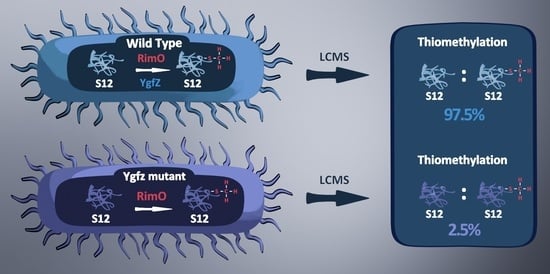
Essentiality of the Escherichia coli YgfZ Protein for the In Vivo Thiomethylation of Ribosomal Protein S12 by the RimO Enzyme
Abstract
:1. Introduction
2. Results
2.1. Growth Characteristics of the Bacterial Strains
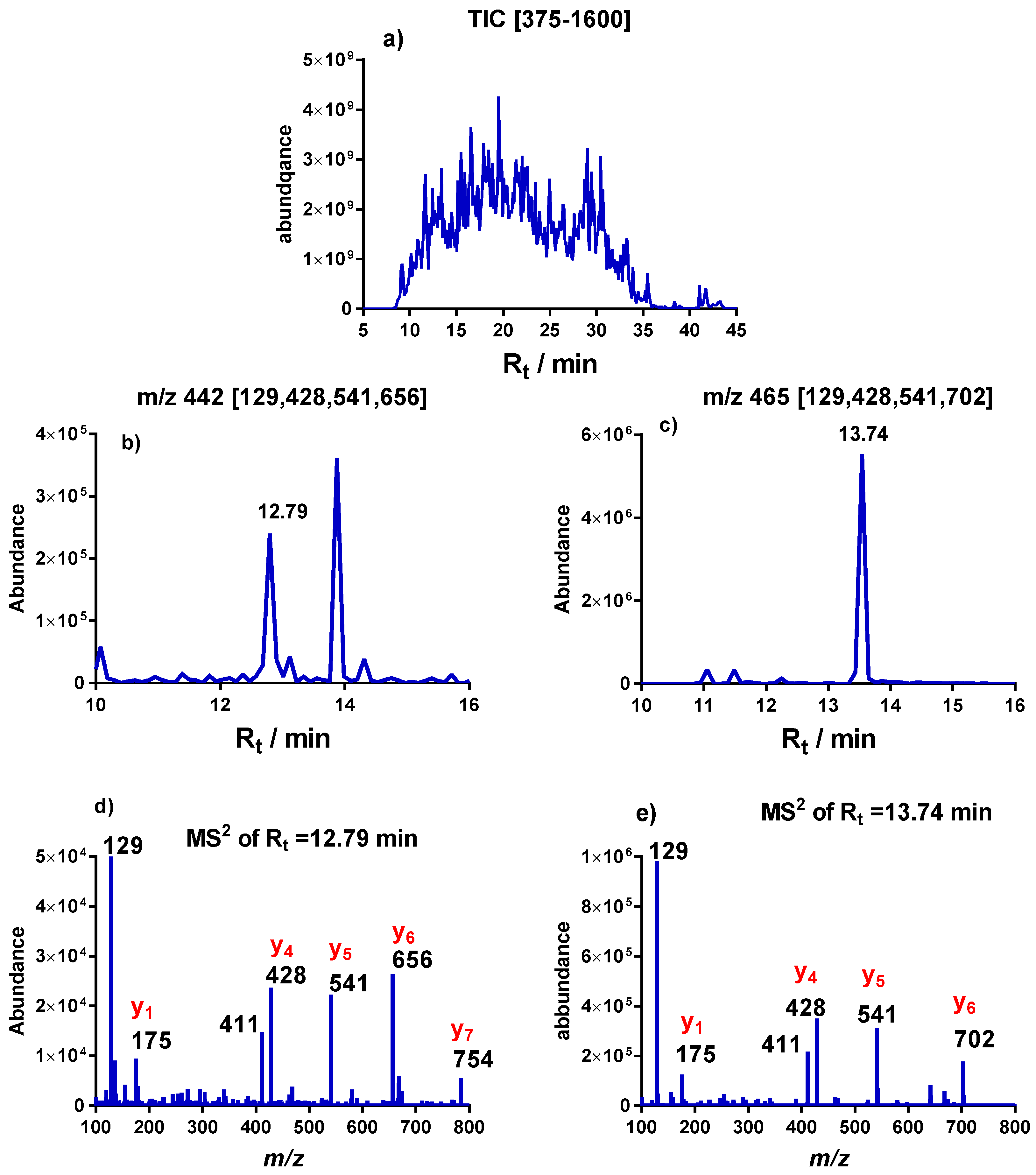
2.2. MS2 Analysis of Thiomethylation of S12
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Bacterial Strains and Growth Conditions
4.3. Extraction of Bacterial Protein
4.4. LC-MS Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Py, B.; Barras, F. Building Fe–S Proteins: Bacterial Strategies. Nat. Rev. Microbiol. 2010, 8, 436–446. [Google Scholar] [CrossRef] [PubMed]
- Braymer, J.J.; Freibert, S.A.; Rakwalska-Bange, M.; Lill, R. Mechanistic Concepts of Iron-Sulfur Protein Biogenesis in Biology. Biochim. Biophys. Acta-Mol. Cell Res. 2021, 1868, 118863. [Google Scholar] [CrossRef] [PubMed]
- Keseler, I.M.; Collado-Vides, J.; Santos-Zavaleta, A.; Peralta-Gil, M.; Gama-Castro, S.; Muniz-Rascado, L.; Bonavides-Martinez, C.; Paley, S.; Krummenacker, M.; Altman, T.; et al. EcoCyc: A Comprehensive Database of Escherichia Coli Biology. Nucleic Acids Res. 2011, 39, D583–D590. [Google Scholar] [CrossRef] [PubMed]
- Esquilin-Lebron, K.; Dubrac, S.; Barras, F.; Boyd, J.M. Bacterial Approaches for Assembling Iron-Sulfur Proteins. Mbio 2021, 12, e02425-21. [Google Scholar] [CrossRef]
- Mühlenhoff, U.; Weiler, B.D.; Nadler, F.; Millar, R.; Kothe, I.; Freibert, S.A.; Altegoer, F.; Bange, G.; Lill, R. The Iron-Sulfur Cluster Assembly (ISC) Protein Iba57 Executes a Tetrahydrofolate-Independent Function in Mitochondrial [4Fe-4S] Protein Maturation. J. Biol. Chem. 2022, 298, 102465. [Google Scholar] [CrossRef]
- Gourdoupis, S.; Nasta, V.; Calderone, V.; Ciofi-Baffoni, S.; Banci, L. IBA57 Recruits ISCA2 to Form a [2Fe-2S] Cluster-Mediated Complex. J. Am. Chem. Soc. 2018, 140, 14401–14412. [Google Scholar] [CrossRef] [Green Version]
- Gelling, C.; Dawes, I.W.; Richhardt, N.; Lill, R.; Mühlenhoff, U. Mitochondrial Iba57p Is Required for Fe/S Cluster Formation on Aconitase and Activation of Radical SAM Enzymes. Mol. Cell. Biol. 2008, 28, 1851–1861. [Google Scholar] [CrossRef] [Green Version]
- Sheftel, A.D.; Wilbrecht, C.; Stehling, O.; Niggemeyer, B.; Elsässer, H.P.; Mühlenhoff, U.; Lill, R. The Human Mitochondrial ISCA1, ISCA2, and IBA57 Proteins Are Required for [4Fe-4S] Protein Maturation. Mol. Biol. Cell 2012, 23, 1157–1166. [Google Scholar] [CrossRef]
- Teplyakov, A.; Obmolova, G.; Sarikaya, E.; Pullalarevu, S.; Krajewski, W.; Galkin, A.; Howard, A.J.; Herzberg, O.; Gilliland, G.L. Crystal Structure of the YgfZ Protein from Escherichia coli Suggests a Folate-Dependent Regulatory Role in One-Carbon Metabolism. J. Bacteriol. 2004, 186, 7134–7140. [Google Scholar] [CrossRef] [Green Version]
- Waller, J.C.; Alvarez, S.; Naponelli, V.; Lara-Nuñez, A.; Blaby, I.K.; Da Silva, V.; Ziemak, M.J.; Vickers, T.J.; Beverley, S.M.; Edison, A.S.; et al. A Role for Tetrahydrofolates in the Metabolism of Iron-Sulfur Clusters in All Domains of Life. Proc. Natl. Acad. Sci. USA 2010, 107, 10412–10417. [Google Scholar] [CrossRef] [Green Version]
- Ote, T.; Hashimoto, M.; Ikeuchi, Y.; Su’etsugu, M.; Suzuki, T.; Katayama, T.; Kato, J.I. Involvement of the Escherichia coli Folate-Binding Protein YgfZ in RNA Modification and Regulation of Chromosomal Replication Initiation. Mol. Microbiol. 2006, 59, 265–275. [Google Scholar] [CrossRef]
- Hasnain, G.; Waller, J.C.; Alvarez, S.; Ravilious, G.E.; Jez, J.M.; Hanson, A.D. Mutational Analysis of YgfZ, a Folate-Dependent Protein Implicated in Iron/Sulphur Cluster Metabolism. FEMS Microbiol. Lett. 2012, 326, 168–172. [Google Scholar] [CrossRef] [Green Version]
- Cheek, J.; Broderick, J.B. Adenosylmethionine-Dependent Iron-Sulfur Enzymes: Versatile Clusters in a Radical New Role. JBIC J. Biol. Inorg. Chem. 2001, 6, 209–226. [Google Scholar] [CrossRef]
- Lanz, N.D.; Booker, S.J. Identification and Function of Auxiliary Iron-Sulfur Clusters in Radical SAM Enzymes. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2012, 1824, 1196–1212. [Google Scholar] [CrossRef] [PubMed]
- Anton, B.P.; Saleh, L.; Benner, J.S.; Raleigh, E.A.; Kasif, S.; Roberts, R.J. RimO, a MiaB-like Enzyme, Methylthiolates the Universally Conserved Asp88 Residue of Ribosomal Protein S12 in Escherichia coli. Proc. Natl. Acad. Sci. USA 2008, 105, 1826–1831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boutigny, S.; Saini, A.; Baidoo, E.E.K.; Yeung, N.; Keasling, J.D.; Butland, G. Physical and Functional Interactions of a Monothiol Glutaredoxin and an Iron Sulfur Cluster Carrier Protein with the Sulfur-Donating Radical S-Adenosyl-L-Methionine Enzyme MiaB. J. Biol. Chem. 2013, 288, 14200–14211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.H.; Saleh, L.; Anton, B.P.; Madinger, C.L.; Benner, J.S.; Iwig, D.F.; Roberts, R.J.; Krebs, C.; Booker, S.J. Characterization of RimO, a New Member of the Methylthiotransferase Subclass of the Radical SAM Superfamily. Biochemistry 2009, 48, 10162–10174. [Google Scholar] [CrossRef] [Green Version]
- Mulliez, E.; Duarte, V.; Arragain, S.; Fontecave, M.; Atta, M. On the Role of Additional [4Fe-4S] Clusters with a Free Coordination Site in Radical-SAM Enzymes. Front. Chem. 2017, 5, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forouhar, F.; Arragain, S.; Atta, M.; Gambarelli, S.; Mouesca, J.M.; Hussain, M.; Xiao, R.; Kieffer-Jaquinod, S.; Seetharaman, J.; Acton, T.B.; et al. Two Fe-S Clusters Catalyse Sulfur Insertion by Radical-SAM Methylthiotransferases. Nat. Chem. Biol. 2013, 9, 333. [Google Scholar] [CrossRef] [Green Version]
- Landgraf, B.J.; Arcinas, A.J.; Lee, K.H.; Booker, S.J. Identification of an Intermediate Methyl Carrier in the Radical S-Adenosylmethionine Methylthiotransferases RimO and MiaB. J. Am. Chem. Soc. 2013, 135, 15404–15416. [Google Scholar] [CrossRef] [Green Version]
- Esakova, O.A.; Grove, T.L.; Yennawar, N.H.; Arcinas, A.J.; Wang, B.; Krebs, C.; Almo, S.C.; Booker, S.J. Structural Basis for TRNA Methylthiolation by the Radical SAM Enzyme MiaB. Nature 2021, 597, 566–570. [Google Scholar] [CrossRef]
- Strader, M.B.; Costantino, N.; Elkins, C.A.; Chen, C.Y.; Patel, I.; Makusky, A.J.; Choy, J.S.; Court, D.L.; Markey, S.P.; Kowalak, J.A. A Proteomic and Transcriptomic Approach Reveals New Insight into β-Methylthiolation of Escherichia Coli Ribosomal Protein S12. Mol. Cell. Proteom. 2011, 10, M110.005199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kowalak, J.A.; Walsh, K.A. β-Methylthio-Aspartic Acid: Identification of a Novel Posttranslational Modification in Ribosomal Protein S12 from Escherichia coli. Protein Sci. 1996, 5, 1625–1632. [Google Scholar] [CrossRef] [PubMed]
- Waller, J.C.; Ellens, K.W.; Hasnain, G.; Alvarez, S.; Rocca, J.R.; Hanson, A.D. Evidence That the Folate-Dependent Proteins YgfZ and MnmEG Have Opposing Effects on Growth and on Activity of the Iron-Sulfur Enzyme MiaB. J. Bacteriol. 2012, 194, 362–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mühlenhoff, U.; Richter, N.; Pines, O.; Pierik, A.J.; Lill, R. Specialized Function of Yeast Isa1 and Isa2 Proteins in the Maturation of Mitochondrial [4Fe-4S] Proteins. J. Biol. Chem. 2011, 286, 41205–41216. [Google Scholar] [CrossRef] [Green Version]
- McCarthy, E.L.; Booker, S.J. Destruction and Reformation of an Iron-Sulfur Cluster during Catalysis by Lipoyl Synthase. Science 2017, 358, 373–377. [Google Scholar] [CrossRef] [Green Version]
- McCarthy, E.L.; Rankin, A.N.; Dill, Z.R.; Booker, S.J. The A-Type Domain in Escherichia Coli NfuA Is Required for Regenerating the Auxiliary [4Fe– 4S] Cluster in Escherichia coli Lipoyl Synthase. J. Biol. Chem. 2019, 294, 1609–1617. [Google Scholar] [CrossRef] [Green Version]
- Esberg, B.; Leung, H.C.E.; Tsui, H.C.T.; Bjork, G.R.; Winkler, M.E. Identification of the miaB Gene, Involved in Methylthiolation of Isopentenylated A37 Derivatives in the tRNA of Salmonella typhimurium and Escherichia coli. J. Bacteriol. 1999, 181, 7256–7265. [Google Scholar] [CrossRef] [Green Version]
- Cronan, J.E. Assembly of Lipoic Acid on Its Cognate Enzymes: An Extraordinary and Essential Biosynthetic Pathway. Microbiol. Mol. Biol. Rev. 2016, 80, 429–450. [Google Scholar] [CrossRef] [Green Version]
- Miller, J.H. Experiments in Molecular Genetics; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1972. [Google Scholar]
- Guyer, M.S.; Reed, R.R.; Steitz, J.A.; Low, K.B. Identification of a Sex-Factor-Affinity Site in E. coli as Gamma Delta. Cold Spring Harb. Symp. Quant. Biol. 1981, 45 Pt 1, 135–140. [Google Scholar] [CrossRef]



| wt | rimO | ygfZ | |
|---|---|---|---|
| 30 °C | 36.1 ± 1.1 min | NT a | 59 ± 6.1 min b |
| 37 °C | 21.5 ± 0.5 min | 22.2 ± 1.0 min | NT |
| 42 °C | 18.3 ± 0.2 min | NT | 40 ± 1.8 min |
| VKDLPGVR | VKDLPGVR | |
|---|---|---|
| Rt/min | 12.79 | 13.74 |
| Mw | 882.5297 | 928.6117 |
| [M+2H]+ | 442.2727 | 465.3136 |
| Immonium ion of R | 129.1022 | 129.1022 |
| [R+H]+ (y1) | 175.1190 | 175.1189 |
| [PGVR+H]+ (y4) | 428.2615 | 428.2614 |
| [LPGVR+H]+ (y5) | 541.3453 | 541.3455 |
| [DLPGVR+H]+ (y6) | 656.3723 | 702.3597 |
| Bacterial Strain | Geno- Type | OD600 | t/°C | Biological Replicates | % S12 Thiomethylation | |
|---|---|---|---|---|---|---|
| TC5540 | wt | 0.2 | 37 | 3 | 20 ± 6 | 95 ± 2 |
| 0.7 | 37 | 3 | 31 ± 15 | 96 ± 2 | ||
| 2.5 | 37 | 3 | 83 ± 17 | 99 ± 1 | ||
| TC5542 | rimO | 0.2; 0.7; 2.5 | 37 | 3 | 0 | 0 |
| TC5540 | wt | 0.2 | 30 | 3 | 18 ± 5 | 94 ± 2 |
| 0.2 | 42 | 3 | 40 ± 20 | 96 ± 3 | ||
| TC5541 | ygfZ | 0.2 | 30 | 6 | 0.02 ± 0.03 | 1.5 ± 3 |
| 0.2 | 42 | 6 | 0.03 ± 0.04 | 3 ± 4 |
| E. coli Strain | ||
|---|---|---|
| Wild type | 40 | 30 |
| rimO or miaB (knock-out) | 0 | 0 |
| ygfZ (knock out) rimO+ miaB+ | 0.03 | 2 |
| Strain | Genotype | Reference/Source |
|---|---|---|
| MG1655 | F− λ− rph-1 | [31] |
| TC5540 | F− λ− rph-1 tonA | Spontaneous T1R mutant of MG1655 obtained from Løbner-Olesen |
| JW2866 | ΔlacZ4787::(rrnB-3) hsdR514 ΔaraBAD567 ΔrhaBAD568 ygfZ::aphA | The Keio collection NBRP (NIG, Japan) |
| JD21892 | F− lacIq lacZΔM15 galK2 galT22 lambda- in (rrnD-rrnE)1 rimO::(mini-Tn10 KanR) | Transposon insertion disruptant collection NBRP (NIG, Japan) |
| TC5541 | F− λ− rph-1 tonA ygfZ::aphA | This work |
| TC5542 | F− λ− rph-1 tonA rimO::(mini-Tn10 KanR) | This work |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lund, T.; Kulkova, M.Y.; Jersie-Christensen, R.; Atlung, T. Essentiality of the Escherichia coli YgfZ Protein for the In Vivo Thiomethylation of Ribosomal Protein S12 by the RimO Enzyme. Int. J. Mol. Sci. 2023, 24, 4728. https://doi.org/10.3390/ijms24054728
Lund T, Kulkova MY, Jersie-Christensen R, Atlung T. Essentiality of the Escherichia coli YgfZ Protein for the In Vivo Thiomethylation of Ribosomal Protein S12 by the RimO Enzyme. International Journal of Molecular Sciences. 2023; 24(5):4728. https://doi.org/10.3390/ijms24054728
Chicago/Turabian StyleLund, Torben, Maria Yohanna Kulkova, Rosa Jersie-Christensen, and Tove Atlung. 2023. "Essentiality of the Escherichia coli YgfZ Protein for the In Vivo Thiomethylation of Ribosomal Protein S12 by the RimO Enzyme" International Journal of Molecular Sciences 24, no. 5: 4728. https://doi.org/10.3390/ijms24054728