary cell number, proliferation, and apoptosis during a bovine lactation: Relation to milk production and effect of bST. J. Dairy Sci. 2001, 84, 2177–2187. [Google Scholar] [CrossRef] [PubMed]
- Inman, J.L.; Robertson, C.; Mott, J.D.; Bissell, M.J. Mammary gland development: Cell fate specification, stem cells and the microenvironment. Development 2015, 142, 1028–1042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boutinaud, M.; Guinard-Flament, J.; Jammes, H. The number and activity of mammary epithelial cells, determining factors for milk production. Reprod. Nutr. Dev. 2004, 44, 499–508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, H.; Zhao, Y.; Shi, X.; Tan, Z.; Chi, X.; He, M.; Jiang, G.; Ji, L.; Li, H. Squalene epoxidase promotes the proliferation and metastasis of lung squamous cell carcinoma cells though extracellular signal-regulated kinase signaling. Thorac. Cancer 2019, 10, 428–436. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wang, Y.; Liu, D.; Wong, C.C.; Coker, O.O.; Zhang, X.; Liu, C.; Zhou, Y.; Liu, Y.; Kang, W.; et al. Squalene epoxidase drives cancer cell proliferation and promotes gut dysbiosis to accelerate colorectal carcinogenesis. Gut 2022, 71, 2253–2265. [Google Scholar] [CrossRef] [PubMed]
- Sui, Z.; Zhou, J.; Cheng, Z.; Lu, P. Squalene epoxidase (SQLE) promotes the growth and migration of the hepatocellular carcinoma cells. Tumor Biol. 2015, 36, 6173–6179. [Google Scholar] [CrossRef]
- Brown, D.N.; Caffa, I.; Cirmena, G.; Piras, D.; Garuti, A.; Gallo, M.; Alberti, S.; Nencioni, A.; Ballestrero, A.; Zoppoli, G. Squalene epoxidase is a bona fide oncogene by amplification with clinical relevance in breast cancer. Sci. Rep. 2016, 6, 19435. [Google Scholar] [CrossRef] [Green Version]
- Tong, H.-L.; Li, Q.-Z.; Gao, X.-J.; Yin, D.-Y. Establishment and characterization of a lactating dairy goat mammary gland epithelial cell line. Vitr. Cell. Dev. Biol. Anim. 2012, 48, 149–155. [Google Scholar] [CrossRef]
- Nadeem, A.; Maryam, J. Genetic and genomic dissection of Prolactin revealed potential association with milk production traits in riverine buffalo. Trop. Anim. Health Prod. 2016, 48, 1261–1268. [Google Scholar] [CrossRef]
- Ma, Y.; Khan, M.Z.; Xiao, J.; Alugongo, G.M.; Chen, X.; Chen, T.; Liu, S.; He, Z.; Wang, J.; Shah, M.K.; et al. Genetic markers associated with milk production traits in dairy cattle. Agriculture 2021, 11, 1018. [Google Scholar] [CrossRef]
- Han, B.; Yuan, Y.; Shi, L.; Li, Y.; Liu, L.; Sun, D. Identification of single nucleotide polymorphisms of PIK3R1 and DUSP1 genes and their genetic associations with milk production traits in dairy cows. J. Anim. Sci. Biotechnol. 2019, 10, 81. [Google Scholar] [CrossRef] [Green Version]
- Thawee, L.; Elzo, M.A.; Skorn, K.; Thanathip, S.; Danai, J. Genomic-polygenic and polygenic predictions for milk yield, fat yield, and age at first calving in Thai multibreed dairy population using genic and functional sets of genotypes. Livest. Sci. 2019, 219, 17–24. [Google Scholar] [CrossRef]
- Le Hir, H.; Nott, A.; Moore, M.J. How introns influence and enhance eukaryotic gene expression. Trends Biochem. Sci. 2003, 28, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-F.; Feng, L.; Niu, D.-K. Relationship between mRNA stability and intron presence. Biochem. Biophys. Res. Commun. 2007, 354, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Larson, A. Book review: Molecular Markers, Natural-History and Evolution, by Avise, J.C. Science 1995, 267, 115–116. [Google Scholar] [CrossRef]
- Slatkin, M. Linkage disequilibrium—Understanding the evolutionary past and mapping the medical future. Nat. Rev. Genet. 2008, 9, 477–485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, L.; Li, W.; Song, Q. Chromosome-range whole-genome high-throughput experimental haplotyping by single-chromosome microdissection. In Haplotyping: Methods and Protocols; Tiemann-Boege, I., Betancourt, A., Eds.; Springer: New York, NY, USA, 2017; Volume 1551, pp. 161–169. [Google Scholar]
- Xu, W.; Chen, Q.; Jia, Y.; Deng, J.; Jiang, S.; Qin, G.; Qiu, Q.; Wang, X.; Yang, X.; Jiang, H. Isolation, characterization, and SREBP1 functional analysis of mammary epithelial cell in buffalo. J. Food Biochem. 2019, 43, e12997. [Google Scholar] [CrossRef]
- Suzuki, Y.; Chiba, S.; Nishihara, K.; Nakajima, K.; Hagino, A.; Kim, W.-S.; Lee, H.-G.; Nochi, T.; Suzuki, T.; Roh, S.-G. Chemerin Regulates Epithelial Barrier Function of Mammary Glands in Dairy Cows. Animals 2021, 11, 3194. [Google Scholar] [CrossRef]
- Lin, Y.; Sun, H.; Shaukat, A.; Deng, T.; Abdel-Shafy, H.; Che, Z.; Zhou, Y.; Hu, C.; Li, H.; Wu, Q.; et al. Novel Insight Into the Role of ACSL1 Gene in Milk Production Traits in Buffalo. Front. Genet. 2022, 13, 896910. [Google Scholar] [CrossRef]
- He, L.; Li, H.; Pan, C.; Hua, Y.; Peng, J.; Zhou, Z.; Zhao, Y.; Lin, M. Squalene epoxidase promotes colorectal cancer cell proliferation through accumulating calcitriol and activating CYP24A1-mediated MAPK signaling. Cancer Commun. 2021, 41, 726–746. [Google Scholar] [CrossRef]
- You, W.; Ke, J.; Chen, Y.; Cai, Z.; Huang, Z.-P.; Hu, P.; Wu, X. SQLE, A Key Enzyme in Cholesterol Metabolism, Correlates with Tumor Immune Infiltration and Immunotherapy Outcome of Pancreatic Adenocarcinoma. Front. Immunol. 2022, 13, 864244. [Google Scholar] [CrossRef]
- Liu, H.; Ai, J.; Shen, A.; Chen, Y.; Wang, X.; Peng, X.; Chen, H.; Shen, Y.; Huang, M.; Ding, J.; et al. c-Myc Alteration Determines the Therapeutic Response to FGFR Inhibitors. Clin. Cancer Res. 2017, 23, 974–984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soucek, L.; Whitfield, J.; Martins, C.P.; Finch, A.J.; Murphy, D.J.; Sodir, N.M.; Karnezis, A.N.; Swigart, L.B.; Nasi, S.; Evan, G.I. Modelling Myc inhibition as a cancer therapy. Nature 2008, 455, 679–683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, K.; Atoyan, R.; Borek, M.A.; Dellarocca, S.; Samson, M.E.S.; Ma, A.W.; Xu, G.-X.; Patterson, T.; Tuck, D.P.; Viner, J.L.; et al. Dual HDAC and PI3K Inhibitor CUDC-907 Downregulates MYC and Suppresses Growth of MYC-dependent Cancers. Mol. Cancer Ther. 2017, 16, 285–299. [Google Scholar] [CrossRef] [Green Version]
- Ushmorov, A.; Debatin, K.M.; Beltinger, C. Growth inhibition of murine neuroblastoma cells by c-myc with cell cycle arrest in G(2)/M. Cancer Biol. Ther. 2005, 4, 181–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dotto, G.P. p21(WAF1/Cip1): More than a break to the cell cycle? Biochim. Biophys. Acta Rev. Cancer 2000, 1471, M43–M56. [Google Scholar] [CrossRef]
- Warbrick, E.; Lane, D.P.; Glover, D.M.; Cox, L.S. A small peptide inhibitor of dna-replication defines the site of interaction between the cyclin-dependent kinase inhibitor p21(waf1) and proliferating cell nuclear antigen. Curr. Biol. 1995, 5, 275–282. [Google Scholar] [CrossRef] [Green Version]
- Moldovan, G.-L.; Pfander, B.; Jentsch, S. PCNA, the maestro of the replication fork. Cell 2007, 129, 665–679. [Google Scholar] [CrossRef] [Green Version]
- Liu, K.; Zhang, Q.; Lan, H.; Wang, L.; Mou, P.; Shao, W.; Liu, D.; Yang, W.; Lin, Z.; Lin, Q.; et al. GCN5 Potentiates Glioma Proliferation and Invasion via STAT3 and AKT Signaling Pathways. Int. J. Mol. Sci. 2015, 16, 21897–21910. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.-Y.; Zhang, X.-P.; Wang, W. Coordination of miR-192 and miR-22 in p53-Mediated Cell Fate Decision. Int. J. Mol. Sci. 2019, 20, 4768. [Google Scholar] [CrossRef] [Green Version]
- Miller, N.; Delbecchi, L.; Petitclerc, D.; Wagner, G.F.; Talbot, B.G.; Lacasse, P. Effect of stage of lactation and parity on mammary gland cell renewal. J. Dairy Sci. 2006, 89, 4669–4677. [Google Scholar] [CrossRef] [Green Version]
- Zhichen, T.; Yuanyuan, Z.; Huiming, Z.; Yujia, S.; Yongjiang, M.; Zhangping, Y.; Mingxun, L. Transcriptional regulation of milk fat synthesis in dairy cattle. J. Funct. Foods 2022, 96, 105208. [Google Scholar] [CrossRef]
- Zhang, D.-D.; Wang, D.-D.; Wang, Z.; Wang, Y.-B.; Li, G.-X.; Sun, G.-R.; Tian, Y.-D.; Han, R.-L.; Li, Z.-J.; Jiang, R.-R.; et al. Estrogen Abolishes the Repression Role of gga-miR-221-5p Targeting ELOVL6 and SQLE to Promote Lipid Synthesis in Chicken Liver. Int. J. Mol. Sci. 2020, 21, 1624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, S.M.; Rudolph, M.C.; Mcmanaman, J.L.; Neville, M.C. Secretory activation in the mammary gland: It’s not just about milk protein synthesis! Breast Cancer Res. 2007, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zhao, F.; Wei, C.; Liang, M.; Zhang, N.; Wang, C.; Li, Q.-Z.; Gao, X.-J. Function of SREBP1 in the Milk Fat Synthesis of Dairy Cow Mammary Epithelial Cells. Int. J. Mol. Sci. 2014, 15, 16998–17013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- St-Gelais, D.; Hache, S. Effect of β-casein concentration in cheese milk on rennet coagulation properties, cheese composition and cheese ripening. Food Res. Int. 2005, 38, 523–531. [Google Scholar] [CrossRef]
- Farrell, H.M.J.R.; Jimenez-Flores, R.; Bleck, G.T.; Brown, E.M.; Swaisgood, H.E. Nomenclature of the Proteins of Cows’ Milk—Sixth Revision. J. Dairy Sci. 2004, 87, 1641–1674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.C.; Zhao, S.G.; Wang, S.S.; Luo, C.C.; Gao, H.N.; Zheng, N.; Wang, J.Q. d-Glucose and amino acid deficiency inhibits casein synthesis through JAK2/STAT5 and AMPK/mTOR signaling pathways in mammary epithelial cells of dairy cows. J. Dairy Sci. 2018, 101, 1737–1746. [Google Scholar] [CrossRef]
- Rius, A.G.; Appuhamy, J.A.D.R.N.; Cyriac, J.; Kirovski, D.; Becvar, O.; Escobar, J.; McGilliard, M.L.; Bequette, B.J.; Akers, R.M.; Hanigan, M.D. Regulation of protein synthesis in mammary glands of lactating dairy cows by starch and amino acids. J. Dairy Sci. 2010, 93, 3114–3127. [Google Scholar] [CrossRef] [Green Version]
- Burnett, P.E.; Barrow, R.K.; Cohen, N.A.; Snyder, S.H.; Sabatini, D.M. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc. Natl. Acad. Sci. USA 1998, 95, 1432–1437. [Google Scholar] [CrossRef] [Green Version]
- Schindler, C.; Levy, D.E.; Decker, T. JAK-STAT signaling: From interferons to cytokines. J. Biol. Chem. 2007, 282, 20059–20063. [Google Scholar] [CrossRef] [Green Version]
- Ye, T.; Deng, T.; Hosseini, S.M.; Raza, S.H.A.; Du, C.; Chen, C.; Zhang, X.; Hu, X.; Yang, L. Association analysis between FASN genotype and milk traits in Mediterranean buffalo and its expression among different buffalo tissues. Trop. Anim. Health Prod. 2021, 53, 366. [Google Scholar] [CrossRef] [PubMed]
- Baldi, F.; Massolini Laureano, M.M.; Mansan Gordo, D.G.; Bignardi, A.B.; Aspilcueta Borquis, R.R.; de Albuquerque, L.G.; Tonhati, H. Effect of lactation length adjustment procedures on genetic parameter estimates for buffalo milk yield. Genet. Mol. Biol. 2011, 34, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, J.; Liu, S.; Campanile, G.; Salzano, A.; Gasparrini, B.; Plastow, G.; Zhang, C.; Wang, Z.; Liang, A.; et al. Genome-wide association study for buffalo mammary gland morphology. J. Dairy Res. 2020, 87, 27–31. [Google Scholar] [CrossRef]
- Anand, V.; Dogra, N.; Singh, S.; Kumar, S.N.; Jena, M.K.; Malakar, D.; Dang, A.K.; Mishra, B.P.; Mukhopadhyay, T.K.; Kaushik, J.K.; et al. Establishment and Characterization of a Buffalo (Bubalus bubalis) Mammary Epithelial Cell Line. PLoS ONE 2012, 7, e40469. [Google Scholar] [CrossRef] [Green Version]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Deng, T.; Hua, L.; Zhao, X.; Wu, H.; Sun, P.; Liu, M.; Zhang, S.; Yang, L.; Liang, A. Novel functional mutation of the PDIA3 gene affects milk composition traits in Chinese Holstein cattle. J. Dairy Sci. 2022, 105, 5153–5166. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Ahmad, M.J.; Ye, T.; Du, C.; Zhang, X.; Liang, A.; Yang, L. Cathepsin B Regulates Mice Granulosa Cells’ Apoptosis and Proliferation In Vitro. Int. J. Mol. Sci. 2021, 22, 11827. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, W.; Zhang, F.; Zhong, S.; Sun, Y.; Huo, J.; Zhu, J.; Wu, C. The Gut Microbiota-Produced Indole-3-Propionic Acid Confers the Antihyperlipidemic Effect of Mulberry-Derived 1-Deoxynojirimycin. Msystems 2020, 5, e00313-20. [Google Scholar] [CrossRef] [PubMed]




| Snps | Location | Genotypes | Genotype Frequency | Allele | Allele Frequencies | Heterozygosity | PIC |
|---|---|---|---|---|---|---|---|
| g.14238G > T | intron6 | GT | 0.244 | G | 0.870 | 0.226 | 0.201 |
| GG | 0.748 | T | 0.130 | ||||
| TT | 0.008 | ||||||
| g.16409C > T | intron7 | CC | 0.207 | C | 0.462 | 0.497 | 0.374 |
| TT | 0.283 | T | 0.538 | ||||
| TC | 0.509 | ||||||
| g.18858G > A | Intron7 | GG | 0.061 | G | 0.220 | 0.343 | 0.284 |
| AA | 0.620 | A | 0.780 | ||||
| GA | 0.319 | ||||||
| g.22146C > A | Intron9 | CA | 0.020 | A | 0.973 | 0.053 | 0.052 |
| AA | 0.963 | C | 0.027 | ||||
| CC | 0.017 | ||||||
| g.22834C > T | Intron9 | CC | 0.518 | C | 0.510 | 0.480 | 0.365 |
| TT | 0.318 | T | 0.490 | ||||
| CT | 0.163987 |
| SNPs | Genotype | PM (kg) | MY (kg) | FY (kg) | FP (%) | PY (kg) | PP (%) |
|---|---|---|---|---|---|---|---|
| g.14238G > T | GG | 14.55 ± 0.0.35 | 2792.28 ± 77.98 | 214.44 ± 7.00 | 7.69 ± 0.15 | 125.74 ± 3.57 | 4.53 ± 0.04 |
| GT | 14.78 ± 0.38 | 2855.74 ± 83.00 | 218.69 ± 7.35 | 7.68 ± 0.15 | 128.01 ± 3.82 | 4.51 ± 0.04 | |
| TT | 14.36 ± 1.16 | 2763.13 ± 249.56 | 205.24 ± 21.04 | 7.44 ± 0.42 | 128.04 ± 11.71 | 4.65 ± 0.12 | |
| p-value | 0.60 | 0.44 | 0.53 | 0.81 | 0.63 | 0.39 | |
| g.16409C > T | CC | 15.03 ± 0.24 | 2896.56 ± 51.01 | 228.32 ± 4.29 | 7.89 ± 0.09 | 131.17 ± 2.40 | 4.56 ± 0.03 |
| TC | 15.01 ± 0.17 | 2898.93 ± 35.63 | 229.30 ± 3.01 | 7.95 ± 0.06 | 131.88 ± 1.67 | 4.57 ± 0.02 | |
| TT | 14.85 ± 0.23 | 2865.60 ± 48.61 | 228.47 ± 4.09 | 8.03 ± 0.08 | 129.69 ± 2.28 | 4.56 ± 0.02 | |
| p-value | 0.77 | 0.81 | 0.97 | 0.39 | 0.68 | 0.86 | |
| g.18858G > A | AA | 14.89 ± 0.16 | 2861.68 ± 35.11 | 225.47 ± 2.93 a | 7.89 ± 0.06 | 129.75 ± 1.65 | 4.57 ± 0.02 |
| AG | 15.20 ± 0.20 | 2929.74 ± 44.07 | 235.45 ± 3.67 b | 8.01 ± 0.07 | 133.35 ± 2.07 | 4.57 ± 0.02 | |
| GG | 15.23 ± 0.48 | 2924.20 ± 105.49 | 233.81 ± 8.73 ab | 8.00 ± 0.18 | 133.14 ± 4.95 | 4.58 ± 0.05 | |
| p-value | 0.36 | 0.37 | 0.04 | 0.35 | 0.28 | 0.98 | |
| g.22146C > A | AA | 15.03 ± 0.14 | 2894.36 ± 30.88 | 228.22 ± 2.61 | 7.91 ± 0.05 | 131.15 ± 1.45 | 4.56 ± 0.02 |
| CA | 15.46 ± 0.84 | 3016.19 ± 182.20 | 234.68 ± 15.38 | 7.86 ± 0.31 | 137.92 ± 8.54 | 4.59 ± 0.09 | |
| CC | 15.12 ± 0.80 | 2924.92 ± 171.91 | 233.06 ± 14.33 | 7.93 ± 0.28 | 132.17 ± 8.09 | 4.53 ± 0.09 | |
| p-value | 0.87 | 0.79 | 0.87 | 0.98 | 0.73 | 0.90 | |
| g.22834C > T | CC | 14.70 ± 0.19 a | 2821.10 ± 40.20 a | 222.50 ± 3.36 a | 7.92 ± 0.07 | 128.07 ± 1.90 a | 4.56 ± 0.02 |
| CT | 15.23 ± 0.30 ab | 2939.96 ± 64.98 ab | 227.13 ± 5.41 ab | 7.75 ± 0.11 | 132.27 ± 3.08 ab | 4.53 ± 0.03 | |
| TT | 15.31 ± 0.22 b | 2951.77 ± 46.75 b | 235.56 ± 3.86 b | 8.01 ± 0.08 | 134.53 ± 2.22 b | 4.57 ± 0.02 | |
| p-value | 0.04 | 0.03 | 0.02 | 0.11 | 0.04 | 0.46 |
| Block | H1H1 | H1H3 | H2H3 | p-Value |
|---|---|---|---|---|
| Sequence | CA/CA | CA/TG | TA/TG | |
| Number | 187 | 67 | 53 | |
| Frequencies (%) | 56.50 | 20.24 | 16.01 | |
| PM (kg) | 15.22 ± 0.19 | 15.07 ± 0.28 | 14.76 ± 0.17 | 0.13 |
| MY (kg) | 2933.75 ± 41.55 | 2923.90 ± 60.25 | 2842.81 ± 37.15 | 0.15 |
| FY (kg) | 235.78 ± 3.44 a | 226.97 ± 5.00 ab | 225.02 ± 3.09 b | 0.03 |
| FP (%) | 8.03 ± 0.07 a | 7.73 ± 0.10 b | 7.94 ± 0.06 ab | 0.04 |
| PY (kg) | 133.33 ± 1.88 a | 131.63 ± 2.73 ab | 127.80 ± 1.69 b | 0.04 |
| PP (%) | 4.57 ± 0.02 | 4.53 ± 0.03 | 4.57 ± 0.02 | 0.44 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, C.; Hu, X.; Ahmad, M.J.; Niu, K.; Ye, T.; Liang, A.; Yang, L. Novel Insight into the Role of Squalene Epoxidase (SQLE) Gene in Determining Milk Production Traits in Buffalo. Int. J. Mol. Sci. 2023, 24, 2436. https://doi.org/10.3390/ijms24032436
Chen C, Hu X, Ahmad MJ, Niu K, Ye T, Liang A, Yang L. Novel Insight into the Role of Squalene Epoxidase (SQLE) Gene in Determining Milk Production Traits in Buffalo. International Journal of Molecular Sciences. 2023; 24(3):2436. https://doi.org/10.3390/ijms24032436
Chicago/Turabian StyleChen, Chao, Xiangwei Hu, Muhammad Jamil Ahmad, Kaifeng Niu, Tingzhu Ye, Aixin Liang, and Liguo Yang. 2023. "Novel Insight into the Role of Squalene Epoxidase (SQLE) Gene in Determining Milk Production Traits in Buffalo" International Journal of Molecular Sciences 24, no. 3: 2436. https://doi.org/10.3390/ijms24032436






