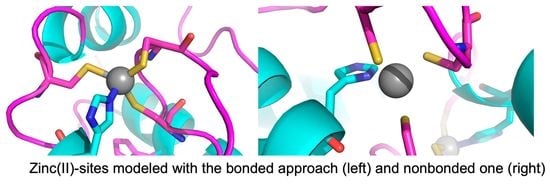
A Comparison of Bonded and Nonbonded Zinc(II) Force Fields with NMR Data
Abstract
:1. Introduction
2. Results
2.1. Background
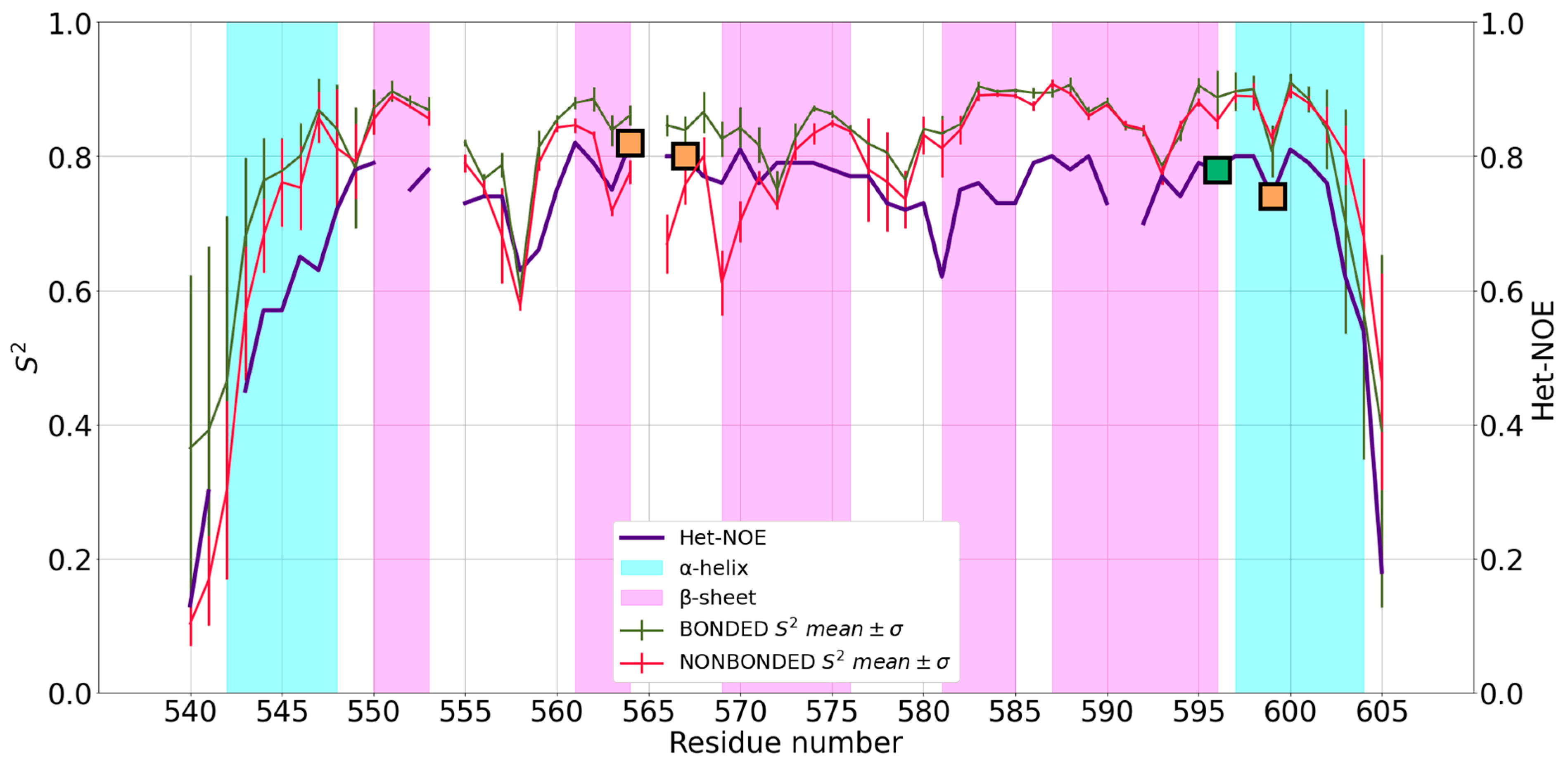
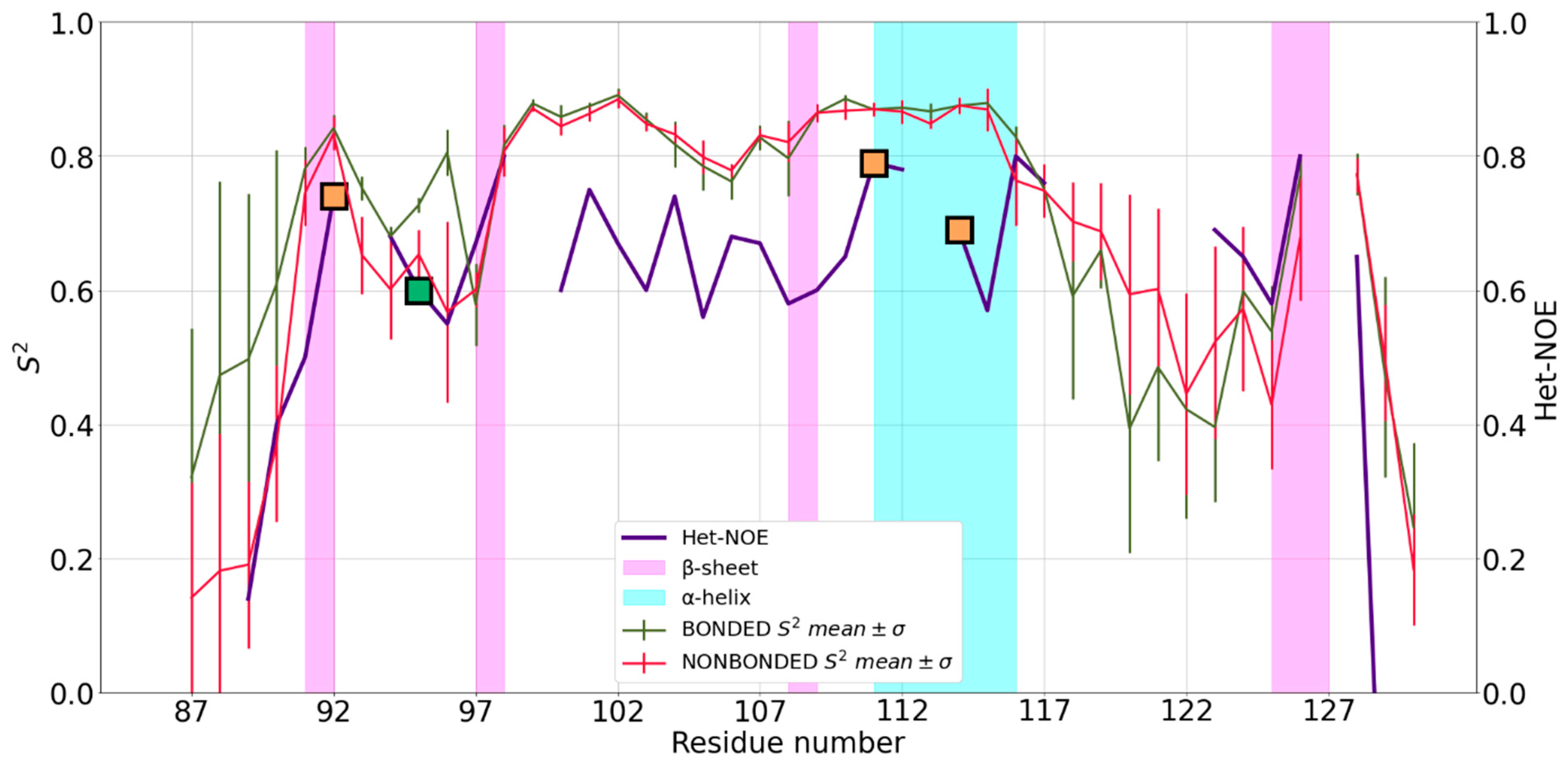
2.2. Analysis of the MD Simulations and Comparison of Simulated vs. Experimental Dynamics

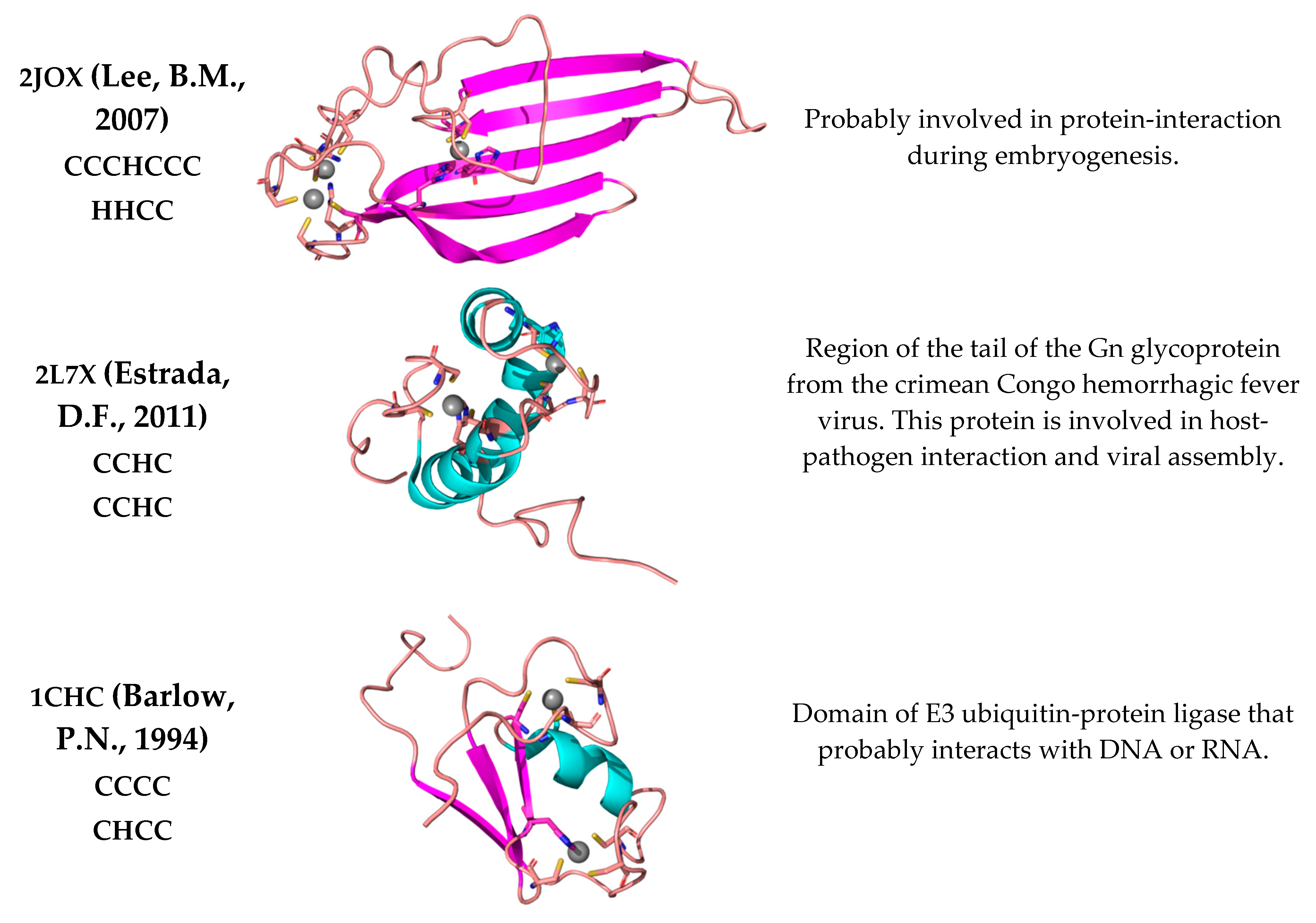
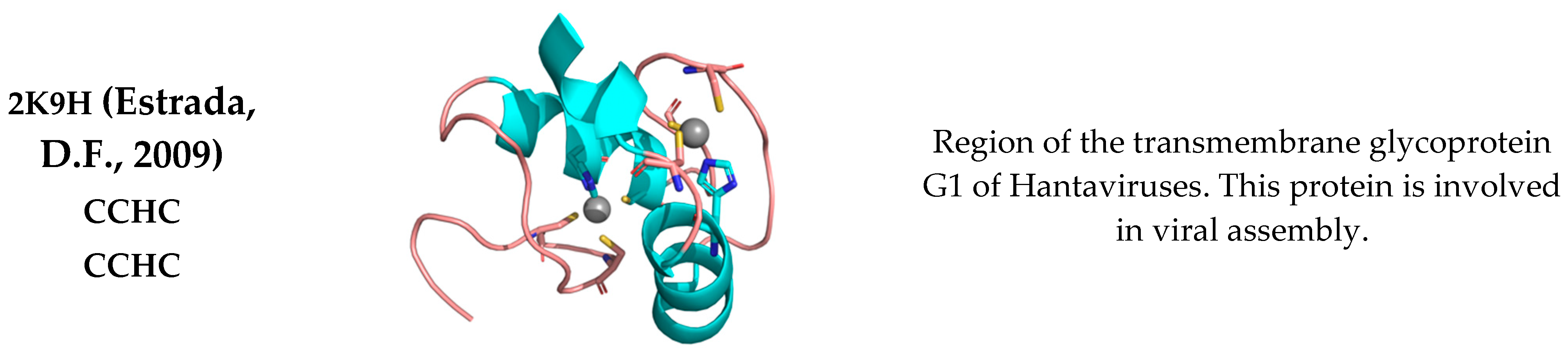
3. Discussion
4. Methods
4.1. Molecular Dynamics Simulations
4.2. Order Parameters
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Andreini, C.; Bertini, I. A bioinformatics view of zinc enzymes. J. Inorg. Biochem. 2012, 111, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Bertini, I.; Sigel, A.; Sigel, H. Handbook on Metalloproteins, 1st ed.; Bertini, I., Sigel, A., Sigel, H., Eds.; Marcel Dekker: New York, NY, USA, 2001; Volume 1. [Google Scholar]
- Frausto da Silva, J.J.R.; Williams, R.J.P. The Biological Chemistry of the Elements: The Inorganic Chemistry of Life; Oxford University Press: New York, NY, USA, 2001. [Google Scholar]
- Boehr, D.D.; Dyson, H.J.; Wright, P.E. An NMR Perspective on Enzyme Dynamics. Chem. Rev. 2006, 106, 3055–3079. [Google Scholar] [CrossRef] [PubMed]
- Palmer, A.G., III. NMR Characterization of the Dynamics of Biomacromolecules. Chem. Rev. 2004, 104, 3623–3640. [Google Scholar] [CrossRef] [PubMed]
- Karplus, M. Molecular dynamics simulations of biomolecules. Acc. Chem. Res. 2002, 35, 321–323. [Google Scholar] [CrossRef] [Green Version]
- Klepeis, J.L.; Lindorff-Larsen, K.; Dror, R.O.; Shaw, D.E. Long-timescale molecular dynamics simulations of protein structure and function. Curr. Opin. Struct. Biol. 2009, 19, 120–127. [Google Scholar] [CrossRef]
- Stone, J.E.; Phillips, J.C.; Freddolino, P.L.; Hardy, D.J.; Trabuco, L.G.; Schulten, K. Accelerating molecular modeling applications with graphics processors. J. Comput. Chem. 2007, 28, 2618–2640. [Google Scholar] [CrossRef] [Green Version]
- Lindorff-Larsen, K.; Maragakis, P.; Piana, S.; Eastwood, M.P.; Dror, R.O.; Shaw, D.E. Systematic validation of protein force fields against experimental data. PLoS ONE 2012, 7, e32131. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Merz, K.M., Jr. Metal Ion Modeling Using Classical Mechanics. Chem. Rev. 2017, 117, 1564–1686. [Google Scholar] [CrossRef]
- Sala, D.; Musiani, F.; Rosato, A. Application of Molecular Dynamics to the Investigation of Metalloproteins Involved in Metal Homeostasis. Eur. J. Inorg. Chem. 2018, 2018, 4661–4677. [Google Scholar] [CrossRef]
- Bayly, C.I.; Cieplak, P.; Cornell, W.D.; Kollman, P.A. A well-behaved elettronics potential based method using charge restraints deriving atomic charges: The RESP model. J. Phys. Chem. 2002, 97, 10269–10280. [Google Scholar] [CrossRef]
- Peters, M.B.; Yang, Y.; Wang, B.; Füsti-Molnár, L.; Weaver, M.N.; Merz, K.M., Jr. Structural Survey of Zinc Containing Proteins and the Development of the Zinc AMBER Force Field (ZAFF). J. Chem. Theory Comput. 2010, 6, 2935–2947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macchiagodena, M.; Pagliai, M.; Andreini, C.; Rosato, A.; Procacci, P. Upgrading and Validation of the AMBER Force Field for Histidine and Cysteine Zinc(II)-Binding Residues in Sites with Four Protein Ligands. J. Chem. Inf. Model. 2019, 59, 3803–3816. [Google Scholar] [CrossRef] [PubMed]
- Macchiagodena, M.; Pagliai, M.; Andreini, C.; Rosato, A.; Procacci, P. Upgraded AMBER Force Field for Zinc-Binding Residues and Ligands for Predicting Structural Properties and Binding Affinities in Zinc-Proteins. ACS Omega 2020, 5, 15301–15310. [Google Scholar] [CrossRef] [PubMed]
- Heinz, H.; Suter, U.W. Atomic Charges for Classical Simulations of Polar Systems. J. Phys. Chem. B 2004, 108, 18341–18352. [Google Scholar] [CrossRef]
- Chaboy, J.; Muñoz-Páez, A.; Merkling, P.J.; Sánchez Marcos, E. The hydration of Cu2+: Can the Jahn-Teller effect be detected in liquid solution? J. Chem. Phys. 2006, 124, 64509. [Google Scholar] [CrossRef] [Green Version]
- Warshel, A.; Levitt, M. Theoretical studies of enzymic reactions: Dielectric, electrostatic and steric stabilization of the carbonium ion in the reaction of lysozyme. J. Mol. Biol. 1976, 103, 227–249. [Google Scholar] [CrossRef]
- Bottaro, S.; Lindorff-Larsen, K. Biophysical experiments and biomolecular simulations: A perfect match? Science 2018, 361, 355–360. [Google Scholar] [CrossRef] [Green Version]
- Kay, L.E.; Torchia, D.A.; Bax, A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: Application to staphylococcal nuclease. Biochemistry 1989, 28, 8972–8979. [Google Scholar] [CrossRef]
- Kluska, K.; Adamczyk, J.; Krężel, A. Metal binding properties, stability and reactivity of zinc fingers. Coord. Chem. Rev. 2018, 367, 18–64. [Google Scholar] [CrossRef]
- Karplus, M.; McCammon, J.A. Molecular dynamics simulations of biomolecules. Nat. Struct. Biol. 2002, 9, 646–652. [Google Scholar] [CrossRef]
- Yang, F.; Hsu, P.; Lee, S.D.; Yang, W.; Hoskinson, D.; Xu, W.; Moore, C.; Varani, G. The C terminus of Pcf11 forms a novel zinc-finger structure that plays an essential role in mRNA 3′-end processing. RNA 2017, 23, 98–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wallenhammar, A.; Anandapadamanaban, M.; Lemak, A.; Mirabello, C.; Lundström, P.; Wallner, B.; Sunnerhagen, M. Solution NMR structure of the TRIM21 B-box2 and identification of residues involved in its interaction with the RING domain. PLoS ONE 2017, 12, e0181551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, B.M.; Buck-Koehntop, B.A.; Martinez-Yamout, M.A.; Dyson, H.J.; Wright, P.E. Embryonic neural inducing factor churchill is not a DNA-binding zinc finger protein: Solution structure reveals a solvent-exposed beta-sheet and zinc binuclear cluster. J. Mol. Biol. 2007, 371, 1274–1289. [Google Scholar] [CrossRef] [Green Version]
- Estrada, D.F.; De Guzman, R.N. Structural characterization of the Crimean-Congo hemorrhagic fever virus Gn tail provides insight into virus assembly. J. Biol. Chem. 2011, 286, 21678–21686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barlow, P.N.; Luisi, B.; Milner, A.; Elliott, M.; Everett, R. Structure of the C3HC4 domain by 1H-nuclear magnetic resonance spectroscopy. A new structural class of zinc-finger. J. Mol. Biol. 1994, 237, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Estrada, D.F.; Boudreaux, D.M.; Zhong, D.; St Jeor, S.C.; De Guzman, R.N. The Hantavirus Glycoprotein G1 Tail Contains Dual CCHC-type Classical Zinc Fingers. J. Biol. Chem. 2009, 284, 8654–8660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prompers, J.J.; Bruschweiler, R. General framework for studying the dynamics of folded and nonfolded proteins by NMR relaxation spectroscopy and MD simulation. J. Am. Chem. Soc. 2002, 124, 4522–4534. [Google Scholar] [CrossRef] [PubMed]
- Showalter, S.A.; Brüschweiler, R. Validation of Molecular Dynamics Simulations of Biomolecules Using NMR Spin Relaxation as Benchmarks: Application to the AMBER99SB Force Field. J. Chem. Theory Comput. 2007, 3, 961–975. [Google Scholar] [CrossRef]
- Ishima, R.; Torchia, D.A. Protein dynamics from NMR. Nat. Struct. Biol. 2000, 7, 740–743. [Google Scholar] [CrossRef]
- Li, C.; Tang, C.; Liu, M. Protein dynamics elucidated by NMR technique. Protein Cell 2013, 4, 726–730. [Google Scholar] [CrossRef] [Green Version]
- Palmer, A.G., III. NMR Probes of Molecular Dynamics: Overview and Comparison with Other Techniques. Ann. Rev. Biophys. Biomol. Struct. 2001, 30, 129–155. [Google Scholar] [CrossRef] [PubMed]
- Lipari, G.; Szabo, A. Model-Free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J. Am. Chem. Soc. 1982, 104, 4546–4559. [Google Scholar] [CrossRef]
- Charlier, C.; Cousin, S.F.; Ferrage, F. Protein dynamics from nuclear magnetic relaxation. Chem. Soc. Rev. 2016, 45, 2410–2422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melse, O.; Antes, I.; Kaila, V.R.I.; Zacharias, M. Benchmarking biomolecular force field-based Zn(2+) for mono- and bimetallic ligand binding sites. J. Comput. Chem. 2023, 44, 912–926. [Google Scholar] [CrossRef] [PubMed]
- Andreini, C.; Cavallaro, G.; Lorenzini, S.; Rosato, A. MetalPDB: A database of metal sites in biological macromolecular structures. Nucleic Acids Res. 2013, 41, D312–D319. [Google Scholar] [CrossRef] [Green Version]
- Putignano, V.; Rosato, A.; Banci, L.; Andreini, C. MetalPDB in 2018: A database of metal sites in biological macromolecular structures. Nucleic Acids Res. 2018, 46, D459–D464. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Merz, K.M., Jr. MCPB.py: A Python Based Metal Center Parameter Builder. J. Chem. Inf. Model. 2016, 56, 599–604. [Google Scholar] [CrossRef]
- Montelione, G.T.; Nilges, M.; Bax, A.; Guntert, P.; Herrmann, T.; Richardson, J.S.; Schwieters, C.D.; Vranken, W.F.; Vuister, G.W.; Wishart, D.S.; et al. Recommendations of the wwPDB NMR Validation Task Force. Structure 2013, 21, 1563–1570. [Google Scholar] [CrossRef] [Green Version]
- Kufareva, I.; Abagyan, R. Methods of protein structure comparison. Methods Mol. Biol. 2012, 857, 231–257. [Google Scholar]
- Roe, D.R.; Cheatham, T.E., III. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- Kabsch, W.; Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef] [PubMed]
- McKinney, W. Data Structures for Statistical Computing in Python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 28 June–3 July 2010. [Google Scholar]
- McKinney, W. Pandas: A Foundational Python Library for Data Analysis and Statistics. Python High Perform. Sci. Comput. 2011, 14, 1–9. [Google Scholar]
- The PyMOL Molecular Graphics System; Version 2.0; Schrödinger, LLC: New York, NY, USA, 2015.
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]


| Zinc-Fingers | Pearson Coefficient for NBFF | Pearson Coefficient for ZAFF |
|---|---|---|
| 2NAX | 0.82 | 0.89 |
| 5JPX | 0.68 | 0.69 |
| 2JOX | 0.77 | n.a. |
| 2L7X | 0.79 | 0.84 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bazayeva, M.; Giachetti, A.; Pagliai, M.; Rosato, A. A Comparison of Bonded and Nonbonded Zinc(II) Force Fields with NMR Data. Int. J. Mol. Sci. 2023, 24, 5440. https://doi.org/10.3390/ijms24065440
Bazayeva M, Giachetti A, Pagliai M, Rosato A. A Comparison of Bonded and Nonbonded Zinc(II) Force Fields with NMR Data. International Journal of Molecular Sciences. 2023; 24(6):5440. https://doi.org/10.3390/ijms24065440
Chicago/Turabian StyleBazayeva, Milana, Andrea Giachetti, Marco Pagliai, and Antonio Rosato. 2023. "A Comparison of Bonded and Nonbonded Zinc(II) Force Fields with NMR Data" International Journal of Molecular Sciences 24, no. 6: 5440. https://doi.org/10.3390/ijms24065440