Circulating miR-122-5p, miR-92a-3p, and miR-18a-5p as Potential Biomarkers in Human Liver Transplantation Follow-Up
Abstract
:1. Introduction
2. Results
2.1. Discovery Phase and miR Profiling
2.2. Validation Phase of Selected miRs by RT-qPCR
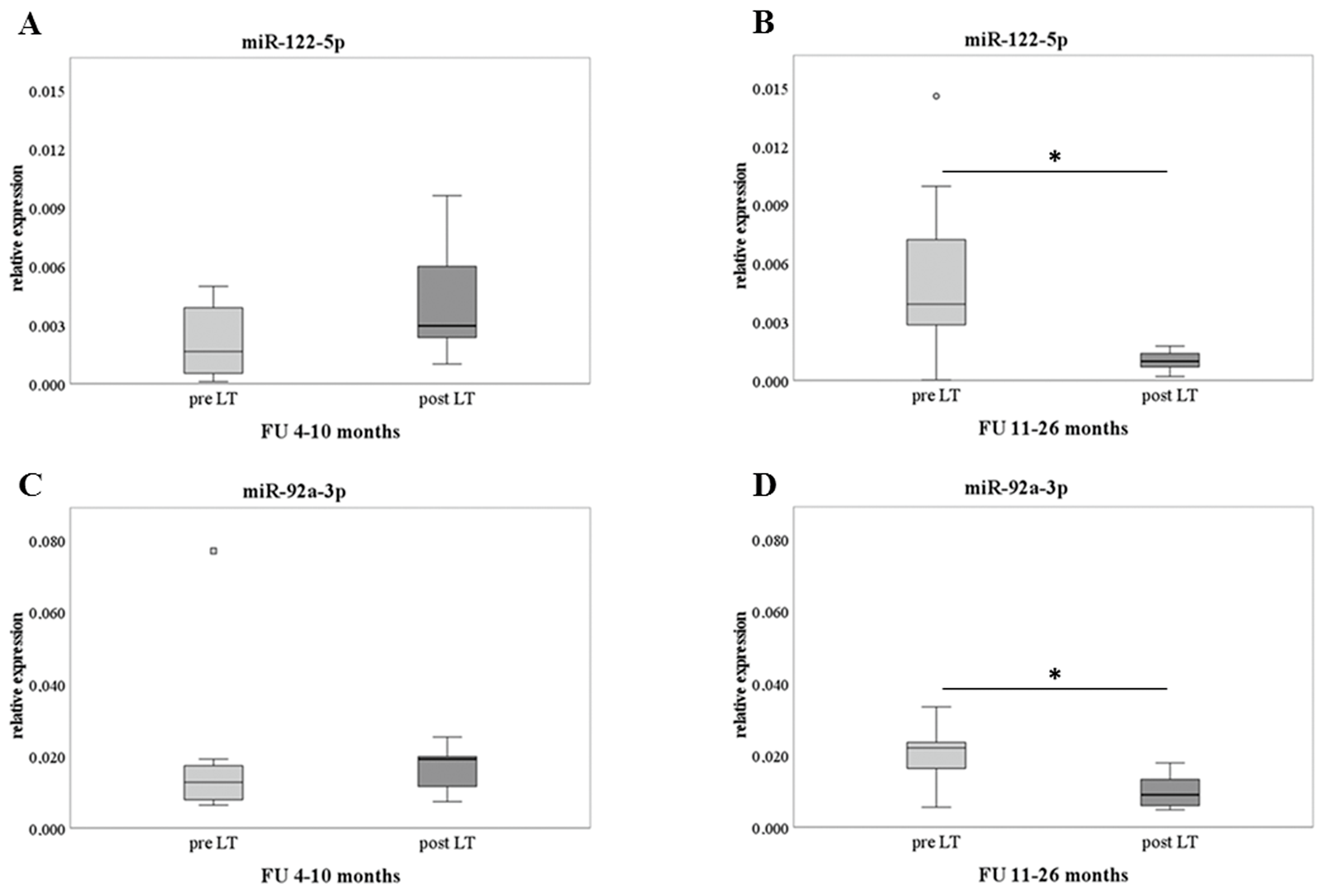
2.3. Pre-Post LT and Analysis of Delta Difference
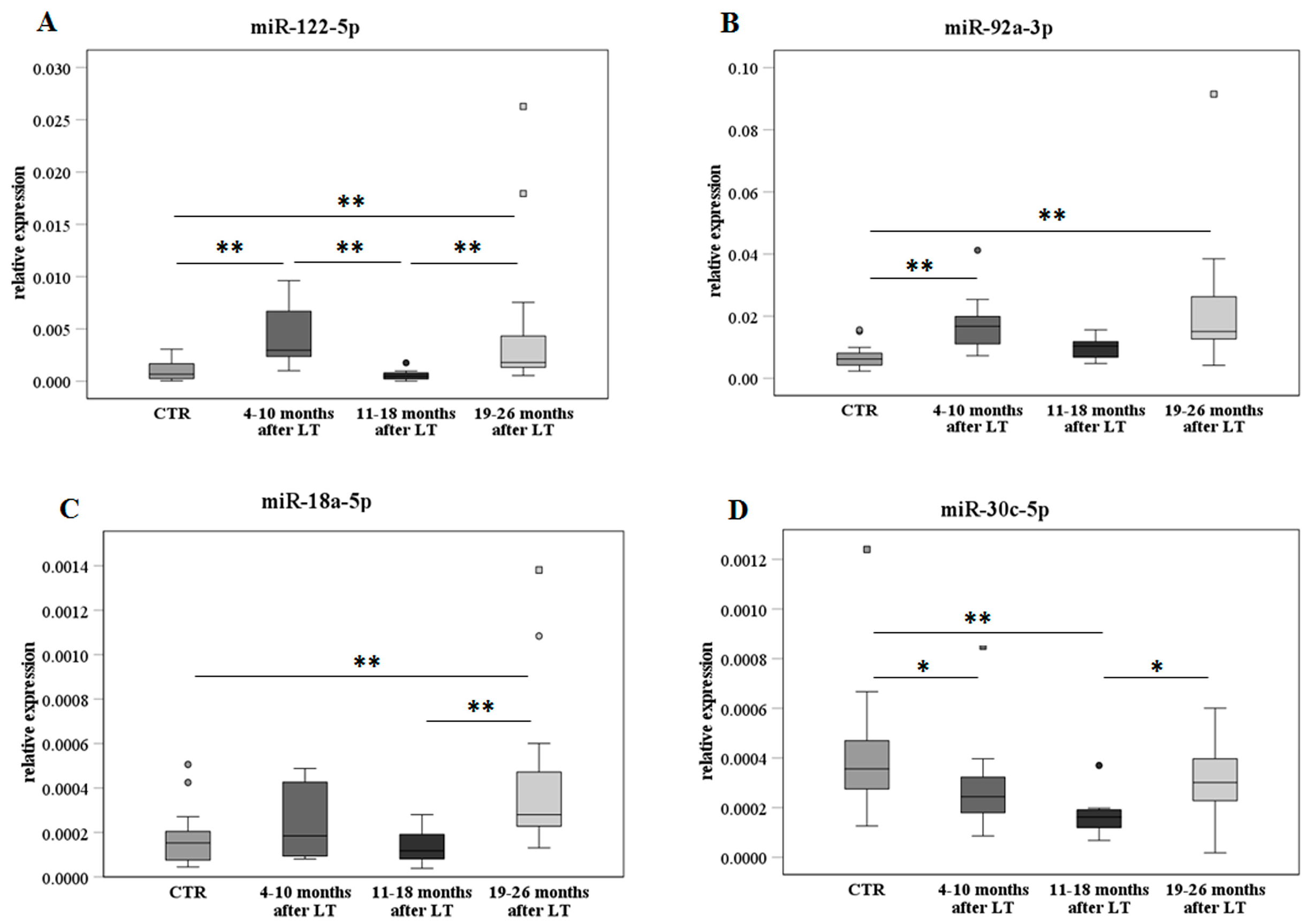
2.4. MiR Analysis after Transplant on the Extended Sample Cohort
2.5. Liver Function Tests (LFTs) along FU Times and Correlation with miR Expression
2.6. Levels of the Selected miRs with or without LT Complications
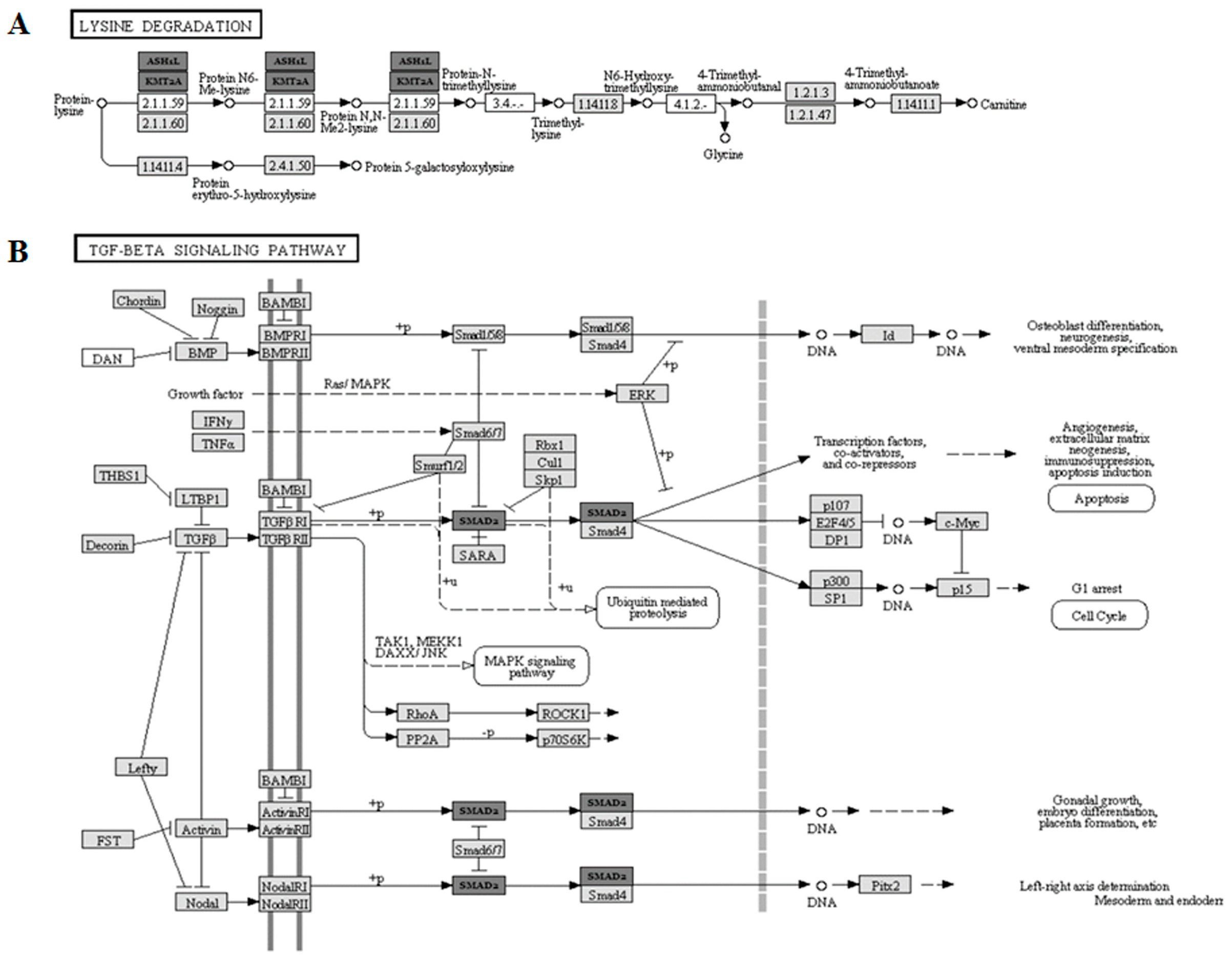
2.7. Bioinformatic Pathway Analysis
2.8. MiRs of Liver Donor–Recipient Age Mismatches
3. Discussion
4. Materials and Methods
4.1. Experimental Design and Study Participants
4.2. RNA Extraction
4.3. MicroRNA Profiling
4.4. Validation in RT-qPCR
4.5. Statistical Analysis
4.6. Bioinformatic Pathway Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Liver transplantation. J. Hepatol. 2016, 64, 433–485. [Google Scholar] [CrossRef]
- Capri, M.; Franceschi, C.; Cescon, M. Biological age of transplanted livers. Aging 2018, 10, 156. [Google Scholar] [CrossRef]
- Craig, E.V.; Heller, M.T. Complications of liver transplant. Abdom. Radiol. 2021, 46, 43–67. [Google Scholar] [CrossRef]
- Watt, K.D. Keys to long-term care of the liver transplant recipient. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 639–648. [Google Scholar] [CrossRef]
- Lim, T.S.; Kim, J.K. Is liver biopsy still useful in the era of non-invasive tests? Clin. Mol. Hepatol. 2020, 26, 302–304. [Google Scholar] [CrossRef]
- Germani, G.; Rodriguez-Castro, K.; Russo, F.P.; Senzolo, M.; Zanetto, A.; Ferrarese, A.; Burra, P. Markers of acute rejection and graft acceptance in liver transplantation. World J. Gastroenterol. WJG 2015, 21, 1061–1068. [Google Scholar] [CrossRef]
- Roedder, S.; Vitalone, M.; Khatri, P.; Sarwal, M.M. Biomarkers in solid organ transplantation: Establishing personalized transplantation medicine. Genome Med. 2011, 3, 37. [Google Scholar] [CrossRef]
- Gowda, S.; Desai, P.B.; Hull, V.V.; Math, A.A.K.; Vernekar, S.N.; Kulkarni, S.S. A review on laboratory liver function tests. Pan Afr. Med. J. 2009, 3, 17. [Google Scholar]
- Badrick, T.; Turner, P. Review and Recommendations for the Component Tests in the Liver Function Test Profile. Indian J. Clin. Biochem. IJCB 2016, 31, 21–29. [Google Scholar] [CrossRef]
- Kim, H.; Kisseleva, T.; Brenner, D.A. Aging and liver disease. Curr. Opin. Gastroenterol. 2015, 31, 184–191. [Google Scholar] [CrossRef]
- Capri, M.; Olivieri, F.; Lanzarini, C.; Remondini, D.; Borelli, V.; Lazzarini, R.; Graciotti, L.; Albertini, M.C.; Bellavista, E.; Santoro, A.; et al. Identification of miR-31-5p, miR-141-3p, miR-200c-3p, and GLT1 as human liver aging markers sensitive to donor-recipient age-mismatch in transplants. Aging Cell 2017, 16, 262–272. [Google Scholar] [CrossRef]
- Lin, H.; Ewing, L.E.; Koturbash, I.; Gurley, B.J.; Miousse, I.R. MicroRNAs as biomarkers for liver injury: Current knowledge, challenges and future prospects. Food Chem. Toxicol. 2017, 110, 229–239. [Google Scholar] [CrossRef]
- Mohr, R.; Özdirik, B.; Lambrecht, J.; Demir, M.; Eschrich, J.; Geisler, L.; Hellberg, T.; Loosen, S.H.; Luedde, T.; Tacke, F.; et al. From Liver Cirrhosis to Cancer: The Role of Micro-RNAs in Hepatocarcinogenesis. Int. J. Mol. Sci. 2021, 22, 1492. [Google Scholar] [CrossRef]
- Zhu, S.-F.; Yuan, W.; Du, Y.-L.; Wang, B.-L. Research progress of lncRNA and miRNA in hepatic ischemia-reperfusion injury. Hepatobiliary Pancreat. Dis. Int. HBPD INT 2022, 22, 45–53. [Google Scholar] [CrossRef]
- Dongiovanni, P.; Meroni, M.; Longo, M.; Fargion, S.; Fracanzani, A.L. miRNA Signature in NAFLD: A Turning Point for a Non-Invasive Diagnosis. Int. J. Mol. Sci. 2018, 19, 3966. [Google Scholar] [CrossRef]
- Laterza, O.F.; Lim, L.; Garrett-Engele, P.W.; Vlasakova, K.; Muniappa, N.; Tanaka, W.K.; Johnson, J.M.; Sina, J.F.; Fare, T.L.; Sistare, F.D.; et al. Plasma MicroRNAs as sensitive and specific biomarkers of tissue injury. Clin. Chem. 2009, 55, 1977–1983. [Google Scholar] [CrossRef]
- Farid, W.R.R.; Pan, Q.; van der Meer, A.J.P.; de Ruiter, P.E.; Ramakrishnaiah, V.; de Jonge, J.; Kwekkeboom, J.; Janssen, H.L.A.; Metselaar, H.J.; Tilanus, H.W.; et al. Hepatocyte-derived microRNAs as serum biomarkers of hepatic injury and rejection after liver transplantation. Liver Transplant. Off. Publ. Am. Assoc. Study Liver Dis. Int. Liver Transplant. Soc. 2012, 18, 290–297. [Google Scholar] [CrossRef]
- Zhou, M.; Hara, H.; Dai, Y.; Mou, L.; Cooper, D.K.C.; Wu, C.; Cai, Z. Circulating Organ-Specific MicroRNAs Serve as Biomarkers in Organ-Specific Diseases: Implications for Organ Allo- and Xeno-Transplantation. Int. J. Mol. Sci. 2016, 17, 1232. [Google Scholar] [CrossRef]
- Loosen, S.H.; Lurje, G.; Wiltberger, G.; Vucur, M.; Koch, A.; Kather, J.N.; Paffenholz, P.; Tacke, F.; Ulmer, F.T.; Trautwein, C.; et al. Serum levels of miR-29, miR-122, miR-155 and miR-192 are elevated in patients with cholangiocarcinoma. PLoS ONE 2019, 14, e0210944. [Google Scholar] [CrossRef]
- Shigoka, M.; Tsuchida, A.; Matsudo, T.; Nagakawa, Y.; Saito, H.; Suzuki, Y.; Aoki, T.; Murakami, Y.; Toyoda, H.; Kumada, T.; et al. Deregulation of miR-92a expression is implicated in hepatocellular carcinoma development. Pathol. Int. 2010, 60, 351–357. [Google Scholar] [CrossRef]
- Mendell, J.T. miRiad Roles for the miR-17-92 Cluster in Development and Disease. Cell 2008, 133, 217–222. [Google Scholar] [CrossRef] [Green Version]
- Mogilyansky, E.; Rigoutsos, I. The miR-17/92 cluster: A comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013, 20, 1603–1614. [Google Scholar] [CrossRef]
- Morsiani, C.; Terlecki-Zaniewicz, L.; Skalicky, S.; Bacalini, M.G.; Collura, S.; Conte, M.; Sevini, F.; Garagnani, P.; Salvioli, S.; Hackl, M.; et al. Circulating miR-19a-3p and miR-19b-3p characterize the human aging process and their isomiRs associate with healthy status at extreme ages. Aging Cell 2021, 20, e13409. [Google Scholar] [CrossRef]
- Bobbili, M.R.; Mader, R.M.; Grillari, J.; Dellago, H. OncomiR-17-5p: Alarm signal in cancer? Oncotarget 2017, 8, 71206–71222. [Google Scholar] [CrossRef]
- Tan, W.; Li, Y.; Lim, S.-G.; Tan, T.M. miR-106b-25/miR-17-92 clusters: Polycistrons with oncogenic roles in hepatocellular carcinoma. World J. Gastroenterol. 2014, 20, 5962–5972. [Google Scholar] [CrossRef]
- Zhu, H.; Han, C.; Wu, T. MiR-17-92 cluster promotes hepatocarcinogenesis. Carcinogenesis 2015, 36, 1213–1222. [Google Scholar] [CrossRef]
- Zhu, H.; Han, C.; Lu, D.; Wu, T. miR-17-92 Cluster Promotes Cholangiocarcinoma Growth. Am. J. Pathol. 2014, 184, 2828–2839. [Google Scholar] [CrossRef]
- Oda, S.; Yokoi, T. Recent progress in the use of microRNAs as biomarkers for drug-induced toxicities in contrast to traditional biomarkers: A comparative review. Drug Metab. Pharmacokinet. 2021, 37, 100372. [Google Scholar] [CrossRef]
- Oksuz, Z.; Serin, M.S.; Kaplan, E.; Dogen, A.; Tezcan, S.; Aslan, G.; Emekdas, G.; Sezgin, O.; Altintas, E.; Tiftik, E.N. Serum microRNAs; miR-30c-5p, miR-223-3p, miR-302c-3p and miR-17-5p could be used as novel non-invasive biomarkers for HCV-positive cirrhosis and hepatocellular carcinoma. Mol. Biol. Rep. 2015, 42, 713–720. [Google Scholar] [CrossRef]
- Fan, J.; Li, H.; Nie, X.; Yin, Z.; Zhao, Y.; Chen, C.; Wen Wang, D. MiR-30c-5p ameliorates hepatic steatosis in leptin receptor-deficient (db/db) mice via down-regulating FASN. Oncotarget 2017, 8, 13450–13463. [Google Scholar] [CrossRef]
- Hu, S.; Zhang, J.; Fang, X.; Guo, G.; Dai, J.; Sheng, Z.; Li, D.; Chen, J.; Zhang, L.; Liu, C.; et al. Identification of microRNA hsa-miR-30c-5p as an inhibitory factor in the progression of hepatocellular carcinoma and investigation of its regulatory network via comprehensive analysis. Bioengineered 2021, 12, 7165–7177. [Google Scholar] [CrossRef]
- Wong, V.C.-L.; Wong, M.-I.; Lam, C.-T.; Lung, M.L.; Lam, K.-O.; Lee, V.H.-F. Hallmark microRNA signature in liquid biopsy identifies hepatocellular carcinoma and differentiates it from liver metastasis. J. Cancer 2021, 12, 4585–4594. [Google Scholar] [CrossRef]
- Akuta, N.; Kawamura, Y.; Suzuki, F.; Saitoh, S.; Arase, Y.; Kunimoto, H.; Sorin, Y.; Fujiyama, S.; Sezaki, H.; Hosaka, T.; et al. Impact of circulating miR-122 for histological features and hepatocellular carcinoma of nonalcoholic fatty liver disease in Japan. Hepatol. Int. 2016, 10, 647–656. [Google Scholar] [CrossRef]
- Arataki, K.; Hayes, C.N.; Akamatsu, S.; Akiyama, R.; Abe, H.; Tsuge, M.; Miki, D.; Ochi, H.; Hiraga, N.; Imamura, M.; et al. Circulating microRNA-22 correlates with microRNA-122 and represents viral replication and liver injury in patients with chronic hepatitis B. J. Med. Virol. 2013, 85, 789–798. [Google Scholar] [CrossRef]
- Moayedi, J.; Hashempour, T.; Musavi, Z.; Arefian, E.; Naderi, M.; Heidari, M.R.; Dehghani, B.; Hasanshahi, Z.; Merat, S. Evaluation of miR-122 Serum Level and IFN-λ3 Genotypes in Patients with Chronic HCV and HCV-Infected Liver Transplant Candidate. Microrna 2021, 10, 58–65. [Google Scholar] [CrossRef]
- Zheng, Y.; Tang, L.; Chen, G.; Liu, Z. Comprehensive Bioinformatics Analysis of Key Methyltransferases and Demethylases for Histone Lysines in Hepatocellular Carcinoma. Technol. Cancer Res. Treat. 2020, 19, 1533033820983284. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Katoh, Y.; Matsumoto, M.; Sato, M.; Ebina, M.; Itoh-Nakadai, A.; Funayama, R.; Nakayama, K.; Unno, M.; Igarashi, K. Regulatory signatures of liver regeneration distilled by integrative analysis of mRNA, histone methylation, and proteomics. J. Biol. Chem. 2017, 292, 8019–8037. [Google Scholar] [CrossRef]
- Bacalini, M.G.; Franceschi, C.; Gentilini, D.; Ravaioli, F.; Zhou, X.; Remondini, D.; Pirazzini, C.; Giuliani, C.; Marasco, E.; Gensous, N.; et al. Molecular Aging of Human Liver: An Epigenetic/Transcriptomic Signature. J. Gerontol. Ser. A 2018. [Google Scholar] [CrossRef]
- Morsiani, C.; Bacalini, M.G.; Santoro, A.; Garagnani, P.; Collura, S.; D’Errico, A.; de Eguileor, M.; Grazi, G.L.; Cescon, M.; Franceschi, C.; et al. The peculiar aging of human liver: A geroscience perspective within transplant context. Ageing Res. Rev. 2019, 51, 24–34. [Google Scholar] [CrossRef]
- Fabregat, I.; Moreno-Càceres, J.; Sánchez, A.; Dooley, S.; Dewidar, B.; Giannelli, G.; ten Dijke, P.; IT-LIVER Consortium. TGF-β signalling and liver disease. FEBS J. 2016, 283, 2219–2232. [Google Scholar] [CrossRef]
- Liu, M.; Chen, P. Proliferation-inhibiting pathways in liver regeneration (Review). Mol. Med. Rep. 2017, 16, 23–35. [Google Scholar] [CrossRef] [Green Version]
- Regateiro, F.S.; Howie, D.; Cobbold, S.P.; Waldmann, H. TGF-β in transplantation tolerance. Curr. Opin. Immunol. 2011, 23, 660–669. [Google Scholar] [CrossRef]



| Sample Code | Donor Age | Recipient Age | Donor Sex | Recipient Sex | Recipient Disease | FU (Months) |
|---|---|---|---|---|---|---|
| 78 | 71 | 40 | M | M | HCV-cirrhosis | 8 |
| 84 | 49 | 49 | M | M | HCC with HBV-cirrhosis | 7 |
| 88 | 12 | 45 | M | M | HCC with HCV-cirrhosis | 4 |
| 51 | 70 | 26 | M | M | HBV and HDV-cirrhosis | 7 |
| 54 | 58 | 46 | F | M | HBV and HDV-cirrhosis | 12 |
| 55 | 68 | 50 | M | M | HCV-cirrhosis | 5 |
| 56 | 74 | 50 | F | F | Alcoholic cirrhosis | 13 |
| 81 | 89 | 55 | M | M | HCC with HBV and HDV-cirrhosis | 18 |
| 86 | 53 | 69 | M | M | HCC with HCV-cirrhosis | 6 |
| 91 | 87 | 34 | M | M | Budd–Chiari syndrome | 26 |
| 96 | 83 | 63 | M | M | HCC with HCV-cirrhosis | 23 |
| 99 | 73 | 48 | M | F | HCC with HCV-cirrhosis | 7 |
| 114 | 83 | 46 | F | M | HBV and HDV-cirrhosis | 20 |
| 121 | 12 | 54 | F | M | Primary biliary cirrhosis | 14 |
| Sample Code | Donor Age | Recipient Age | Donor Sex | Recipient Sex | Recipient Disease | FU (Months) |
|---|---|---|---|---|---|---|
| 1 | 59 | 62 | M | M | HCC with HCV-cirrhosis | 20 |
| 3 | 69 | 51 | F | M | Cryptogenic cirrhosis | 8 |
| 6 | 43 | 57 | M | M | Alcoholic cirrhosis | 23 |
| 9 | 76 | 42 | F | F | Alcoholic cirrhosis | 19 |
| 10 | 37 | 52 | M | M | HCV-cirrhosis | 19 |
| 17 | 23 | 46 | M | M | HCC with HCV and alcoholic cirrhosis | 22 |
| 19 | 87 | 60 | M | M | Alcoholic cirrhosis with HCC | 20 |
| 22A | 29 | 36 | M | M | HCC with HBV and HDV-cirrhosis | 17 |
| 22B | 29 | 56 | M | M | Polycystic liver and kidney disease | 17 |
| 24 | 50 | 66 | M | M | HCC with HCV-cirrhosis | 18 |
| 39 | 20 | 49 | M | M | Amyloidosis | 8 |
| 40 | 74 | 66 | M | F | HCC with HCV-cirrhosis | 11 |
| 41 | 69 | 64 | F | M | HCC with HCV-cirrhosis | 22 |
| 43 | 74 | 56 | F | M | Alcoholic cirrhosis | 22 |
| 45 | 45 | 45 | M | M | HCV-cirrhosis | 26 |
| 46 | 75 | 57 | M | M | Alcoholic cirrhosis with HCC | 10 |
| 48 | 78 | 25 | F | M | Primary sclerosing cholangitis | 8 |
| 104 | 88 | 54 | F | F | HBV and HDV-cirrhosis | 25 |
| 107 | 64 | 52 | M | F | Polycystic liver and kidney disease | 23 |
| Sperman’s Rho | miR-122-5p | miR-92a-3p | miR-18a-5p | miR-30c-5p | |
|---|---|---|---|---|---|
| TBIL | Correlation coefficient | 0.52 | −0.10 | 0.26 | −0.19 |
| Sig. (2-tailed) | 0.10 | 0.77 | 0.44 | 0.58 | |
| N | 11 | 11 | 11 | 11 | |
| IBIL | Correlation coefficient | 0.49 | 0.10 | 0.44 | −0.14 |
| Sig. (2-tailed) | 0.13 | 0.76 | 0.17 | 0.67 | |
| N | 11 | 11 | 11 | 11 | |
| AST | Correlation coefficient | 0.71 | −0.44 | −0.19 | −0.54 |
| Sig. (2-tailed) | 0.01 | 0.17 | 0.58 | 0.09 | |
| N | 11 | 11 | 11 | 11 | |
| ALT | Correlation coefficient | 0.50 | −0.20 | −0.12 | −0.72 |
| Sig. (2-tailed) | 0.12 | 0.55 | 0.73 | 0.01 | |
| N | 11 | 11 | 11 | 11 | |
| GGT | Correlation coefficient | 0.43 | −0.35 | −0.35 | −0.71 |
| Sig. (2-tailed) | 0.22 | 0.33 | 0.32 | 0.02 | |
| N | 10 | 10 | 10 | 10 | |
| ALP | Correlation coefficient | 0.49 | 0.21 | −0.20 | −0.33 |
| Sig. (2-tailed) | 0.15 | 0.56 | 0.59 | 0.35 | |
| N | 10 | 10 | 10 | 10 | |
| ALB | Correlation coefficient | −0.35 | −0.91 | 0.55 | −0.07 |
| Sig. (2-tailed) | 0.45 | 0.00 | 0.20 | 0.88 | |
| N | 7 | 7 | 7 | 7 | |
| Sperman’s Rho | miR-122-5p | miR-92a-3p | miR-18a-5p | miR-30c-5p | |
|---|---|---|---|---|---|
| TBIL | Correlation coefficient | 0.15 | −0.20 | −0.12 | −0.73 |
| Sig. (2-tailed) | 0.62 | 0.51 | 0.69 | 0.00 | |
| N | 13 | 13 | 13 | 13 | |
| IBIL | Correlation coefficient | 0.07 | −0.32 | 0.01 | −0.53 |
| Sig. (2-tailed) | 0.82 | 0.29 | 0.96 | 0.06 | |
| N | 13 | 13 | 13 | 13 | |
| AST | Correlation coefficient | 0.45 | 0.09 | −0.33 | 0.15 |
| Sig. (2-tailed) | 0.13 | 0.77 | 0.27 | 0.63 | |
| N | 13 | 13 | 13 | 13 | |
| ALT | Correlation coefficient | 0.53 | −0.03 | −0.11 | 0.34 |
| Sig. (2-tailed) | 0.06 | 0.92 | 0.72 | 0.26 | |
| N | 13 | 13 | 13 | 13 | |
| GGT | Correlation coefficient | 0.57 | −0.40 | −0.04 | 0.10 |
| Sig. (2-tailed) | 0.04 | 0.18 | 0.90 | 0.75 | |
| N | 13 | 13 | 13 | 13 | |
| ALP | Correlation coefficient | 0.65 | −0.24 | −0.05 | −0.37 |
| Sig. (2-tailed) | 0.02 | 0.42 | 0.88 | 0.22 | |
| N | 13 | 13 | 13 | 13 | |
| ALB | Correlation coefficient | 0.29 | −0.23 | 0.46 | −0.44 |
| Sig. (2-tailed) | 0.58 | 0.66 | 0.36 | 0.38 | |
| N | 6 | 6 | 6 | 6 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morsiani, C.; Collura, S.; Sevini, F.; Ciurca, E.; Bertuzzo, V.R.; Franceschi, C.; Grazi, G.L.; Cescon, M.; Capri, M. Circulating miR-122-5p, miR-92a-3p, and miR-18a-5p as Potential Biomarkers in Human Liver Transplantation Follow-Up. Int. J. Mol. Sci. 2023, 24, 3457. https://doi.org/10.3390/ijms24043457
Morsiani C, Collura S, Sevini F, Ciurca E, Bertuzzo VR, Franceschi C, Grazi GL, Cescon M, Capri M. Circulating miR-122-5p, miR-92a-3p, and miR-18a-5p as Potential Biomarkers in Human Liver Transplantation Follow-Up. International Journal of Molecular Sciences. 2023; 24(4):3457. https://doi.org/10.3390/ijms24043457
Chicago/Turabian StyleMorsiani, Cristina, Salvatore Collura, Federica Sevini, Erika Ciurca, Valentina Rosa Bertuzzo, Claudio Franceschi, Gian Luca Grazi, Matteo Cescon, and Miriam Capri. 2023. "Circulating miR-122-5p, miR-92a-3p, and miR-18a-5p as Potential Biomarkers in Human Liver Transplantation Follow-Up" International Journal of Molecular Sciences 24, no. 4: 3457. https://doi.org/10.3390/ijms24043457





