Quantitative Analysis of Transcription Termination via Position-Selective Labeling of RNA (PLOR) Method
Abstract
1. Introduction
2. Results
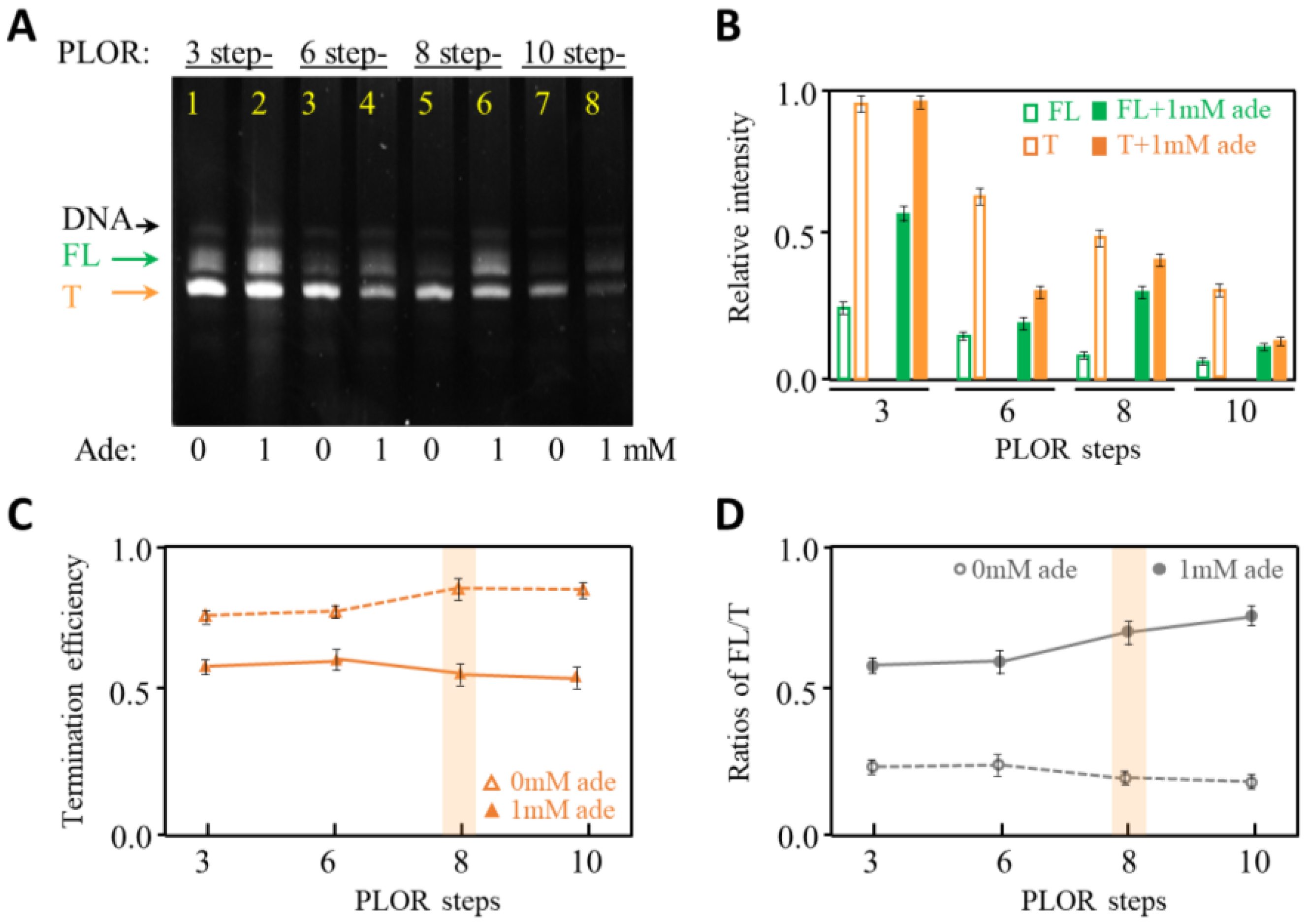
2.1. The Effects of Pausing Modes on Transcription Termination of Adenine Riboswitch RNA
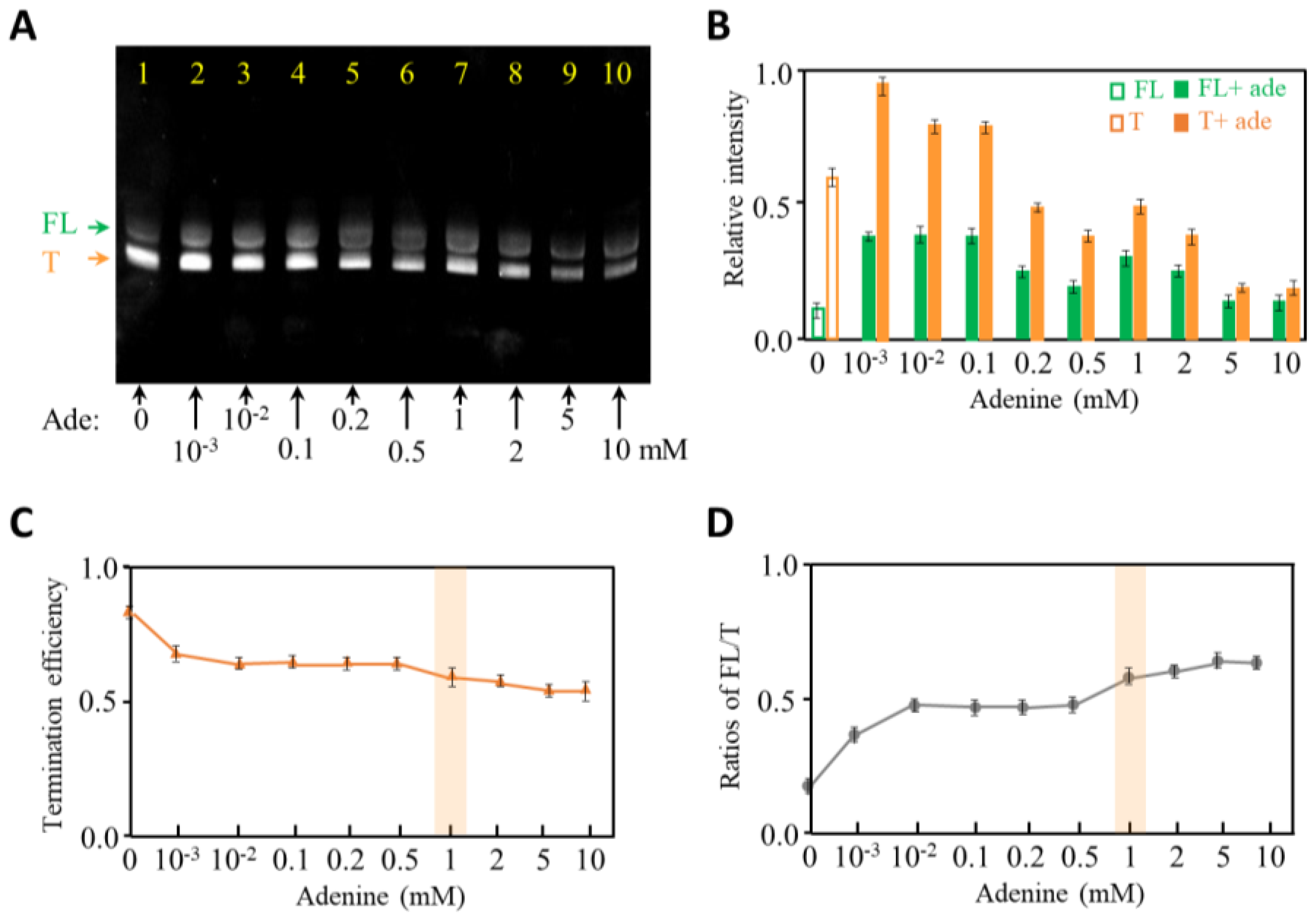
2.2. The Effects of Adenine Concentration on Transcription Termination of Adenine Riboswitch RNA
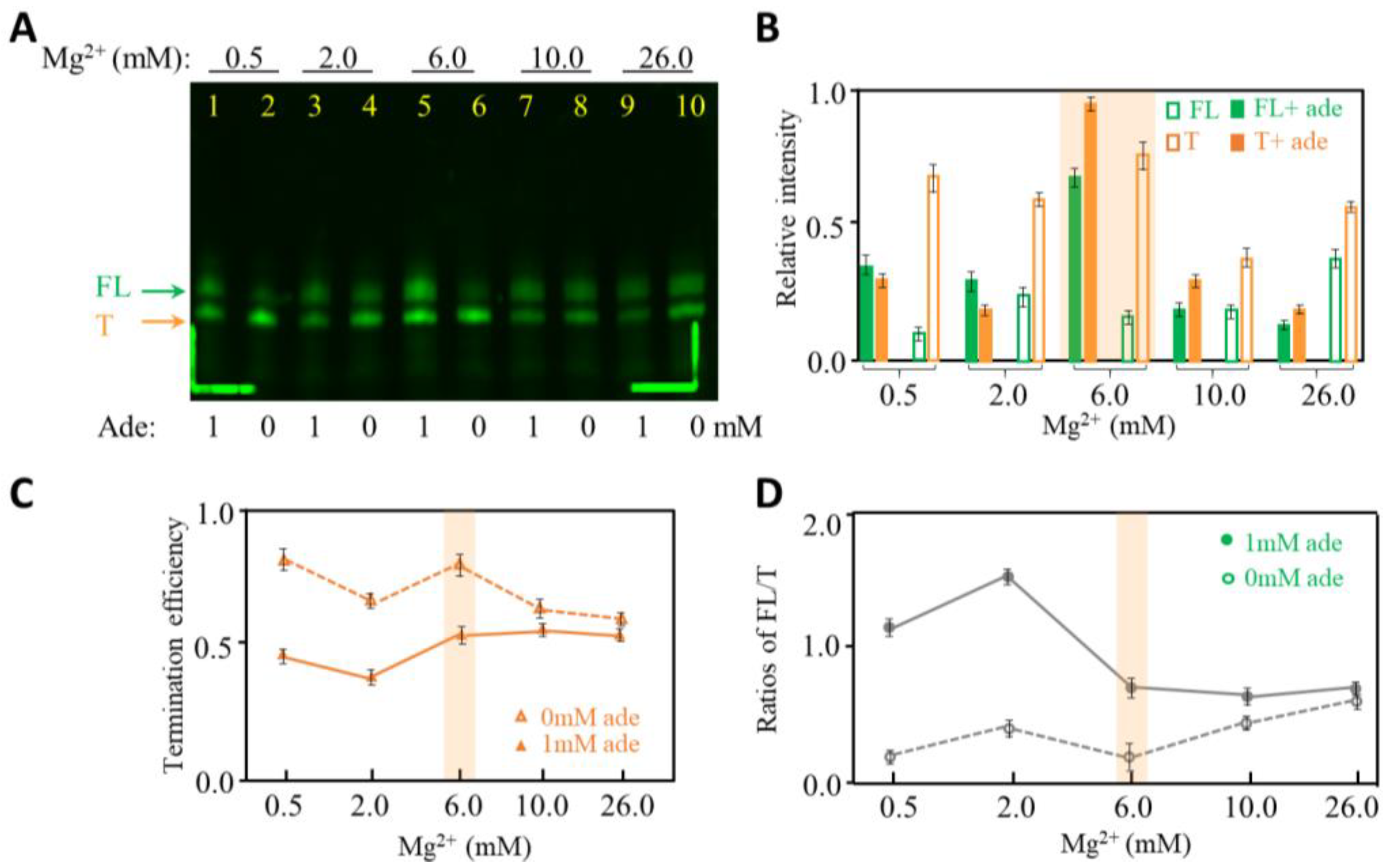
2.3. The Effects of Mg2+ Concentration on Transcription Termination of Adenine Riboswitch RNA
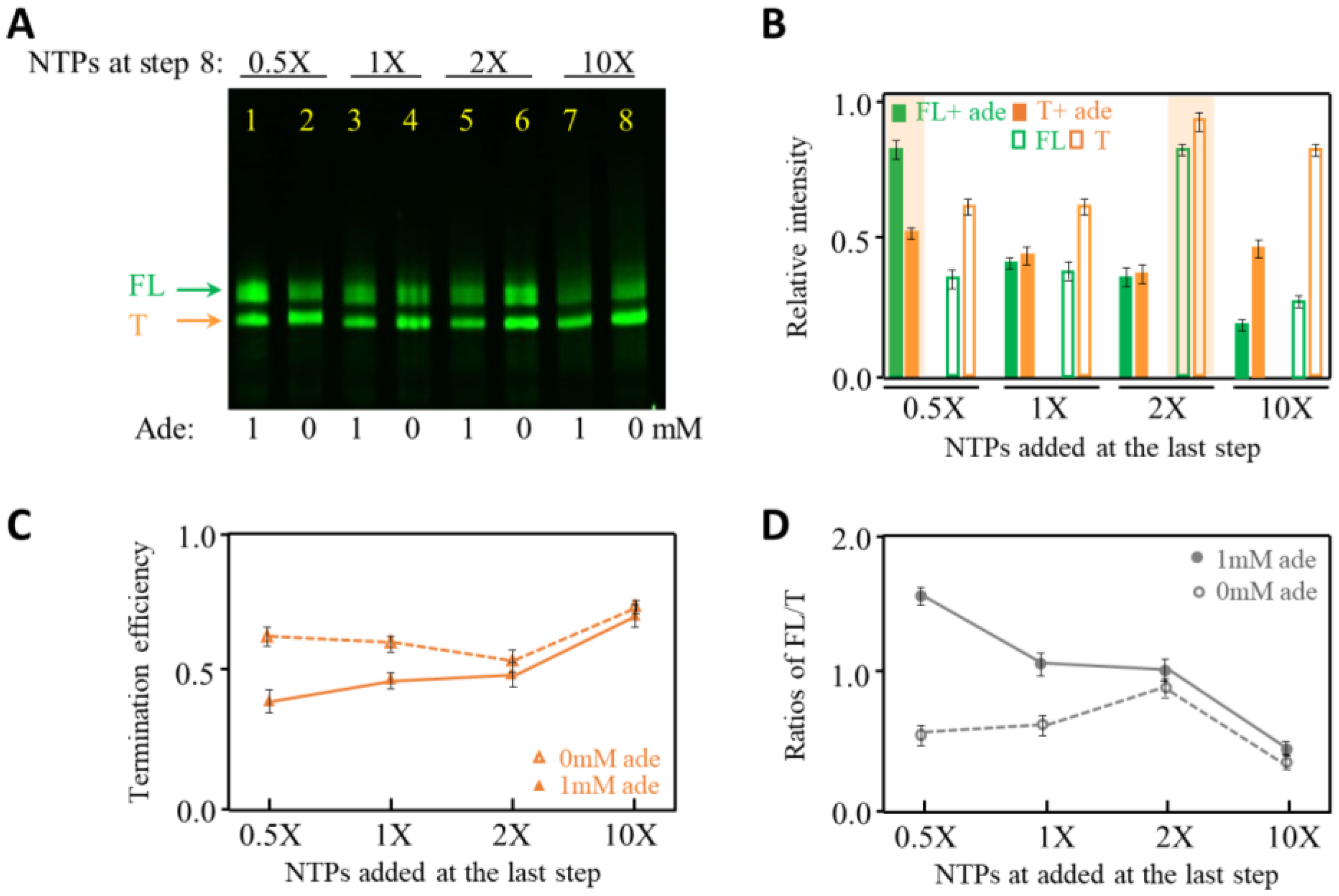
2.4. The Effects of NTP Concentration on Transcription Termination of Adenine Riboswitch RNA
3. Discussion
4. Materials and Methods
4.1. Preparation of Solid-Phase DNA Templates for PLOR
4.2. PLOR Reactions to Generate Terminated and Full-Length RNA
4.3. Quantification of Terminated and Full-Length RNA Products
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Higgs, P.G.; Lehman, N. The RNA World: Molecular cooperation at the origins of life. Nat. Rev. Genet. 2015, 16, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Chen, L. Circular RNAs: Characterization, cellular roles, and applications. Cell 2022, 185, 2016–2034. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Meng, Q.; Qian, J.; Li, M.; Gu, C.; Yang, Y. Review: RNA-based diagnostic markers discovery and therapeutic targets development in cancer. Pharmacol. Therapeut. 2022, 234, 108123. [Google Scholar] [CrossRef] [PubMed]
- Deogharia, M.; Gurha, P. The “guiding” principles of noncoding RNA function. WIREs RNA 2022, 13, e1704. [Google Scholar] [CrossRef]
- Wang, D.; Ye, R.; Cai, Z.; Xue, Y. Emerging roles of RNA-RNA interactions in transcriptional regulation. WIREs RNA 2022, 13, e1712. [Google Scholar] [CrossRef]
- Singh, S.; Shyamal, S.; Panda, A.C. Detecting RNA-RNA interactome. WIREs RNA 2022, 13, e1715. [Google Scholar] [CrossRef]
- Zogg, H.; Singh, R.; Ro, S. Current advances in RNA therapeutics for human diseases. Int. J. Mol. Sci. 2022, 23, 2736. [Google Scholar] [CrossRef]
- Santangelo, T.J.; Artsimovitch, I. Termination and antitermination: RNA polymerase runs a stop sign. Nat. Rev. Microbiol. 2011, 9, 319–329. [Google Scholar] [CrossRef]
- Porrua, O.; Boudvillain, M.; Libri, D. Transcription termination: Variations on common themes. Trends Genet. 2016, 32, 508–522. [Google Scholar] [CrossRef]
- Gusarov, I.; Nudler, E. The mechanism of intrinsic transcription termination. Mol. Cell 1999, 3, 495–504. [Google Scholar] [CrossRef]
- Binas, O.; Schamber, T.; Schwalbe, H. The conformational landscape of transcription intermediates involved in the regulation of the ZMP-sensing riboswitch from Thermosinus carboxydivorans. Nucleic Acids Res. 2020, 48, 6970–6979. [Google Scholar] [CrossRef] [PubMed]
- Machtel, P.; Wasilewska-Burczyk, A.; Zacharjasz, J.; Grzywacz, K. PTT-quant: A new method for direct identification and absolute quantification of premature transcription termination events, following the example of bacterial riboswitches. Appl. Microbiol. Biotechnol. 2022, 106, 1557–1570. [Google Scholar] [CrossRef] [PubMed]
- Marcano-Velázquez, J.G.; Batey, R.T. Structure-guided mutational analysis of gene regulation by the Bacillus subtilis pbuE adenine-responsive riboswitch in a cellular context. J. Biol. Chem. 2015, 290, 4464–4475. [Google Scholar] [CrossRef] [PubMed]
- Chauvier, A.; Ajmera, P.; Yadav, R.; Walter, N.G. Dynamic competition between a ligand and transcription factor NusA governs riboswitch-mediated transcription regulation. Proc. Natl. Acad. Sci. USA 2021, 118, e2109026118. [Google Scholar] [CrossRef] [PubMed]
- Strobel, E.J.; Cheng, L.; Berman, K.E.; Carlson, P.D.; Lucks, J.B. A ligand-gated strand displacement mechanism for ZTP riboswitch transcription control. Nat. Chem. Biol. 2019, 15, 1067–1076. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kang, C. Opposite consequences of two transcription pauses caused by an intrinsic terminator oligo(U): Antitermination versus termination by bacteriophage T7 RNA polymerase. J. Biol. Chem. 2011, 286, 15738–15746. [Google Scholar] [CrossRef] [PubMed]
- Mentesana, P.E.; Chin-Bow, S.T.; Sousa, R.; McAllister, W.T. Characterization of halted T7 RNA polymerase elongation complexes reveals multiple factors that contribute to stability. J. Mol. Biol. 2000, 302, 1049–1062. [Google Scholar] [CrossRef]
- Maraia, R.J. Transcription termination factor La is also an initiation factor for RNA polymerase III. Proc. Natl. Acad. Sci. USA 1996, 93, 3383–3387. [Google Scholar] [CrossRef]
- Passalacqua, L.F.M.; Dingilian, A.I.; Lupták, A. Single-pass transcription by T7 RNA polymerase. RNA 2020, 26, 2062–2071. [Google Scholar] [CrossRef]
- Chung, S.; Lerner, E.; Jin, Y.; Kim, S.; Alhadid, Y.; Grimaud, L.W.; Zhang, I.X.; Knobler, C.M.; Gelbart, W.M.; Weiss, S. The effect of macromolecular crowding on single-round transcription by Escherichia coli RNA polymerase. Nucleic Acids Res. 2019, 47, 1440–1450. [Google Scholar] [CrossRef]
- Lim, G.; Hohng, S. Single-molecule fluorescence studies on cotranscriptional G-quadruplex formation coupled with R-loop formation. Nucleic Acids Res. 2020, 48, 9195–9203. [Google Scholar] [CrossRef]
- Liu, Y.; Yu, P.; Dyba, M.; Sousa, R.; Stagno, J.R.; Wang, Y.X. Applications of PLOR in labeling large RNAs at specific sites. Methods 2016, 103, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Holmstrom, E.; Yu, P.; Tan, K.; Zuo, X.; Nesbitt, D.J.; Sousa, R.; Stagno, J.R.; Wang, Y.X. Incorporation of isotopic, fluorescent, and heavy-atom-modified nucleotides into RNAs by position-selective labeling of RNA. Nat. Protoc. 2018, 13, 987–1005. [Google Scholar] [CrossRef]
- Liu, Y.; Holmstrom, E.; Zhang, J.; Yu, P.; Wang, J.; Dyba, M.A.; Chen, D.; Ying, J.; Lockett, S.; Nesbitt, D.J.; et al. Synthesis and applications of RNAs with position-selective labeling and mosaic composition. Nature 2015, 522, 368–372. [Google Scholar] [CrossRef]
- Frieda, K.L.; Block, S.M. Direct observation of cotranscriptional folding in an adenine riboswitch. Science 2012, 338, 397–400. [Google Scholar] [CrossRef] [PubMed]
- Chinnappan, R.; Dubé, A.; Lemay, J.; Lafontaine, D.A. Fluorescence monitoring of riboswitch transcription regulation using a dual molecular beacon assay. Nucleic Acids Res. 2013, 41, e106. [Google Scholar] [CrossRef] [PubMed]
- Lyakhov, D.L.; He, B.; Zhang, X.; Studier, F.W.; Dunn, J.J.; McAllister, W.T. Pausing and termination by bacteriophage T7 RNA polymerase. J. Mol. Biol. 1998, 280, 201–213. [Google Scholar] [CrossRef]
- Mairhofer, J.; Wittwer, A.; Cserjan-Puschmann, M.; Striedner, G. Preventing T7 RNA polymerase read-through transcription-A synthetic termination signal capable of improving bioprocess stability. ACS Synth. Biol. 2015, 4, 265–273. [Google Scholar] [CrossRef]
- Milligan, J.F.; Groebe, D.R.; Witherell, G.W.; Uhlenbeck, O.C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987, 15, 8783–8798. [Google Scholar] [CrossRef]
- McCown, P.J.; Corbino, K.A.; Stav, S.; Sherlock, M.E.; Breaker, R.R. Riboswitch diversity and distribution. RNA 2017, 23, 995–1011. [Google Scholar] [CrossRef]
- Serganov, A.; Nudler, E. A decade of riboswitches. Cell 2013, 152, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Daube, S.S.; von Hippel, P.H. Functional transcription elongation complexes from synthetic RNA-DNA bubble duplexes. Science 1992, 258, 1320–1324. [Google Scholar] [CrossRef] [PubMed]
- Woodson, S.A. Metal ions and RNA folding: A highly charged topic with a dynamic future. Curr. Opin. Chem. Biol. 2005, 9, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Sala, A.; Shoaib, M.; Anufrieva, O.; Mutharasu, G.; Hoque, R.J.; Yli-Harja, O.; Kandhavelu, M. Transcription closed and open complex dynamics studies reveal balance between genetic determinants and co-factors. Phys. Biol. 2015, 12, 036003. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, X.; Hong, L.; Liu, Y. Entropy driving the Mg2+-induced folding of TPP riboswitch. J. Phys. Chem. B 2022, 126, 9457–9464. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chien, P.-Y.; Gao, L.; Liu, Y. Quantitative Analysis of Transcription Termination via Position-Selective Labeling of RNA (PLOR) Method. Int. J. Mol. Sci. 2023, 24, 4934. https://doi.org/10.3390/ijms24054934
Chien P-Y, Gao L, Liu Y. Quantitative Analysis of Transcription Termination via Position-Selective Labeling of RNA (PLOR) Method. International Journal of Molecular Sciences. 2023; 24(5):4934. https://doi.org/10.3390/ijms24054934
Chicago/Turabian StyleChien, Ping-Yi, Lingzhi Gao, and Yu Liu. 2023. "Quantitative Analysis of Transcription Termination via Position-Selective Labeling of RNA (PLOR) Method" International Journal of Molecular Sciences 24, no. 5: 4934. https://doi.org/10.3390/ijms24054934
APA StyleChien, P.-Y., Gao, L., & Liu, Y. (2023). Quantitative Analysis of Transcription Termination via Position-Selective Labeling of RNA (PLOR) Method. International Journal of Molecular Sciences, 24(5), 4934. https://doi.org/10.3390/ijms24054934








