Epigenetic Regulation of Driver Genes in Testicular Tumorigenesis
Abstract
:1. Introduction: Epigenetics, Differentiation, and Testicular Germ Cell Tumors
2. Testicular Germ Cell Tumors Type II
2.1. Pathology
2.2. Tumorigenesis
2.3. Cytogenetics
2.4. Genetics
3. Epigenetics and TGCT
3.1. Tumorigenesis
3.2. DNA Methylations
3.3. Histone Modifications and Modulations
3.4. Driver Genes
3.5. Epidrugs and TGCT
4. iPSC in Oncology
4.1. TGCT
4.2. Other Malignancies
5. Perspectives and Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AFP | alfa fetoprotein |
| 5-aza | 5-azacytidine |
| 5-aza-cD | 5-aza-2′-deoxycytidine |
| BEP | bleomycin, etoposide, and (cis)platin |
| bromodomaini | bromodomain inhibitor |
| CC | choriocarcinoma |
| CDK | cyclin D kinase |
| CDKi | CDK inhibitor |
| CYT | spermatocyte |
| DNMT | DNA methyltransferase |
| DATECA | the Danish testis cancer study group |
| DNMT | DNA methyl transferase |
| DNMTi | DNA methyltransferase inhibitor |
| EC | embryonal carcinoma |
| EGFR | epidermal growth factor receptor |
| ESC | embryonic stem cells |
| EZH2 | enhancer of Zeste-2 |
| EZH2i | EZH2 inhibitor |
| GCNIS | germ cell neoplasia in situ |
| GON | gonocytes |
| HAT | histone acetylase |
| hCG | human chorionic gonadotropin |
| HDAC | histone deacetylase |
| HDACi | histone deacetylase inhibitor |
| HDMT | histone demethylase |
| HMT | histone methyltransferase |
| iPSC | induced pluri- to totipotent stem cells |
| KDM | lysine demethylase |
| KDMi | lysine demethylase inhibitor |
| Kme | methylated histone 3 lysine |
| LDH | Lactate dehydrogenase |
| LSD1i | LSD1 inhibitor |
| 5mC | 5-methyl cytosine |
| 5mhC | 5-methylhydroxy cytosine |
| MGCT | microinvasive germ cell tumor |
| NGC | normal germ cells |
| NSCLC | non-small cell lung cancer |
| NST | non-seminomatous germ cell tumor |
| OCT4 | the protein of POU5F1 |
| OSKM | the OCT4, SOX2, KLF4, and MYC panel |
| OSLN | the OCT4, SOX2, LIN28, and NANOG panel |
| PGC | primordial germ cells |
| PRC2 | polycomb repressor complex 2 |
| SCLC | small cell lung cancer |
| SCEC | small cell esophageal cancer |
| SE | seminoma |
| SPA | spermatogonia type A |
| SPB | spermatogonia type B |
| TER | teratoma |
| TET | DNA demethylase |
| TGCA | The Genomic Cancer Atlas |
| TKI | tyrosine kinase inhibitor |
| TNM | international tumor, nodes, and metastases classification |
| TSS | transcription start site. |
| WHO | World Health Organization |
| YST | yolk sac tumor |
References
- Waddington, C.H. Evolutionary adaptation. Perspect. Biol. Med. 1959, 2, 379–401. [Google Scholar] [CrossRef]
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007, 318, 1917–1920. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [Green Version]
- Gutierrez-Aranda, I.; Ramos-Mejia, V.; Bueno, C.; Munoz-Lopez, M.; Real, P.J.; Macia, A.; Sanchez, L.; Ligero, G.; Garcia-Parez, J.L.; Menendez, P. Human induced pluripotent stem cells develop teratoma more efficiently and faster than human embryonic stem cells regardless the site of injection. Stem Cells 2010, 28, 1568–1570. [Google Scholar] [CrossRef] [Green Version]
- Chu, Y.-H.; Hang, W.; Hu, R. Exclusively intertubular seminoma arising in undescendedn testes: Report of two cases. Hum. Pathol. Case Rep. 2018, 11, 15–18. [Google Scholar] [CrossRef]
- Von Eyben, F.E.; Parraga-Alava, J. Meta-analysis of gene expressions in testicular germ cell tumor histologies. Int. J. Mol. Sci. 2020, 21, 4487. [Google Scholar] [CrossRef]
- Jostes, S.V.; Fellermeyer, M.; Arevalo, L.; Merges, G.E.; Kristiansen, G.; Nettersheim, D.; Schorle, H. Unique and redundant roles of SOX2 and SOX17 in regulating the germ cell tumor fate. Int. J. Cancer 2020, 146, 1592–1605. [Google Scholar] [CrossRef] [Green Version]
- Nettersheim, D.; Heimsoeth, A.; Jostes, S.; Schneider, S.; Fellermeyer, M.; Hofmann, A.; Schorle, H. SOX2 is essential for in vivo reprogramming of seminoma-like TCam-2 cells to an embryonal carcinoma-like fate. Oncotarget 2016, 7, 47095–47110. [Google Scholar] [CrossRef] [Green Version]
- Nonaka, D. Differential expression of SOX2 and SOX17 in testicular germ cell tumors. Am. J. Clin. Pathol. 2009, 131, 731–736. [Google Scholar] [CrossRef]
- Eldar-Geva, T.; Gross-Tsur, V.; Hirsch, H.J.; Altarescu, G.; Segal, R.; Zeligson, S.; Golomb, E.; Epsztejn-Litman, S.; Eiges, R. Incomplete methylation of a germ cell tumor (Seminoma) in a Prader-Willi male. Mol. Genet. Genomic Med. 2018, 6, 811–818. [Google Scholar] [CrossRef]
- Schulz, W.A.; Hoffmann, M.J. Transcription factor networks in embryonic stem cells and testicular cancer and the definition of epigenetics. Epigenetics 2007, 2, 37–42. [Google Scholar] [CrossRef] [Green Version]
- Nogales, F.F.; Prat, J.; Schuldt, M.; Cruz-Viruel, N.; Kaur, B.; D’Angelo, E.; Matias-Guiu, X.; Vidal, A.; McCluggage, W.G.; Oosterhuis, J.W. Germ cell tumour growth patterns originating from clear cell carcinomas of the ovary and endometrium: A comparative immunohistochemical study favouring their origin from somatic stem cells. Histopathology 2018, 72, 634–647. [Google Scholar] [CrossRef]
- Oosterhuis, J.W.; Looijenga, L.H.J. Human germ cell tumours from a developmental perspective. Nat. Rev. Cancer 2019, 19, 522–537. [Google Scholar] [CrossRef]
- Wilms, M. Die teratoiden Geschwülste des Hodens, mit einschluss der sogenante Cystoide und Encondrome. Beitr. Pathol. Anat. 1896, 19, 233–366. [Google Scholar]
- Atkin, N.B.; Baker, M.C. Specific chromosome change, i(12p), in testicular tumours? Lancet 1982, 2, 1349. [Google Scholar] [CrossRef]
- Castedo, S.M.; de Jong, B.; Oosterhuis, J.W.; Seruca, R.; te Meerman, G.J.; Dam, A.; Schraffordt Koops, H. Cytogenetic analysis of ten human seminomas. Cancer Res. 1989, 49, 439–443. [Google Scholar]
- Castedo, S.M.; de Jong, B.; Oosterhuis, J.W.; Seruca, R.; Idenburg, V.J.; Dam, A.; te Meerman, G.; Koops, H.S.; Sleijfer, D.T. Chromosomal changes in human primary testicular nonseminomatous germ cell tumors. Cancer Res. 1989, 49, 5696–5701. [Google Scholar]
- Castedo, S.M.; de Jong, B.; Oosterhuis, J.W.; Idenburg, V.J.; Seruca, R.; Buist, J.; te Meerman, G.J.; Schraffordt Koops, H.; Sleijfer, D.T. Chromosomal changes in mature residual teratomas following polychemotherapy. Cancer Res. 1989, 49, 672–676. [Google Scholar]
- Fichtner, A.; Richter, A.; Filmar, S.; Gaisa, N.T.; Schweyer, S.; Reis, H.; Nettersheim, D.; Oing, C.; Gayer, F.A.; Leha, A.; et al. The detection of isochromosome i(12p) in malignant germ cell tumours and tumours with somatic malignant transformation by the use of quantitative real-time polymerase chain reaction. Histopathology 2021, 78, 593–606. [Google Scholar] [CrossRef]
- von Eyben, F.E. Chromosomes, genes, and development of testicular germ cell tumors. Cancer Genet. Cytogenet. 2004, 151, 93–138. [Google Scholar] [CrossRef]
- Zondag, H.A. Enzyme activity in dysgerminoma and seminoma. A study of lactic dehydrogenase isoenzymes in malignant diseases. R. I. Med. J. 1964, 47, 273–281. [Google Scholar]
- von Eyben, F.E.; Blaabjerg, O.; Petersen, P.H.; Horder, M.; Nielsen, H.V.; Andersen, S.K.; Parlev, E. Lactate dehydrogenase isoenzyme 1 in testis cancer. Lancet 1987, 2, 1035–1036. [Google Scholar] [CrossRef]
- von Eyben, F.E. A systematic review of lactate dehydrogenase isoenzyme 1 and germ cell tumors. Clin. Biochem. 2001, 34, 441–454. [Google Scholar] [CrossRef]
- von Eyben, F.E.; Parraga-Alava, J.; Tu, S.M. Testicular germ cell tumors type 2 have high RNA expression of LDHB, the gene for lactate dehydrogenase subunit B. Asian J. Androl. 2021, 23, 357–362. [Google Scholar] [CrossRef]
- Wittekind, C. TNM classification of testicular tumors. Definitions and prerequisites for correct application. Pathologe 2014, 35, 252–255. [Google Scholar] [CrossRef]
- International Germ Cell Consensus Classification Group. International Germ Cell Consensus Classification: A prognostic factor-based staging system for metastatic germ cell cancers. J. Clin. Oncol. 1997, 15, 594–603. [Google Scholar] [CrossRef]
- Einhorn, L.H. Testicular cancer: An oncological success story. Clin. Cancer Res. 1997, 3, 2630–2632. [Google Scholar]
- Gurney, J.K.; Florio, A.A.; Znaor, A.; Ferlay, J.; Laversanne, M.; Sarfati, D.; Bray, F.; McGlynn, K.A. International trends in the incidence of testicular cancer: Lessons from 35 years and 41 countries. Eur. Urol. 2019, 76, 615–623. [Google Scholar] [CrossRef]
- Toner, G.C.; Geller, N.L.; Tan, C.; Nisselbaum, J.; Bosl, G.J. Serum tumor marker half-life during chemotherapy allows early prediction of complete response and survival in nonseminomatous germ cell tumors. Cancer Res. 1990, 50, 5904–5910. [Google Scholar]
- Clemmesen, J. Testis cancer incidence—Suggestion of a world pattern. Int. J. Androl. 1981, 4 (Suppl. S4), 111–120. [Google Scholar] [CrossRef]
- von Eyben, F.E.; Jensen, M.B.; Hoyer, S. Frequency and markers of precursor lesions and implications for the pathogenesis of testicular germ cell tumors. Clin. Genitourin. Cancer 2017, 18, E211–E221. [Google Scholar] [CrossRef] [Green Version]
- Krag Jacobsen, G.; Barlebo, H.; Olsen, J.; Schultz, H.P.; Starklint, H.; Sogaard, H.; Vaeth, M. Testicular germ cell tumours in Denmark 1976–1980. Pathology of 1058 consecutive cases. Acta Radiol. Oncol. 1984, 23, 239–247. [Google Scholar] [CrossRef]
- Norgaard-Pedersen, B.; Schultz, H.; Arends, J.; Brincker, H.; Jacobsen, G.K.; Lindelov, B.; Rorth, M.; Svennekjaer, I.L. Biochemical markers for testicular germ-cell tumors in relation to histology and stage: Some experiences from the Danish Testicular Cancer (DATECA) study from 1976 through 1981. Ann. N. Y. Acad. Sci. 1983, 417, 390–399. [Google Scholar] [CrossRef]
- Trabert, B.; Sigurdson, A.J.; Sweeney, A.M.; Amato, R.J.; Strom, S.S.; McGlynn, K.A. Baldness, acne and testicular germ cell tumours. Int. J. Androl. 2011, 34, e59–e67. [Google Scholar] [CrossRef]
- Dixon, F.J.; Moore, R.A. Testicular tumors. A clinicopathological study. Cancer 1953, 6, 427–454. [Google Scholar] [CrossRef]
- Teilum, G. Endodermal sinus tumors of the ovary and testis. Comparative morphogenesis of the so-called mesoephroma ovarii (Schiller) and extraembryonic (yolk sac-allantoic) structures of the rat’s placenta. Cancer 1959, 12, 1092–1105. [Google Scholar] [CrossRef]
- de Jong, B.; Oosterhuis, J.W.; Castedo, S.M.; Vos, A.; te Meerman, G.J. Pathogenesis of adult testicular germ cell tumors. A cytogenetic model. Cancer Genet. Cytogenet. 1990, 48, 143–167. [Google Scholar] [CrossRef]
- Schulze, C.; Holstein, A.F. On the histology of human seminoma: Development of the solid tumor from intratubular seminoma cells. Cancer 1977, 39, 1090–1100. [Google Scholar] [CrossRef]
- von Eyben, F.E.; Jacobsen, G.K.; Skotheim, R.I. Microinvasive germ cell tumor of the testis. Virchows Arch. 2005, 447, 610–625. [Google Scholar] [CrossRef]
- Holstein, A.F.; Schutte, B.; Becker, H.; Hartman, M. Morphology of normal and malignant germ cells. Int. J. Androl. 1987, 10, 1–18. [Google Scholar] [CrossRef]
- Harrison, N.J.; Baker, D.; Andrews, P.W. Culture adaptation of embryonic stem cells echoes germ cell malignancy. Int. J. Androl. 2007, 30, 275–281, discussion 281. [Google Scholar] [CrossRef]
- Ben-David, U.; Benvenisty, N. The tumorigenicity of human embryonic and induced pluripotent stem cells. Nat. Rev. Cancer 2011, 11, 268–277. [Google Scholar] [CrossRef]
- Blum, B.; Benvenisty, N. The tumorigenicity of human embryonic stem cells. Adv. Cancer Res. 2008, 100, 133–158. [Google Scholar]
- Baker, D.E.; Harrison, N.J.; Maltby, E.; Smith, K.; Moore, H.D.; Shaw, P.J.; Heath, P.R.; Holden, H.; Andrews, P.W. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat. Biotechnol. 2007, 25, 207–215. [Google Scholar] [CrossRef]
- Kawakami, T.; Okamoto, K.; Sugihara, H.; Hattori, T.; Reeve, A.E.; Ogawa, O.; Okada, Y. The roles of supernumerical X chromosomes and XIST expression in testicular germ cell tumors. J. Urol. 2003, 169, 1546–1552. [Google Scholar] [CrossRef]
- Walt, H.; Emmerich, P.; Jauch, A.; DeLozier-Blanchet, C.D. Characterization of precancerous and neoplastic human testicular germ cells. Recent Results Cancer Res. 1991, 123, 37–44. [Google Scholar]
- Bates, S.; Ryan, K.M.; Phillips, A.C.; Vousden, K.H. Cell cycle arrest and DNA endoreduplication following p21Waf1/Cip1 expression. Oncogene 1998, 17, 1691–1703. [Google Scholar] [CrossRef]
- Baldini, E.; Arlot-Bonnemains, Y.; Mottolese, M.; Sentinelli, S.; Antoniani, B.; Sorrenti, S.; Salducci, M.; Comini, E.; Ulisse, S.; D’Armiento, M. Deregulation of Aurora kinase gene expression in human testicular germ cell tumours. Andrologia 2010, 42, 260–267. [Google Scholar] [CrossRef]
- Yan, X.; Cao, L.; Li, Q.; Wu, Y.; Zhang, H.; Saiyin, H.; Liu, X.; Zhang, X.; Shi, Q.; Yu, L. Aurora C is directly associated with Survivin and required for cytokinesis. Genes Cells 2005, 10, 617–626. [Google Scholar] [CrossRef]
- de Graaff, W.E.; Oosterhuis, J.W.; de Jong, B.; Dam, A.; van Putten, W.L.; Castedo, S.M.; Sleijfer, D.T.; Schraffordt Koops, H. Ploidy of testicular carcinoma in situ. Lab. Investig. 1992, 66, 166–168. [Google Scholar]
- Shen, H.; Shih, J.; Hollern, D.P.; Wang, L.; Bowlby, R.; Tickoo, S.K.; Thorsson, V.; Mungall, A.J.; Newton, Y.; Hegde, A.M.; et al. Integrated molecular characterization of testicular germ cell tumors. Cell. Rep. 2018, 23, 3392–3406. [Google Scholar] [CrossRef]
- Looijenga, L.H.; Rosenberg, C.; van Gurp, R.J.; Geelen, E.; van Echten-Arends, J.; de Jong, B.; Mostert, M.; Wolter Oosterhuis, J. Comparative genomic hybridization of microdissected samples from different stages in the development of a seminoma and a non-seminoma. J. Pathol. 2000, 191, 187–192. [Google Scholar] [CrossRef]
- Cutcutache, I.; Suzuki, Y.; Tan, I.B.; Ramgopal, S.; Zhang, S.; Ramnarayanan, K.; Gan, A.; Lee, H.H.; Tay, S.T.; Ooi, A.; et al. Exome-wide sequencing shows low mutation rates and identifies novel mutated genes in seminomas. Eur. Urol. 2015, 68, 77–83. [Google Scholar] [CrossRef] [Green Version]
- Litchfield, K.; Summersgill, B.; Yost, S.; Sultana, R.; Labreche, K.; Dudakia, D.; Renwick, A.; Seal, S.; Al-Saadi, R.; Broderick, P.; et al. Whole-exome sequencing reveals the mutational spectrum of testicular germ cell tumours. Nat. Commun. 2015, 6, 5973. [Google Scholar] [CrossRef] [Green Version]
- Alagaratnam, S.; Lind, G.E.; Kraggerud, S.M.; Lothe, R.A.; Skotheim, R.I. The testicular germ cell tumour transcriptome. Int. J. Androl. 2011, 34, e133–e151. [Google Scholar] [CrossRef]
- Okamoto, K. Epigenetics: A way to understand the origin and biology of testicular germ cell tumors. Int. J. Urol. 2012, 19, 504–511. [Google Scholar] [CrossRef]
- Ushida, H.; Kawakami, T.; Minami, K.; Chano, T.; Okabe, H.; Okada, Y.; Okamoto, K. Methylation profile of DNA repetitive elements in human testicular germ cell tumor. Mol. Carcinog. 2012, 51, 711–722. [Google Scholar] [CrossRef]
- Smiraglia, D.J.; Szymanska, J.; Kraggerud, S.M.; Lothe, R.A.; Peltomaki, P.; Plass, C. Distinct epigenetic phenotypes in seminomatous and nonseminomatous testicular germ cell tumors. Oncogene 2002, 21, 3909–3916. [Google Scholar] [CrossRef] [Green Version]
- Peltomaki, P. DNA methylation changes in human testicular cancer. Biochim. Biophys. Acta 1991, 1096, 187–196. [Google Scholar] [CrossRef]
- Sicinski, P.; Donaher, J.L.; Geng, Y.; Parker, S.B.; Gardner, H.; Park, M.Y.; Robker, R.L.; Richards, J.S.; McGinnis, L.K.; Biggers, J.D.; et al. Cyclin D2 is an FSH-responsive gene involved in gonadal cell proliferation and oncogenesis. Nature 1996, 384, 470–474. [Google Scholar] [CrossRef]
- Godmann, M.; Gashaw, I.; Eildermann, K.; Schweyer, S.; Bergmann, M.; Skotheim, R.I.; Behr, R. The pluripotency transcription factor Kruppel-like factor 4 is strongly expressed in intratubular germ cell neoplasia unclassified and seminoma. Mol. Hum. Reprod. 2009, 15, 479–488. [Google Scholar] [CrossRef] [Green Version]
- Sikora, K.; Evan, G.; Stewart, J.; Watson, J.V. Detection of the c-myc oncogene product in testicular cancer. Br. J. Cancer 1985, 52, 171–176. [Google Scholar] [CrossRef] [Green Version]
- Strohmeyer, T.; Reissmann, P.; Cordon-Cardo, C.; Hartmann, M.; Ackermann, R.; Slamon, D. Correlation between retinoblastoma gene expression and differentiation in human testicular tumors. Proc. Natl. Acad. Sci. USA 1991, 88, 6662–6666. [Google Scholar] [CrossRef] [Green Version]
- Boublikova, L.; Buchler, T.; Stary, J.; Abrahamova, J.; Trka, J. Molecular biology of testicular germ cell tumors: Unique features awaiting clinical application. Crit. Rev. Oncol. Hematol. 2014, 89, 366–385. [Google Scholar] [CrossRef]
- Goldberg, E.; Eddy, G.; Stewart, J.; Watson, J.V. LDHC: The ultimate testis-specific gene. J. Androl. 2010, 31, 86–94. [Google Scholar] [CrossRef]
- Skude, G.; von Eyben, F.E.; Kristiansen, P. Additional lactate dehydrogenase (LDH) isoenzymes in normal testis and spermatozoa of adult man. Mol. Gen. Genet. 1984, 198, 172–174. [Google Scholar] [CrossRef]
- Saeed, B.A.; Barband, R.S.; Alnasiri, U.S. Lactate dehydrogenase C4 (LDH-C4) is essential for the sperm count and motility: A case-control study. Baghdad J. Biochem. Appl. Biol. Sci. 2021, 2, 146–159. [Google Scholar] [CrossRef]
- Nettersheim, D.; Arndt, I.; Sharma, R.; Riesenberg, S.; Jostes, S.; Schneider, S.; Holzel, M.; Kristiansen, G.; Schorle, H. The cancer/testis-antigen PRAME supports the pluripotency network and represses somatic and germ cell differentiation programs in seminomas. Br. J. Cancer 2016, 115, 454–464. [Google Scholar] [CrossRef] [Green Version]
- Ricci, C.; Franceschini, T.; Giunchi, F.; Grillini, M.; Ambrosi, F.; Massari, F.; Mollica, V.; Colecchia, M.; Fiorentino, M. Immunohistochemical expression of preferentially expressed antigen in melanoma (PRAME) in the uninvolved background testis, germ cell neoplasia in situ, and germ cell tumors of the testis. Am. J. Clin. Pathol. 2022, 157, 644–648. [Google Scholar] [CrossRef]
- Orsatti, A.; Sirolli, M.; Ambrosi, F.; Franceschini, T.; Giunchi, F.; Franchini, E.; Grillini, M.; Massari, F.; Mollica, V.; Bianchi, F.M.; et al. SOX2 and PRAME in the “reprogramming” of seminoma cells. Pathol. Res. Pract. 2022, 237, 154044. [Google Scholar] [CrossRef]
- Cheung, H.H.; Yang, Y.; Lee, T.L.; Rennert, O.; Chan, W.Y. Hypermethylation of genes in testicular embryonal carcinomas. Br. J. Cancer 2016, 114, 230–236. [Google Scholar] [CrossRef] [Green Version]
- Juric, D.; Sale, S.; Hromas, R.A.; Yu, R.; Wang, Y.; Duran, G.E.; Tibshirani, R.; Einhorn, L.H.; Sikic, B.I. Gene expression profiling differentiates germ cell tumors from other cancers and defines subtype-specific signatures. Proc. Natl. Acad. Sci. USA 2005, 102, 17763–17768. [Google Scholar] [CrossRef] [Green Version]
- Sperger, J.M.; Chen, X.; Draper, J.S.; Antosiewicz, J.E.; Chon, C.H.; Jones, S.B.; Brooks, J.D.; Andrews, P.W.; Brown, P.O.; Thomson, J.A. Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc. Natl. Acad. Sci. USA 2003, 100, 13350–13355. [Google Scholar] [CrossRef] [Green Version]
- Gilbert, D.; Rapley, E.; Shipley, J. Testicular germ cell tumours: Predisposition genes and the male germ cell niche. Nat. Rev. Cancer 2011, 11, 278–288. [Google Scholar] [CrossRef]
- Rosenberg, C.; Van Gurp, R.J.; Geelen, E.; Oosterhuis, J.W.; Looijenga, L.H. Overrepresentation of the short arm of chromosome 12 is related to invasive growth of human testicular seminomas and nonseminomas. Oncogene 2000, 19, 5858–5862. [Google Scholar] [CrossRef] [Green Version]
- Ottesen, A.M.; Skakkebaek, N.E.; Lundsteen, C.; Leffers, H.; Larsen, J.; Rajpert-De Meyts, E. High-resolution comparative genomic hybridization detects extra chromosome arm 12p material in most cases of carcinoma in situ adjacent to overt germ cell tumors, but not before the invasive tumor development. Genes Chromosomes Cancer 2003, 38, 117–125. [Google Scholar] [CrossRef]
- Datta, M.W.; Macri, E.; Signoretti, S.; Renshaw, A.A.; Loda, M. Transition from in situ to invasive testicular germ cell neoplasia is associated with the loss of p21 and gain of mdm-2 expression. Mod. Pathol. 2001, 14, 437–442. [Google Scholar] [CrossRef] [Green Version]
- Gashaw, I.; Dushaj, O.; Behr, R.; Biermann, K.; Brehm, R.; Rubben, H.; Grobholz, R.; Schmid, K.W.; Bergmann, M.; Winterhager, E. Novel germ cell markers characterize testicular seminoma and fetal testis. Mol. Hum. Reprod. 2007, 13, 721–727. [Google Scholar] [CrossRef]
- Skotheim, R.I.; Lind, G.E.; Monni, O.; Nesland, J.M.; Abeler, V.M.; Fossa, S.D.; Duale, N.; Brunborg, G.; Kallioniemi, O.; Andrews, P.W.; et al. Differentiation of human embryonal carcinomas in vitro and in vivo reveals expression profiles relevant to normal development. Cancer Res. 2005, 65, 5588–5598. [Google Scholar] [CrossRef] [Green Version]
- Korkola, J.E.; Houldsworth, J.; Chadalavada, R.S.; Olshen, A.B.; Dobrzynski, D.; Reuter, V.E.; Bosl, G.J.; Chaganti, R.S. Down-regulation of stem cell genes, including those in a 200-kb gene cluster at 12p13.31, is associated with in vivo differentiation of human male germ cell tumors. Cancer Res. 2006, 66, 820–827. [Google Scholar] [CrossRef] [Green Version]
- Nettersheim, D.; Gillis, A.J.; Looijenga, L.H.; Schorle, H. TGF-beta1, EGF and FGF4 synergistically induce differentiation of the seminoma cell line TCam-2 into a cell type resembling mixed non-seminoma. Int. J. Androl. 2011, 34, e189–e203. [Google Scholar] [CrossRef]
- Benesova, M.; Trejbalova, K.; Kucerova, D.; Vernerova, Z.; Hron, T.; Szabo, A.; Amouroux, R.; Klezl, P.; Hajkova, P.; Hejnar, J. Overexpression of TET dioxygenases in seminomas associates with low levels of DNA methylation and hydroxymethylation. Mol. Carcinog. 2017, 56, 1837–1850. [Google Scholar] [CrossRef] [Green Version]
- Spiller, C.M.; Gillis, A.J.; Burnet, G.; Stoop, H.; Koopman, P.; Bowles, J.; Looijenga, L.H. Cripto: Expression, epigenetic regulation and potential diagnostic use in testicular germ cell tumors. Mol. Oncol. 2016, 10, 526–537. [Google Scholar] [CrossRef] [Green Version]
- Norgaard-Pedersen, B.; Albrechtsen, R.; Teilum, G. Serum alpha-foetoprotein as a marker for endodermal sinus tumour (yolk sac tumour) or a vitelline component of “teratocarcinoma”. Acta Pathol. Microbiol. Scand. A 1975, 83, 573–589. [Google Scholar] [CrossRef]
- Lempiainen, A.; Sankila, A.; Hotakainen, K.; Haglund, C.; Blomqvist, C.; Stenman, U.H. Expression of human chorionic gonadotropin in testicular germ cell tumors. Urol. Oncol. 2014, 32, 727–734. [Google Scholar] [CrossRef]
- Jacobsen, C.; Honecker, F. Cisplatin resistance in germ cell tumours: Models and mechanisms. Andrology 2015, 3, 111–121. [Google Scholar] [CrossRef]
- Christophersen, N.S.; Helin, K. Epigenetic control of embryonic stem cell fate. J. Exp. Med. 2010, 207, 2287–2295. [Google Scholar] [CrossRef] [Green Version]
- Nettersheim, D.; Heukamp, L.C.; Fronhoffs, F.; Grewe, M.J.; Haas, N.; Waha, A.; Honecker, F.; Waha, A.; Kristiansen, G.; Schorle, H. Analysis of TET expression/activity and 5mC oxidation during normal and malignant germ cell development. PLoS ONE 2013, 8, e82881. [Google Scholar] [CrossRef]
- Liu, J.; Shi, H.; Li, X.; Chen, G.; Larsson, C.; Lui, W.O. miR2233p regulates cell growth and apoptosis via FBXW7 suggesting an oncogenic role in human testicular germ cell tumors. Int. J. Oncol. 2017, 50, 356–364. [Google Scholar] [CrossRef] [Green Version]
- Kristensen, D.G.; Nielsen, J.E.; Jørgensen, A.; Skakkebæk, N.E.; Rajpert-De Meyts, E.; Almstrup, K. Evidence that active demethylation mechanisms maintain the genome of carcinoma in situ cells hypomethylated in the adult testis. Br. J. Cancer 2014, 110, 668–678. [Google Scholar] [CrossRef]
- Kristensen, D.G.; Skakkebaek, N.E.; Rajpert-DeMeyts, E.; Almstrup, K. Epigenetic features of testicular germ cell tumours in relation to epigenetic characteristics of foetal germ cells. Int. J. Dev. Biol. 2013, 57, 309–317. [Google Scholar] [CrossRef] [Green Version]
- Aksoy, I.; Jauch, R.; Chen, J.; Dyla, M.; Divakar, U.; Bogu, G.K.; Teo, R.; Leng Ng, C.K.; Herath, W.; Lili, S.; et al. Oct4 switches partnering from Sox2 to Sox17 to reinterpret the enhancer code and specify endoderm. EMBO J. 2013, 32, 938–953. [Google Scholar] [CrossRef] [Green Version]
- Kushwaha, R.; Jagadish, N.; Kustagi, M.; Mendiratta, G.; Seandel, M.; Soni, R.; Korkola, J.E.; Thodima, V.; Califano, A.; Bosl, G.J.; et al. Mechanism and role of SOX2 repression in seminoma: Relevance to human germline specification. Stem Cell Rep. 2016, 6, 772–783. [Google Scholar] [CrossRef] [Green Version]
- Fritzsche, F.R.; Hasler, A.; Bode, P.K.; Adams, H.; Seifert, H.H.; Sulser, T.; Moch, H.; Barghorn, A.; Kristiansen, G. Expression of histone deacetylases 1, 2 and 3 in histological subtypes of testicular germ cell tumours. Histol. Histopathol. 2011, 26, 1555–1561. [Google Scholar]
- de Jong, J.; Stoop, H.; Gillis, A.J.; Hersmus, R.; van Gurp, R.J.; van de Geijn, G.J.; van Drunen, E.; Beverloo, H.B.; Schneider, D.T.; Sherlock, J.K.; et al. Further characterization of the first seminoma cell line TCam-2. Genes Chromosomes Cancer 2008, 47, 185–196. [Google Scholar] [CrossRef]
- van der Zwan, Y.G.; Rijlaarsdam, M.A.; Rossello, F.J.; Notini, A.J.; de Boer, S.; Watkins, D.N.; Gillis, A.J.; Dorssers, L.C.; White, S.J.; Looijenga, L.H. Seminoma and embryonal carcinoma footprints identified by analysis of integrated genome-wide epigenetic and expression profiles of germ cell cancer cell lines. PLoS ONE 2014, 9, e98330. [Google Scholar] [CrossRef] [Green Version]
- Nettersheim, D.; Jostes, S.; Sharma, R.; Schneider, S.; Hofmann, A.; Ferreira, H.J.; Hoffmann, P.; Kristiansen, G.; Esteller, M.B.; Schorle, H. BMP inhibition in seminomas initiates acquisition of pluripotency via NODAL signaling resulting in reprogramming to an embryonal carcinoma. PLoS Genet. 2015, 11, e1005415. [Google Scholar] [CrossRef] [Green Version]
- Lobo, J.; Guimaraes, R.; Miranda-Goncalves, V.; Monteiro-Reis, S.; Cantante, M.; Antunes, L.; Braga, I.; Mauricio, J.; Looijenga, L.H.; Jeronimo, C.; et al. Differential expression of DNA methyltransferases and demethylases among the various testicular germ cell tumor subtypes. Epigenomics 2020, 12, 1579–1592. [Google Scholar] [CrossRef]
- Villodre, E.S.; Felipe, K.B.; Oyama, M.Z.; Oliveira, F.H.; Lopez, P.; Solari, C.; Sevlever, G.; Guberman, A.; Lenz, G. Silencing of the transcription factors Oct4, Sox2, Klf4, c-Myc or Nanog has different effect on teratoma growth. Biochem. Biophys. Res. Commun. 2019, 517, 324–329. [Google Scholar] [CrossRef]
- Eckert, D.; Biermann, K.; Nettersheim, D.; Gillis, A.J.; Steger, K.; Jack, H.M.; Muller, A.M.; Looijenga, L.H.; Schorle, H. Expression of BLIMP1/PRMT5 and concurrent histone H2A/H4 arginine 3 dimethylation in fetal germ cells, CIS/IGCNU and germ cell tumors. BMC Dev. Biol. 2008, 8, 106. [Google Scholar] [CrossRef] [Green Version]
- Miranda-Goncalves, V.; Lobo, J.; Guimaraes-Teixeira, C.; Barros-Silva, D.; Guimaraes, R.; Cantante, M.; Braga, I.; Mauricio, J.; Oing, C.; Honecker, F.; et al. The component of the m(6)A writer complex VIRMA is implicated in aggressive tumor phenotype, DNA damage response and cisplatin resistance in germ cell tumors. J. Exp. Clin. Cancer Res. 2021, 40, 268. [Google Scholar] [CrossRef]
- Wermann, H.; Stoop, H.; Gillis, A.J.; Honecker, F.; van Gurp, R.J.; Ammerpohl, O.; Richter, J.; Oosterhuis, J.W.; Bokemeyer, C.; Looijenga, L.H. Global DNA methylation in fetal human germ cells and germ cell tumours: Association with differentiation and cisplatin resistance. J. Pathol. 2010, 221, 433–442. [Google Scholar] [CrossRef]
- Fazal, Z.; Singh, R.; Fang, F.; Bikorimana, E.; Baldwin, H.; Corbet, A.; Tomlin, M.; Yerby, C.; Adra, N.; Albany, C.; et al. Hypermethylation and global remodelling of DNA methylation is associated with acquired cisplatin resistance in testicular germ cell tumours. Epigenetics 2021, 16, 1071–1084. [Google Scholar] [CrossRef]
- Buljubasic, R.; Buljubasic, M.; Bojanac, A.K.; Ulamec, M.; Vlahovic, M.; Jezek, D.; Bulic-Jakus, F.; Sincic, N. Epigenetics and testicular germ cell tumors. Gene 2018, 661, 22–33. [Google Scholar] [CrossRef]
- De Jong, J.; Weeda, S.; Gillis, A.J.; Oosterhuis, J.W.; Looijenga, L.H. Differential methylation of the OCT3/4 upstream region in primary human testicular germ cell tumors. Oncol. Rep. 2007, 18, 127–132. [Google Scholar] [CrossRef]
- Mallik, S.; Qin, G.; Jia, P.; Zhao, Z. Molecular signatures identified by integrating gene expression and methylation in non-seminoma and seminoma of testicular germ cell tumours. Epigenetics 2021, 16, 162–176. [Google Scholar] [CrossRef]
- Brait, M.; Maldonado, L.; Begum, S.; Loyo, M.; Wehle, D.; Tavora, F.F.; Looijenga, L.H.; Kowalski, J.; Zhang, Z.; Rosenbaum, E.; et al. DNA methylation profiles delineate epigenetic heterogeneity in seminoma and non-seminoma. Br. J. Cancer 2012, 106, 414–423. [Google Scholar] [CrossRef] [Green Version]
- Chu, W.K.; Hung, L.M.; Hou, C.W.; Chen, J.K. MicroRNA 630 represses NANOG expression through transcriptional and post-transcriptional regulation in human embryonal carcinoma Cells. Int. J. Mol. Sci. 2021, 23, 46. [Google Scholar] [CrossRef]
- Agger, K.; Santoni-Rugiu, E.; Holmberg, C.; Karlstrom, O.; Helin, K. Conditional E2F1 activation in transgenic mice causes testicular atrophy and dysplasia mimicking human CIS. Oncogene 2005, 24, 780–789. [Google Scholar] [CrossRef] [Green Version]
- Lambrot, R.; Kimmins, S. Histone methylation is a critical regulator of the abnormal expression of POU5F1 and RASSF1A in testis cancer cell lines. Int. J. Androl. 2011, 34, 110–123. [Google Scholar] [CrossRef]
- Wang, J.; Lu, F.; Ren, Q.; Sun, H.; Xu, Z.; Lan, R.; Liu, Y.; Ward, D.; Quan, J.; Ye, T.; et al. Novel histone demethylase LSD1 inhibitors selectively target cancer cells with pluripotent stem cell properties. Cancer Res. 2011, 71, 7238–7249. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.; Yang, Y.; Li, W.; Chen, Q.; Li, J.; Pan, X.; Zhou, L.; Liu, C.; Chen, C.; He, J.; et al. Reciprocal regulation of Akt and Oct4 promotes the self-renewal and survival of embryonal carcinoma cells. Mol. Cell. 2012, 48, 627–640. [Google Scholar] [CrossRef] [Green Version]
- Hinz, S.; Magheli, A.; Weikert, S.; Schulze, W.; Krause, H.; Schrader, M.; Miller, K.; Kempkensteffen, C. Deregulation of EZH2 expression in human spermatogenic disorders and testicular germ cell tumors. World J. Urol. 2010, 28, 631–635. [Google Scholar] [CrossRef]
- Barrand, S.; Andersen, I.S.; Collas, P. Promoter-exon relationship of H3 lysine 9, 27, 36 and 79 methylation on pluripotency-associated genes. Biochem. Biophys. Res. Commun. 2010, 401, 611–617. [Google Scholar] [CrossRef]
- Singh, R.; Fazal, Z.; Bikorimana, E.; Boyd, R.I.; Yerby, C.; Tomlin, M.; Baldwin, H.; Shokry, D.; Corbet, A.K.; Shahid, K.; et al. Reciprocal epigenetic remodeling controls testicular cancer hypersensitivity to hypomethylating agents and chemotherapy. Mol. Oncol. 2022, 16, 683–698. [Google Scholar] [CrossRef]
- Gillis, A.J.; Stoop, H.; Biermann, K.; van Gurp, R.J.; Swartzman, E.; Cribbes, S.; Ferlinz, A.; Shannon, M.; Oosterhuis, J.W.; Looijenga, L.H. Expression and interdependencies of pluripotency factors LIN28, OCT3/4, NANOG and SOX2 in human testicular germ cells and tumours of the testis. Int. J. Androl. 2011, 34, e160–e174. [Google Scholar] [CrossRef]
- de Jong, J.; Looijenga, L.H. Stem cell marker OCT3/4 in tumor biology and germ cell tumor diagnostics: History and future. Crit. Rev. Oncog. 2006, 12, 171–203. [Google Scholar] [CrossRef]
- Gopalan, A.; Dhall, D.; Olgac, S.; Fine, S.W.; Korkola, J.E.; Houldsworth, J.; Chaganti, R.S.; Bosl, G.J.; Reuter, V.E.; Tickoo, S.K. Testicular mixed germ cell tumors: A morphological and immunohistochemical study using stem cell markers, OCT3/4, SOX2 and GDF3, with emphasis on morphologically difficult-to-classify areas. Mod. Pathol. 2009, 22, 1066–1074. [Google Scholar] [CrossRef] [Green Version]
- Wongtrakoongate, P. Epigenetic therapy of cancer stem and progenitor cells by targeting DNA methylation machineries. World J. Stem Cells 2015, 7, 137–148. [Google Scholar] [CrossRef]
- Boer, B.; Kopp, J.; Mallanna, S.; Desler, M.; Chakravarthy, H.; Wilder, P.J.; Bernadt, C.; Rizzino, A. Elevating the levels of Sox2 in embryonal carcinoma cells and embryonic stem cells inhibits the expression of Sox2:Oct-3/4 target genes. Nucleic Acids Res. 2007, 35, 1773–1786. [Google Scholar] [CrossRef]
- Hoang, N.; Zhang, X.; Zhang, C.; Vo, V.; Leng, F.; Saxena, L.; Yin, F.; Lu, F.; Zheng, G.; Bhowmik, P.; et al. New histone demethylase LSD1 inhibitor selectively targets teratocarcinoma and embryonic carcinoma cells. Bioorg. Med. Chem. 2018, 26, 1523–1537. [Google Scholar] [CrossRef]
- Li, W.; Fan, R.; Sun, M.; Jiang, T.; Gong, Y. Identification of Oct4-activating compounds that enhance reprogramming efficiency. Proc. Natl. Acad. Sci. USA 2012, 109, 20853–20858. [Google Scholar] [CrossRef] [Green Version]
- Eini, R.; Stoop, H.; Gillis, A.J.; Biermann, K.; Dorssers, L.C.; Looijenga, L.H. Role of SOX2 in the etiology of embryonal carcinoma, based on analysis of the NCCIT and NT2 cell lines. PLoS ONE 2014, 9, e83585. [Google Scholar] [CrossRef] [Green Version]
- Greber, B.; Lehrech, H.; Adjaye, I. Silencing of core transcription factors in human EC cells highlights the importance of autocrine FGF signaling for self-renewal. BMC Dev. Biol. 2007, 7, 46. [Google Scholar] [CrossRef] [Green Version]
- Fuchs, H.; Theuser, M.; Wruck, W.; Adjaye, J. miR-27 negatively regulates pluripotency-associated genes in human embryonal carcinoma cells. PLoS ONE 2014, 9, e111637. [Google Scholar] [CrossRef]
- Rijlaarsdam, M.A.; Looijenga, L.H. An oncofetal and developmental perspective on testicular germ cell cancer. Semin. Cancer Biol. 2014, 29, 59–74. [Google Scholar] [CrossRef]
- Muller, M.R.; Skowron, M.A.; Albers, P.; Nettersheim, D. Molecular and epigenetic pathogenesis of germ cell tumors. Asian J. Urol. 2021, 8, 144–154. [Google Scholar] [CrossRef]
- Van Der Zwan, Y.G.; Stoop, H.; Rossello, F.; White, S.J.; Looijenga, L.H. Role of epigenetics in the etiology of germ cell cancer. Int. J. Dev. Biol. 2013, 57, 299–308. [Google Scholar] [CrossRef] [Green Version]
- Hopman, A.N.H.; Moshi, J.M.; Hoogduin, K.J.; Ummelen, M.; Henfling, M.E.R.; van Engeland, M.; Wouters, K.A.D.; Stoop, D.S.; Looijenga, L.H.J.; Ramaekers, F.C.S. SOX17 expression and its downregulation by promoter methylation in cervical adenocarcinoma in situ and adenocarcinoma. Histopathology 2020, 76, 383–393. [Google Scholar] [CrossRef] [Green Version]
- Tan, D.S.; Holzner, M.; Weng, M.; Srivastava, Y.; Jauch, R. Sox17 in cellular reprogramming and cancer. Sem. Cancer Biol. 2020, 67, 383–393. [Google Scholar] [CrossRef]
- Jostes, S.; Nettersheim, D.; Schneider, S.; Schorle, H. Cultivation of testicular germ cell cancer cell lines and establishment of gene-edited subclones using CRISPR/Cas9. Methods Mol. Biol. 2021, 2195, 85–97. [Google Scholar]
- West, J.A.; Viswanathan, S.R.; Yabuuchi, A.; Cunniff, K.; Takeuchi, A.; Park, I.H.; Sero, J.E.; Zhu, H.; Perez-Atayde, A.; Frazier, A.L.; et al. A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature 2009, 460, 909–913. [Google Scholar] [CrossRef] [Green Version]
- Echigoya, K.; Koyama, M.; Negishi, L.; Takizawa, Y.; Mizukami, Y.; Shimabayashi, H.; Kuroda, A.; Kurumizaka, H. Nucleosome binding by the pioneer transcription factor OCT4. Sci. Rep. 2020, 10, 11832. [Google Scholar] [CrossRef]
- Murray, M.J.; Saini, H.K.; Siegler, C.A.; Hanning, J.E.; Barker, E.M.; van Dongen, S.; Ward, D.M.; Raby, K.L.; Groves, I.J.; Scarpini, C.G.; et al. LIN28 Expression in malignant germ cell tumors downregulates let-7 and increases oncogene levels. Cancer Res. 2013, 73, 4872–4884. [Google Scholar] [CrossRef] [Green Version]
- Nettersheim, D.; Biermann, K.; Gillis, A.J.; Steger, K.; Looijenga, L.H.; Schorle, H. NANOG promoter methylation and expression correlation during normal and malignant human germ cell development. Epigenetics 2011, 6, 114–122. [Google Scholar] [CrossRef] [Green Version]
- Rodda, D.J.; Chew, J.L.; Lim, L.H.; Loh, Y.H.; Wang, B.; Ng, H.H.; Robson, P. Transcriptional regulation of nanog by OCT4 and SOX2. J. Biol. Chem. 2005, 280, 24731–24737. [Google Scholar] [CrossRef] [Green Version]
- Kuroda, T.; Tada, M.; Kubota, H.; Kimura, H.; Hatano, S.Y.; Suemori, H.; Nakatsuji, N.; Tada, T. Octamer and sox elements are required for transcriptional cis regulation of Nanog gene expression. Mol. Cell. Biol. 2005, 25, 2475–2485. [Google Scholar] [CrossRef] [Green Version]
- Hart, A.H.; Hartley, L.; Parker, K.; Ibrahim, M.; Looijenga, L.H.; Pauchnik, M.; Chow, C.W.; Robb, L. The pluripotency homeobox gene NANOG is expressed in human germ cell tumors. Cancer 2005, 104, 2092–2098. [Google Scholar] [CrossRef]
- Do, H.J.; Lee, W.Y.; Lim, H.Y.; Oh, J.H.; Kim, D.K.; Kim, J.H.; Kim, T.; Kim, J.H. Two potent transactivation domains in the C-terminal region of human NANOG mediate transcriptional activation in human embryonic carcinoma cells. J. Cell. Biochem. 2009, 106, 1079–1089. [Google Scholar] [CrossRef]
- Quagliana, J.M.; O’Bryan, R.M.; Baker, L.; Gottlieb, J.; Morrison, F.S.; Eyre, H.J.; Tucker, W.G.; Costanzi, J. Phase II study of 5-azacytidine in solid tumors. Cancer Treat. Rep. 1977, 61, 51–54. [Google Scholar]
- Roth, B.J.; Elson, P.; Sledge, G.W., Jr.; Einhorn, L.H.; Trump, D.L. 5-Azacytidine (NSC 102816) in refractory germ cell tumors. A phase II trial of the Eastern Cooperative Oncology Group. Investig. New Drugs 1993, 11, 201–202. [Google Scholar] [CrossRef]
- Minucci, S.; Horn, V.; Bhattacharyya, N.; Russanova, V.; Ogryzko, V.V.; Gabriele, L.; Howard, B.H.; Ozato, K. A histone deacetylase inhibitor potentiates retinoid receptor action in embryonal carcinoma cells. Proc. Natl. Acad. Sci. USA 1997, 94, 11295–11300. [Google Scholar] [CrossRef] [Green Version]
- Albany, C.; Fazal, Z.; Singh, R.; Bikorimana, E.; Adra, N.; Hanna, N.H.; Einhorn, L.H.; Perkins, S.M.; Sandusky, G.E.; Christensen, B.C.; et al. A phase 1 study of combined guadecitabine and cisplatin in platinum refractory germ cell cancer. Cancer Med. 2021, 10, 156–163. [Google Scholar] [CrossRef]
- Oing, C.; Verem, I.; Mansour, W.Y.; Bokemeyer, C.; Dyshlovoy, S.; Honecker, F. 5-Azacitidine exerts prolonged pro-apoptotic effects and overcomes cisplatin-resistance in non-seminomatous germ cell tumor cells. Int. J. Mol. Sci. 2018, 20, 21. [Google Scholar] [CrossRef] [Green Version]
- Clavel, M.; Monfardini, S.; Fossa, S.; Smyth, J.; Renard, J.; Kaye, S.B. 5-Aza-2’-deoxycytidine (NSC 127716) in non-seminomatous testicular cancer. Phase II from the EORTC Early Clinical Trials Cooperative Group and Genito-Urinary Group. Ann. Oncol. 1992, 3, 399–400. [Google Scholar] [CrossRef]
- Biswal, B.K.; Beyrouthy, M.J.; Hever-Jardine, M.P.; Armstrong, D.; Tomlinson, C.R.; Christensen, B.C.; Marsit, C.J.; Spinella, M.J. Acute hypersensitivity of pluripotent testicular cancer-derived embryonal carcinoma to low-dose 5-aza deoxycytidine is associated with global DNA damage-associated p53 activation, anti-pluripotency and DNA demethylation. PLoS ONE 2012, 7, e53003. [Google Scholar] [CrossRef]
- Beyrouthy, M.J.; Garner, K.M.; Hever, M.P.; Freemantle, S.J.; Eastman, A.; Dmitrovsky, E.; Spinella, M.J. High DNA methyltransferase 3B expression mediates 5-aza-deoxycytidine hypersensitivity in testicular germ cell tumors. Cancer Res. 2009, 69, 9360–9366. [Google Scholar] [CrossRef] [Green Version]
- Juttermann, R.; Li, E.; Jaenisch, R. Toxicity of 5-aza-2’-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc. Natl. Acad. Sci. USA 1994, 91, 11797–11801. [Google Scholar] [CrossRef] [Green Version]
- Lind, G.E.; Skotheim, R.I.; Fraga, M.F.; Abeler, V.M.; Esteller, M.; Lothe, R.A. Novel epigenetically deregulated genes in testicular cancer include homeobox genes and SCGB3A1 (HIN-1). J. Pathol. 2006, 210, 441–449. [Google Scholar] [CrossRef]
- Lee, M.G.; Wynder, C.; Schmidt, D.M.; McCafferty, D.G.; Shiekhattar, R. Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications. Chem. Biol. 2006, 13, 563–567. [Google Scholar] [CrossRef] [Green Version]
- Steinemann, G.; Dittmer, A.; Schmidt, J.; Josuttis, D.; Fähling, M.; Biersack, B.; Beindorff, N.; Jolante Koziolek, E.; Schobert, R.; Brenner, W.; et al. Antitumor and antiangiogenic activity of the novel chimeric inhibitor animacroxam in testicular germ cell cancer. Mol. Oncol. 2019, 13, 2679–2696. [Google Scholar] [CrossRef] [Green Version]
- Lobo, J.; Guimaraes-Teixeira, C.; Barros-Silva, D.; Miranda-Goncalves, V.; Camilo, V.; Guimaraes, R.; Cantante, M.; Braga, I.; Mauricio, J.; Oing, C.; et al. Efficacy of HDAC inhibitors belinostat and panobinostat against cisplatin-sensitive and cisplatin-resistant testicular germ cell tumors. Cancers 2020, 12, 2903. [Google Scholar] [CrossRef]
- Nettersheim, D.; Gillis, A.; Biermann, K.; Looijenga, L.H.; Schorle, H. The seminoma cell line TCam-2 is sensitive to HDAC inhibitor depsipeptide but tolerates various other chemotherapeutic drugs and loss of NANOG expression. Genes Chromosomes Cancer 2011, 50, 1033–1042. [Google Scholar] [CrossRef]
- Jostes, S.; Nettersheim, D.; Fellermeyer, M.; Schneider, S.; Hafezi, F.; Honecker, F.; Schumacher, V.; Geyer, M.; Kristiansen, G.; Schorle, H. The bromodomain inhibitor JQ1 triggers growth arrest and apoptosis in testicular germ cell tumours in vitro and in vivo. J. Cell. Mol. Med. 2017, 21, 1300–1314. [Google Scholar] [CrossRef]
- Nettersheim, D.; Jostes, S.; Fabry, M.; Honecker, F.; Schumacher, V.; Kirfel, J.; Kristiansen, G.; Schorle, H. A signaling cascade including ARID1A, GADD45B and DUSP1 induces apoptosis and affects the cell cycle of germ cell cancers after romidepsin treatment. Oncotarget 2016, 7, 74931–74946. [Google Scholar] [CrossRef]
- Muller, M.R.; Burmeister, A.; Skowron, M.A.; Stephan, A.; Bremmer, F.; Wakileh, G.A.; Petzsch, P.; Kohrer, K.; Albers, P.; Nettersheim, D. Therapeutical interference with the epigenetic landscape of germ cell tumors: A comparative drug study and new mechanistical insights. Clin. Epigenetics 2022, 14, 5. [Google Scholar] [CrossRef]
- Funke, K.; Duster, R.; Wilson, P.D.; Arevalo, L.; Geyer, M.; Schorle, H. Transcriptional CDK inhibitors as potential treatment option for testicular germ cell tumors. Cancers 2022, 14, 1690. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Friedman, N.B.; Moore, R.A. Tumors of the testis; a report on 922 cases. Mil. Surg. 1946, 99, 573–593. [Google Scholar] [CrossRef]
- Pugh, R.C.B.; Parkinson, C. The origin and classification of testicular germ cell tumours. Int. J. Androl. 1981, 4 (Suppl. S4), 15–24. [Google Scholar] [CrossRef]
- Skakkebaek, N.E.; Berthelsen, J.G.; Visfeldt, J. Clinical aspects of testicular carcinoma-in-situ. Int. J. Androl. 1981, 4 (Suppl. S4), 153–160. [Google Scholar] [CrossRef]
- Looijenga, L.H.; Van Agthoven, T.; Biermann, K. Development of malignant germ cells—The genvironmental hypothesis. Int. J. Dev. Biol. 2013, 57, 241–253. [Google Scholar] [CrossRef]
- Bredael, J.J.; Vugrin, D.; Whitmore, W.F., Jr. Autopsy findings in 154 patients with germ cell tumors of the testis. Cancer 1982, 50, 548–551. [Google Scholar] [CrossRef]
- Rabes, H.M. Proliferation of human testicular tumours. Int. J. Androl. 1987, 10, 127–137. [Google Scholar] [CrossRef]
- Shen, L.; Huang, X.; Xie, X.; Su, J.; Yuan, J.; Chen, X. High expression of SOX2 and OCT4 indicates radiation resistance and an independent negative prognosis in cervical squamous cell carcinoma. J. Histochem. Cytochem. 2014, 62, 499–509. [Google Scholar] [CrossRef] [Green Version]
- Oosterhuis, J.W.; Looijenga, L. Germ cell tumors from a developmental perspective: Cells of origin, pathogenesis, and molecular biology (merging patterns). In Pathology and Biology of Human Germ Cell Tumors; Nogales, F.F., Jimenez, R.E., Eds.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 23–129. ISBN 978-3-662-53773-2. [Google Scholar]
- von Eyben, F.E. Biochemical markers in advanced testicular tumors: Serum lactate dehydrogenase, urinary chorionic gonadotropin and total urinary estrogens. Cancer 1978, 41, 648–652. [Google Scholar] [CrossRef]
- von Eyben, F.E.; Jacobsen, G.K.; Pedersen, H.; Jacobsen, M.; Clausen, P.P.; Zibrandtsen, P.C.; Gullberg, B. Multivariate analysis of risk factors in patients with metastatic testicular germ cell tumors treated with vinblastine and bleomycin. Invasion Metastasis 1982, 2, 125–135. [Google Scholar]
- Bosl, G.J.; Geller, N.L.; Cirrincione, C.; Vogelzang, N.J.; Kennedy, B.J.; Whitmore, W.F., Jr.; Vugrin, D.; Scher, H.; Nisselbaum, J.; Golbey, R.B. Multivariate analysis of prognostic variables in patients with metastatic testicular cancer. Cancer Res. 1983, 43, 3403–3407. [Google Scholar]
- Seidel, C.; Daugaard, G.; Nestler, T.; Tryakin, A.; Fedyanin, M.; Fankhauser, C.D.; Hermanns, T.; Aparicio, J.; Heinzelbecker, J.; Paffenholz, P.; et al. The prognostic significance of lactate dehydrogenase levels in seminoma patients with advanced disease: An analysis by the Global Germ Cell Tumor Collaborative Group (G3). World J. Urol. 2021, 39, 3407–3414. [Google Scholar] [CrossRef]
- Vogelzang, N.J.; Lange, P.H.; Goldberg, E. Absence of sperm-specific lactate dehydrogenase-x in patients with testis cancer. Oncodev. Biol. Med. 1982, 3, 269–272. [Google Scholar]
- von Eyben, F.E.; de Graaff, W.E.; Marrink, J.; Blaabjerg, O.; Sleijfer, D.T.; Koops, H.S.; Oosterhuis, J.W.; Petersen, P.H.; van Echten-Arends, J.; de Jong, B. Serum lactate dehydrogenase isoenzyme 1 activity in patients with testicular germ cell tumors correlates with the total number of copies of the short arm of chromosome 12 in the tumor. Mol. Gen. Genet. 1992, 235, 140–146. [Google Scholar] [CrossRef]
- von Eyben, F.E.; Blaabjerg, O.; Madsen, E.L.; Petersen, P.H.; Smith-Sivertsen, C.; Gullberg, B. Serum lactate dehydrogenase isoenzyme 1 and tumour volume are indicators of response to treatment and predictors of prognosis in metastatic testicular germ cell tumours. Eur. J. Cancer 1992, 28, 410–415. [Google Scholar] [CrossRef]
- von Eyben, F.E.; Skude, G.; Fossa, S.D.; Klepp, O.; Bormer, O. Serum lactate dehydrogenase (S-LDH) and S-LDH isoenzymes in patients with testicular germ cell tumors. Mol. Gen. Genet. 1983, 189, 326–333. [Google Scholar] [CrossRef]
- von Eyben, F.E.; Blaabjerg, O.; Petersen, P.H.; Mommsen, S.; Madsen, E.L.; Kirpekar, F.; Li, S.S.; Kristiansen, K. Lactate dehydrogenase isoenzyme 1 in testicular germ cell tumors. Recent Results Cancer Res. 1991, 123, 85–92. [Google Scholar]
- Rorth, M.; Jacobsen, G.K.; von der Maase, H.; Madsen, E.L.; Nielsen, O.S.; Pedersen, M.; Schultz, H. Surveillance alone versus radiotherapy after orchiectomy for clinical stage I nonseminomatous testicular cancer. Danish Testicular Cancer Study Group. J. Clin. Oncol. 1991, 9, 1543–1548. [Google Scholar] [CrossRef]
- von der Maase, H.; Specht, L.; Jacobsen, G.K.; Jakobsen, A.; Madsen, E.L.; Pedersen, M.; Rorth, M.; Schultz, H. Surveillance following orchidectomy for stage I seminoma of the testis. Eur. J. Cancer 1993, 29, 1931–1934. [Google Scholar] [CrossRef]
- von Eyben, F.E.; Madsen, E.L.; Blaabjerg, O.; Petersen, P.H.; Jacobsen, G.K.; Specht, L.; Pedersen, B.N.; von der Maase, H. Serum lactate dehydrogenase isoenzyme 1 in patients with seminoma stage I followed with surveillance. Acta Oncol. 2002, 41, 77–83. [Google Scholar] [CrossRef] [Green Version]
- von Eyben, F.E.; Madsen, E.L.; Blaabjerg, O.; Petersen, P.H.; von der Maase, H.; Jacobsen, G.K.; Rorth, M. Serum lactate dehydrogenase isoenzyme 1 and relapse in patients with nonseminomatous testicular germ cell tumors clinical stage I. Acta Oncol. 2001, 40, 536–540. [Google Scholar] [CrossRef]
- Stephenson, A.; Eggener, S.E.; Bass, E.B.; Chelnick, D.M.; Daneshmand, S.; Feldman, D.; Gilligan, T.; Karam, J.A.; Leibovich, B.; Liauw, S.L.; et al. Diagnosis and treatment of early stage testicular cancer: AUA guideline. J. Urol. 2019, 202, 272–281. [Google Scholar] [CrossRef] [Green Version]
- Adra, N.; Einhorn, L.H. Testicular cancer update. Clin. Adv. Hematol. Oncol. 2017, 15, 386–396. [Google Scholar]
- Beyer, J.; Collette, L.; Sauve, N.; Daugaard, G.; Feldman, D.R.; Tandstad, T.; Tryakin, A.; Stahl, O.; Gonzalez-Billalabeitia, E.; De Giorgi, U.; et al. Survival and new prognosticators in metastatic seminoma: Results from the IGCCCG-Update Consortium. J. Clin. Oncol. 2021, 39, 1553–1562. [Google Scholar] [CrossRef]
- Gillessen, S.; Sauve, N.; Collette, L.; Daugaard, G.; de Wit, R.; Albany, C.; Tryakin, A.; Fizazi, K.; Stahl, O.; Gietema, J.A.; et al. Predicting outcomes in men with metastatic nonseminomatous germ cell tumors (NSGCT): Results from the IGCCCG Update Consortium. J. Clin. Oncol. 2021, 39, 1563–1574. [Google Scholar] [CrossRef]
- Fizazi, K.; Delva, R.; Caty, A.; Chevreau, C.; Kerbrat, P.; Rolland, F.; Priou, F.; Geoffrois, L.; Rixe, O.; Beuzeboc, P.; et al. A risk-adapted study of cisplatin and etoposide, with or without ifosfamide, in patients with metastatic seminoma: Results of the GETUG S99 multicenter prospective study. Eur. Urol. 2014, 65, 381–386. [Google Scholar] [CrossRef]
- Fossa, S.D.; Dahl, A.A.; Thorsen, L.; Hellesnes, R.; Kiserud, C.E.; Tandstad, T.; Brydoy, M.; Haugnes, H.S.; Myklebust, T.A. Mortality and second cancer incidence after treatment for testicular cancer: Psychosocial health and lifestyle are modifiable prognostic factors. J. Clin. Oncol. 2022, 40, 12. [Google Scholar] [CrossRef]
- Ishida, H.; Kasajima, A.; Kamei, T.; Miura, T.; Oka, N.; Yazdani, S.; Ozawa, Y.; Fujishima, F.; Sakurada, A.; Nakamura, Y.; et al. SOX2 and Rb1 in esophageal small-cell carcinoma: Their possible involvement in pathogenesis. Mod. Pathol. 2017, 30, 660–671. [Google Scholar] [CrossRef] [Green Version]
- Rudin, C.M.; Durinck, S.; Stawiski, E.W.; Poirier, J.T.; Modrusan, Z.; Shames, D.S.; Bergbower, E.A.; Guan, Y.; Shin, J.; Guillory, J.; et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat. Genet. 2012, 44, 1111–1116. [Google Scholar] [CrossRef]
- Kim, J.; Woo, A.J.; Chu, J.; Snow, J.W.; Fujiwara, Y.; Kim, C.G.; Cantor, A.B.; Orkin, S.H. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell 2010, 143, 313–324. [Google Scholar] [CrossRef] [Green Version]
- Ben-Porath, I.; Thomson, M.W.; Carey, V.J.; Ge, R.; Bell, G.W.; Regev, A.; Weinberg, R.A. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 2008, 40, 499–507. [Google Scholar] [CrossRef]
- Mamun, M.A.; Mannoor, K.; Cao, J.; Qadri, F.; Song, X. SOX2 in cancer stemness: Tumor malignancy and therapeutic potentials. J. Mol. Cell. Biol. 2020, 12, 85–98. [Google Scholar] [CrossRef] [Green Version]
- Hepburn, A.C.; Steele, R.E.; Veeratterapillay, R.; Wilson, L.; Kounatidou, E.E.; Barnard, A.; Berry, P.; Cassidy, J.R.; Moad, M.; El-Sherif, A.; et al. The induction of core pluripotency master regulators in cancers defines poor clinical outcomes and treatment resistance. Oncogene 2019, 38, 4412–4424. [Google Scholar] [CrossRef]
- Hatina, J.; Kripnerova, M.; Houdek, Z.; Pesta, M.; Tichanek, F. Pluripotency stemness and cancer: More questions than answers. Adv. Exp. Med. Biol. 2022, 1376, 77–100. [Google Scholar]
- Feinberg, A.P. The key role of epigenetics in human disease prevention and mitigation. N. Engl. J. Med. 2018, 378, 1323–1334. [Google Scholar] [CrossRef]
- Grubelnik, G.; Bostjancic, E.; Groselj, A.; Zidar, N. Expression of NANOG and its regulation in oral squamous cell carcinoma. Biomed. Res. Int. 2020, 245, 456–464. [Google Scholar] [CrossRef]
- Mansouri, S.; Nejad, R.; Karabork, M.; Ekinci, C.; Solaroglu, I.; Aldape, K.D.; Zadeh, G. Sox2: Regulation of expression and contribution to brain tumors. CNS Oncol. 2016, 5, 159–173. [Google Scholar] [CrossRef]
- Najafzadeh, B.; Asadzadeh, Z.; Motafakker Azad, R.; Mokhtarzadeh, A.; Baghbanzadeh, A.; Alemohammad, H.; Abdoli Shadbad, M.; Vasefifar, P.; Najafi, S.; Baradaran, B. The oncogenic potential of NANOG: An important cancer induction mediator. J. Cell. Physiol. 2021, 236, 2443–2458. [Google Scholar] [CrossRef]
- Egevad, L.; Delahunt, B.; Srigley, J.R.; Samaratunga, H. International Society of Urological Pathology (ISUP) grading of prostate cancer—An ISUP consensus on contemporary grading. APMIS 2016, 124, 433–435. [Google Scholar] [CrossRef]
- Nepali, K.; Liou, J.P. Recent developments in epigenetic cancer therapeutics: Clinical advancement and emerging trends. J. Biomed. Sci. 2021, 28, 27. [Google Scholar] [CrossRef]
- Pojani, E.; Barlocco, D. Romidepsin (FK228), a histone deacetylase inhibitor and its analogues in cancer chemotherapy. Curr. Med. Chem. 2021, 28, 1290–1303. [Google Scholar] [CrossRef]
- De Souza, C.; Chatterji, B.P. HDAC inhibitors as novel anti-cancer therapeutics. Recent Pat. Anticancer Drug Discov. 2015, 10, 145–162. [Google Scholar] [CrossRef]
- Patnaik, M.M.; Lasho, T. Myelodysplastic syndrome/myeloproliferative neoplasm overlap syndromes: A focused review. Hematol. Am. Soc. Hematol. Educ. Program 2020, 2020, 460–464. [Google Scholar] [CrossRef]
- Laubach, J.P.; Moreau, P.; San-Miguel, J.F.; Richarson, P.G. Panobinostat for the treatment of multiple myeloma. Clin. Cancer Res. 2015, 21, 4767–4773. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.Z.; Kwitkowski, V.E.; Del Valle, P.L.; Ricci, M.S.; Saber, H.; Habtemariam, B.A.; Bullock, J.; Bloomquist, E.; Li Shen, Y.; Chen, X.H.; et al. FDA approval: Belinostat for the treatment of patients with relapsed or refractory peripheral T-cell lymphoma. Clin. Cancer Res. 2015, 21, 2666–2670. [Google Scholar] [CrossRef] [Green Version]
- Eckschlager, T.; Plch, J.; Stiborova, M.; Hrabeta, J. Histone deacetylase inhibitors as anticancer drugs. Int. J. Mol. Sci. 2017, 18, 1414. [Google Scholar] [CrossRef] [Green Version]
- Fenaux, P.; Mufti, G.J.; Hellstrom-Lindberg, E.; Santini, V.; Finelli, C.; Giagounidis, A.; Schoch, R.; Gattermann, N.; Sanz, G.; List, A.; et al. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: A randomised, open-label, phase III study. Lancet Oncol. 2009, 10, 223–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhillon, S. Trilaciclib: First approval. Drugs 2021, 81, 867–874. [Google Scholar] [CrossRef]
- von Eyben, F.E. Epidermal growth factor receptor inhibition and non-small cell lung cancer. Crit. Rev. Clin. Lab. Sci. 2006, 43, 291–323. [Google Scholar] [CrossRef]
- Qi, Y.; Xia, X.; Shao, L.; Guo, L.; Dong, Y.; Tian, J.; Xu, L.; Niu, R.; Wei, S. An updated network meta-analysis of EGFR-TKIs and combination therapy in the first-line treatment of advanced EGFR mutation positive non-small cell lung cancer. Front. Oncol. 2022, 12, 616546. [Google Scholar] [CrossRef]
- Festuccia, N.; Osorno, R.; Halbritter, F.; Karwacki-Neisius, V.; Navarro, P.; Colby, D.; Wong, F.; Yates, A.; Tomlinson, S.R.; Chambers, I. Esrrb is a direct Nanog target gene that can substitute for Nanog function in pluripotent cells. Cell. Stem Cell 2012, 11, 477–490. [Google Scholar] [CrossRef] [Green Version]
- Rony, I.K.; Baten, A.; Bloomfield, J.A.; Islam, M.E.; Billah, M.M.; Islam, K.D. Inducing pluripotency in vitro: Recent advances and highlights in induced pluripotent stem cells generation and pluripotency reprogramming. Cell. Prolif. 2015, 48, 140–156. [Google Scholar] [CrossRef]
- Wang, C.; Lin, Y.; Zhu, H.; Zhou, Y.; Mao, F.; Huang, X.; Sun, Q.; Li, C. Efficacy and safety profile of histone deacetylase inhibitors for metastatic breast cancer: A meta-analysis. Front. Oncol. 2022, 12, 901152. [Google Scholar] [CrossRef]
- DiNardo, C.D.; Jonas, B.A.; Pullarkat, V.; Thirman, M.J.; Garcia, J.S.; Wei, A.H.; Konopleva, M.; Dohner, H.; Letai, A.; Fenaux, P.; et al. Azacitidine and venetoclax in previously untreated acute myeloid leukemia. N. Engl. J. Med. 2020, 383, 617–629. [Google Scholar] [CrossRef] [PubMed]
- Blagitko-Dorfs, N.; Schlosser, P.; Greve, G.; Pfeifer, D.; Meier, R.; Baude, A.; Brocks, D.; Plass, C.; Lubbert, M. Combination treatment of acute myeloid leukemia cells with DNMT and HDAC inhibitors: Predominant synergistic gene downregulation associated with gene body demethylation. Leukemia 2019, 33, 945–956. [Google Scholar] [CrossRef]
- Lobo, J.; van Zogchel, L.M.J.; Nuru, M.G.; Gillis, A.J.M.; van der Schoot, C.E.; Tytgat, G.A.M.; Looijenga, L.H.J. Combining hypermethylated RASSF1A detection using ddPCR with miR-371a-3p testing: An improved panel of liquid biopsy biomarkers for testicular germ cell tumor patients. Cancers 2021, 13, 5228. [Google Scholar] [CrossRef]
- Markulin, D.; Vojta, A.; Samarzija, I.; Gamulin, M.; Beceheli, I.; Jukic, I.; Maglov, C.; Zoldos, V.; Fucic, A. Association between RASSF1A promoter methylation and testicular germ cell tumor: A meta-analysis and a cohort study. Cancer Genomics Proteomics 2017, 14, 363–372. [Google Scholar]
- Bartkova, J.; Lukas, C.; Sorensen, C.S.; Rajpert-De Meyts, E.; Skakkebaek, N.E.; Lukas, J.; Bartek, J. Deregulation of the RB pathway in human testicular germ cell tumours. J. Pathol. 2003, 200, 149–156. [Google Scholar] [CrossRef]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- Hoff, A.M.; Kraggerud, S.M.; Alagaratnam, S.; Berg, K.C.G.; Johannessen, B.; Holand, M.; Nilsen, G.; Lingjaerde, O.C.; Andrews, P.W.; Lothe, R.A.; et al. Frequent copy number gains of SLC2A3 and ETV1 in testicular embryonal carcinomas. Endocr. Relat. Cancer 2020, 27, 457–468. [Google Scholar] [CrossRef]



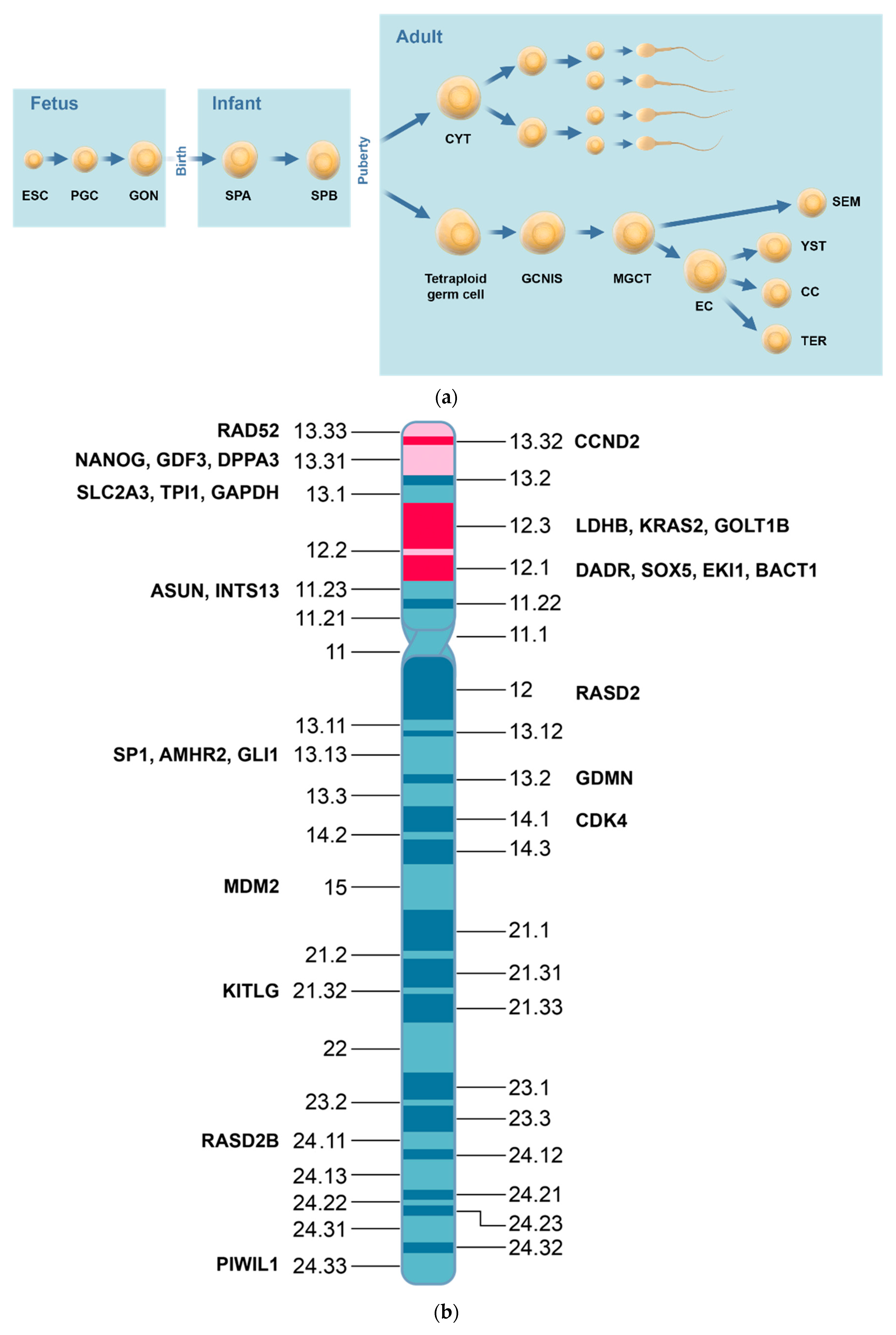
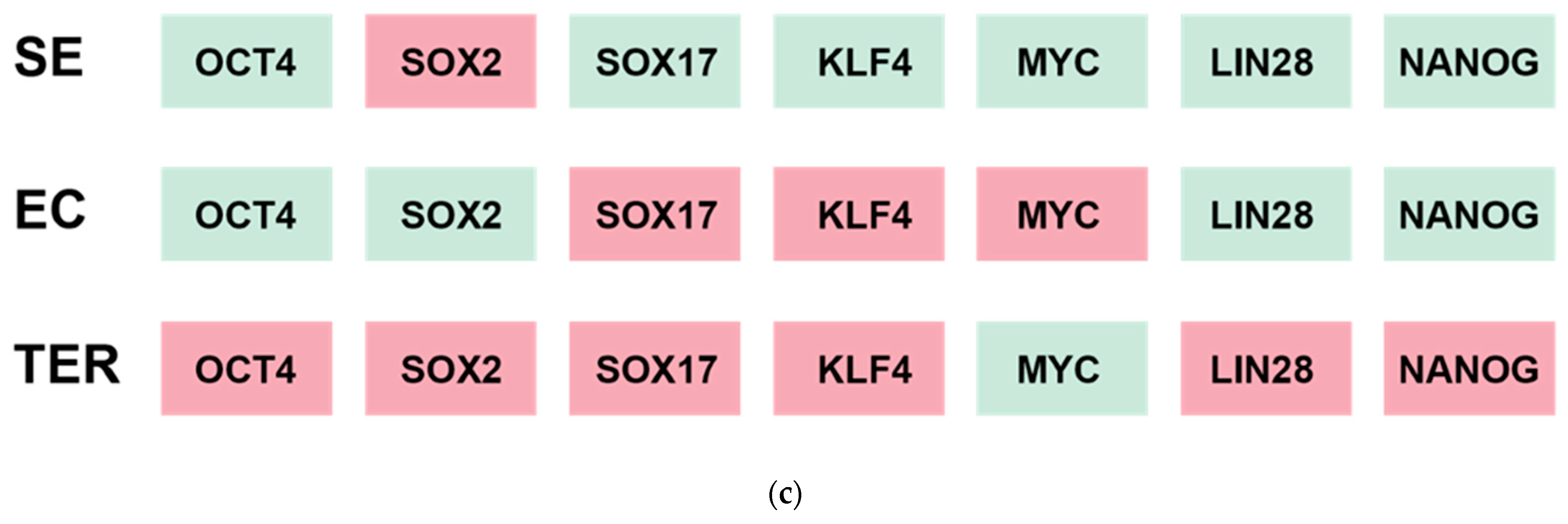
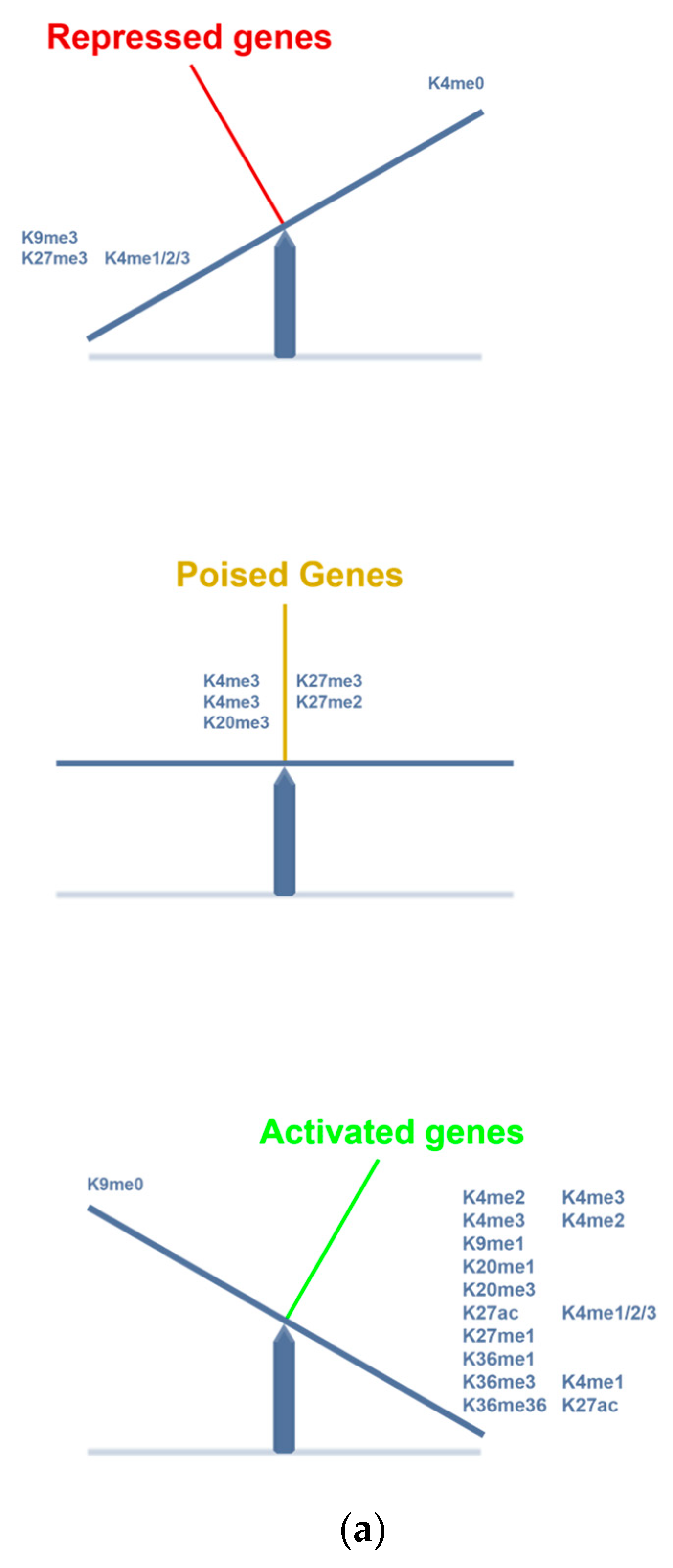

| Expressed Transcription Factors | Histologic Types | NGC | GCNIS | SE | EC | TER | |
|---|---|---|---|---|---|---|---|
| Epigenetic mediators | High | PRAME LDHC | PRAME, KIT, SOX17, NANOG | PRAME, KIT, SOX17, KLF4 POU5F1, NANOG, MYC | NANOG, POU5F1, DNMT3B | RB1, MYC | |
| Low | RB1 | RB1 | RB1, MYC | POU5F1, SO)X2, LIN28, NANOG | |||
| 5mC | +++ | Neg | Neg/++ | +++ | |||
| 5mhC | Neg | Neg | +++ | ||||
| Epigenetic modifiers | H2A.Z | +/+++ | +++ | Neg/+ | Neg/+ | ||
| H3K4 | Me1 +++ Me2/me3 ++ | Me1/me2/me3 +++ | Me1 ++/+++ | Me1 neg/+ Me2/Me3++/+++ | |||
| H3K9me1/me2 | Neg/+ | ++ | ++ | ||||
| H3K9ac | +++ | Neg/+ | Neg/+ | ||||
| H3K27me2 | |||||||
| H3K27me3 | +++ | Ac/+ | +++ | neg | |||
| Epigenetic modulators | DNMT | Low | High | ||||
| TET | Low | High | low | ||||
| HMT | EZD2 | +++ | Neg/+ | neg | |||
| HDMT | UTX | Neg | Neg | Neg | Neg | ||
| JMJDD3 | Neg/+ | Neg | Neg/+ | Neg/+ | |||
| HAT | High | Low | |||||
| HDAC | Low | High |
| Pharmacologic Group | Drugs | TGCT | Ref | Other Malignancies | |
|---|---|---|---|---|---|
| Studies | Indications | ||||
| Cell lines | Patients | ||||
| DNMTi | 5-aza | X | X | Myelodysplastic syndromes | |
| 5-aza-cD | X | X | |||
| Guadecitabine | X | ||||
| KDMi | Chaertocin | [141] | |||
| JIB-04 | [141] | ||||
| EZH2i | Tazemetostat | Follicular NH lymphoma | |||
| Bromodomaini | JQ11 | X | [143] | ||
| LSD1i | CBB3001 | X | |||
| HDACi | belinostat | X | [144] | Cutaneous T cell lymphoma | |
| chicamide | X | ||||
| depsipeptide | X | [144] | |||
| panobinostat | X | [144] | Cutaneous T cell lymphoma | ||
| Romidepsin | X | Cutaneous T cell lymphoma | |||
| Quisinostat | X | [141] | |||
| trichostatin | |||||
| vorinostat | X | ||||
| CDKi | dinaciclib | [140] | |||
| Flavo-piridol | [140] | ||||
| NVP-2 | [140] | ||||
| SY0351 | [140] | ||||
| thal-sns-032 | [140] | ||||
| THZ1 | [140] | ||||
| THZ531 | [140] | ||||
| YKL-5-214 | [140] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
von Eyben, F.E.; Kristiansen, K.; Kapp, D.S.; Hu, R.; Preda, O.; Nogales, F.F. Epigenetic Regulation of Driver Genes in Testicular Tumorigenesis. Int. J. Mol. Sci. 2023, 24, 4148. https://doi.org/10.3390/ijms24044148
von Eyben FE, Kristiansen K, Kapp DS, Hu R, Preda O, Nogales FF. Epigenetic Regulation of Driver Genes in Testicular Tumorigenesis. International Journal of Molecular Sciences. 2023; 24(4):4148. https://doi.org/10.3390/ijms24044148
Chicago/Turabian Stylevon Eyben, Finn E., Karsten Kristiansen, Daniel S. Kapp, Rong Hu, Ovidiu Preda, and Francisco F. Nogales. 2023. "Epigenetic Regulation of Driver Genes in Testicular Tumorigenesis" International Journal of Molecular Sciences 24, no. 4: 4148. https://doi.org/10.3390/ijms24044148





