Role of Adhesion G Protein-Coupled Receptors in Immune Dysfunction and Disorder
Abstract
:1. Introduction
2. ADGRB1/BAI1
2.1. Molecular and Functional Characterization of ADGRB1/BAI1
2.2. Physiopathological role of ADGRB1/BAI1 in the Immune System
3. ADGRE1/EMR1 (Emr1, F4/80)
3.1. Molecular and Functional Characterization of ADGRE1/EMR1
3.2. Physiopathological Role of ADGRE1/EMR1 in the Immune System
4. ADGRE2/EMR2
4.1. Molecular and Functional Characterization of ADGRE2/EMR2
4.2. Physiopathological Role of ADGRE2/EMR2 in the Immune System
5. ADGRE5/CD97
5.1. Molecular and Functional Characterization of ADGRE5/CD97
5.2. Physiopathological Role of ADGRE5/CD97 in the Immune System
6. ADGRG1/GPR56
6.1. Molecular and Functional Characterization of ADGRG1/GPR56
6.2. Physiopathological Role of ADGRG1/GPR56 in the Immune System
7. ADGRG3/GPR97
7.1. Physiopathological Role of Murine ADGRG3/GPR97 in the Immune System
7.2. Physiopathological Role of Human ADGRG3/GPR97 in the Immune System
8. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hamann, J.; Aust, G.; Arac, D.; Engel, F.B.; Formstone, C.; Fredriksson, R.; Hall, R.A.; Harty, B.L.; Kirchhoff, C.; Knapp, B.; et al. International Union of Basic and Clinical Pharmacology. XCIV. Adhesion G protein-coupled receptors. Pharmacol. Rev. 2015, 67, 338–367. [Google Scholar] [CrossRef] [PubMed]
- Morgan, R.K.; Anderson, G.R.; Arac, D.; Aust, G.; Balenga, N.; Boucard, A.; Bridges, J.P.; Engel, F.B.; Formstone, C.J.; Glitsch, M.D.; et al. The expanding functional roles and signaling mechanisms of adhesion G protein-coupled receptors. Ann. N. Y. Acad. Sci. 2019, 1456, 5–25. [Google Scholar] [CrossRef]
- Kishore, A.; Hall, R.A. Versatile Signaling Activity of Adhesion GPCRs. Handb. Exp. Pharmacol. 2016, 234, 127–146. [Google Scholar] [PubMed]
- Lin, H.H.; Chang, G.W.; Davies, J.Q.; Stacey, M.; Harris, J.; Gordon, S. Autocatalytic cleavage of the EMR2 receptor occurs at a conserved G protein-coupled receptor proteolytic site motif. J. Biol. Chem. 2004, 279, 31823–31832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arac, D.; Boucard, A.A.; Bolliger, M.F.; Nguyen, J.; Soltis, S.M.; Sudhof, T.C.; Brunger, A.T. A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis. EMBO J. 2012, 31, 1364–1378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Promel, S.; Langenhan, T.; Arac, D. Matching structure with function: The GAIN domain of adhesion-GPCR and PKD1-like proteins. Trends Pharmacol. Sci. 2013, 34, 470–478. [Google Scholar] [CrossRef]
- Liebscher, I.; Schon, J.; Petersen, S.C.; Fischer, L.; Auerbach, N.; Demberg, L.M.; Mogha, A.; Coster, M.; Simon, K.U.; Rothemund, S.; et al. A Tethered Agonist within the Ectodomain Activates the Adhesion G Protein-Coupled Receptors GPR126 and GPR133. Cell Rep. 2015, 10, 1021. [Google Scholar] [CrossRef] [Green Version]
- Stoveken, H.M.; Hajduczok, A.G.; Xu, L.; Tall, G.G. Adhesion G protein-coupled receptors are activated by exposure of a cryptic tethered agonist. Proc. Natl. Acad. Sci. USA 2015, 112, 6194–6199. [Google Scholar] [CrossRef] [Green Version]
- Liebscher, I.; Schoneberg, T. Tethered Agonism: A Common Activation Mechanism of Adhesion GPCRs. Handb. Exp. Pharmacol. 2016, 234, 111–125. [Google Scholar]
- Lin, H.H.; Ng, K.F.; Chen, T.C.; Tseng, W.Y. Ligands and Beyond: Mechanosensitive Adhesion GPCRs. Pharmaceuticals 2022, 15, 219. [Google Scholar] [CrossRef]
- Wilde, C.; Mitgau, J.; Suchy, T.; Schoneberg, T.; Liebscher, I. Translating the force-mechano-sensing GPCRs. Am. J. Physiol. Cell Physiol. 2022, 322, C1047–C1060. [Google Scholar] [CrossRef]
- Lala, T.; Hall, R.A. Adhesion G protein-coupled receptors: Structure. signaling, physiology, and pathophysiology, Physiol. Rev. 2022, 102, 1587–1624. [Google Scholar] [PubMed]
- Hamann, J.; Hsiao, C.C.; Lee, C.S.; Ravichandran, K.S.; Lin, H.H. Adhesion GPCRs as Modulators of Immune Cell Function. Handb. Exp. Pharmacol. 2016, 234, 329–350. [Google Scholar] [PubMed]
- Lin, H.H.; Hsiao, C.C.; Pabst, C.; Hebert, J.; Schoneberg, T.; Hamann, J. Adhesion GPCRs in Regulating Immune Responses and Inflammation. Adv. Immunol. 2017, 136, 163–201. [Google Scholar]
- Park, D.; Ravichandran, K.S. Emerging roles of brain-specific angiogenesis inhibitor 1. Adv. Exp. Med. Biol. 2010, 706, 167–178. [Google Scholar]
- Cork, S.M.; Van Meir, E.G. Emerging roles for the BAI1 protein family in the regulation of phagocytosis. synaptogenesis, neurovasculature, and tumor development. J. Mol. Med. 2011, 89, 743–752. [Google Scholar] [CrossRef] [Green Version]
- Stephenson, J.R.; Purcell, R.H.; Hall, R.A. The BAI subfamily of adhesion GPCRs: Synaptic regulation and beyond. Trends Pharmacol. Sci. 2014, 35, 208–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishimori, H.; Shiratsuchi, T.; Urano, T.; Kimura, Y.; Kiyono, K.; Tatsumi, K.; Yoshida, S.; Ono, M.; Kuwano, M.; Nakamura, Y.; et al. A novel brain-specific p53-target gene, BAI1, containing thrombospondin type 1 repeats inhibits experimental angiogenesis. Oncogene 1997, 15, 2145–2150. [Google Scholar]
- Kaur, B.; Brat, D.J.; Calkins, C.C.; Van Meir, E.G. Brain angiogenesis inhibitor 1 is differentially expressed in normal brain and glioblastoma independently of p53 expression. Am. J. Pathol. 2003, 162, 19–27. [Google Scholar] [CrossRef] [Green Version]
- Kaur, B.; Brat, D.J.; Devi, N.S.; Van Meir, E.G. Vasculostatin. a proteolytic fragment of brain angiogenesis inhibitor 1, is an antiangiogenic and antitumorigenic factor, Oncogene 2005, 24, 3632–3642. [Google Scholar]
- Kaur, B.; Cork, S.M.; Sandberg, E.M.; Devi, N.S.; Zhang, Z.; Klenotic, P.A.; Febbraio, M.; Shim, H.; Mao, H.; Tucker-Burden, C.; et al. Vasculostatin inhibits intracranial glioma growth and negatively regulates in vivo angiogenesis through a CD36-dependent mechanism. Cancer Res. 2009, 69, 1212–1220. [Google Scholar] [CrossRef] [Green Version]
- Cork, S.M.; Kaur, B.; Devi, N.S.; Cooper, L.; Saltz, J.H.; Sandberg, E.M.; Kaluz, S.; Van Meir, E.G. A proprotein convertase/MMP-14 proteolytic cascade releases a novel 40 kDa vasculostatin from tumor suppressor BAI1. Oncogene 2012, 31, 5144–5152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, D.; Tosello-Trampont, A.C.; Elliott, M.R.; Lu, M.; Haney, L.B.; Ma, Z.; Klibanov, A.L.; Mandell, J.W.; Ravichandran, K.S. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature 2007, 450, 430–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, S.; Owen, K.A.; Ly, K.T.; Park, D.; Black, S.G.; Wilson, J.M.; Sifri, C.D.; Ravichandran, K.S.; Ernst, P.B.; Casanova, J.E. Brain angiogenesis inhibitor 1 (BAI1) is a pattern recognition receptor that mediates macrophage binding and engulfment of Gram-negative bacteria. Proc. Natl. Acad. Sci. USA 2011, 108, 2136–2141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koh, J.T.; Kook, H.; Kee, H.J.; Seo, Y.W.; Jeong, B.C.; Lee, J.H.; Kim, M.Y.; Yoon, K.C.; Jung, S.; Kim, K.K. Extracellular fragment of brain-specific angiogenesis inhibitor 1 suppresses endothelial cell proliferation by blocking alphavbeta5 integrin. Exp. Cell Res. 2004, 294, 172–184. [Google Scholar] [CrossRef]
- Benavente, F.; Piltti, K.M.; Hooshmand, M.J.; Nava, A.A.; Lakatos, A.; Feld, B.G.; Creasman, D.; Gershon, P.D.; Anderson, A. Novel C1q receptor-mediated signaling controls neural stem cell behavior and neurorepair. Elife 2020, 9, e55732. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Miao, Y.; Wicklein, R.; Sun, Z.; Wang, J.; Jude, K.M.; Fernandes, R.A.; Merrill, S.A.; Wernig, M.; Garcia, K.C.; et al. RTN4/NoGo-receptor binding to BAI adhesion-GPCRs regulates neuronal development. Cell 2021, 184, 5869–5885.e25. [Google Scholar] [CrossRef] [PubMed]
- Duman, J.G.; Tzeng, C.P.; Tu, Y.K.; Munjal, T.; Schwechter, B.; Ho, T.S.; Tolias, K.F. The adhesion-GPCR BAI1 regulates synaptogenesis by controlling the recruitment of the Par3/Tiam1 polarity complex to synaptic sites. J. Neurosci. 2013, 33, 6964–6978. [Google Scholar] [CrossRef] [Green Version]
- Zhu, D.; Li, C.; Swanson, A.M.; Villalba, R.M.; Guo, J.; Zhang, Z.; Matheny, S.; Murakami, T.; Stephenson, J.R.; Daniel, S.; et al. BAI1 regulates spatial learning and synaptic plasticity in the hippocampus. J. Clin. Investig. 2015, 125, 1497–1508. [Google Scholar] [CrossRef] [Green Version]
- Rosenblum, M.D.; Remedios, K.A.; Abbas, A.K. Mechanisms of human autoimmunity. J. Clin. Investig. 2015, 125, 2228–2233. [Google Scholar] [CrossRef] [Green Version]
- Das, S.; Sarkar, A.; Ryan, K.A.; Fox, S.; Berger, A.H.; Juncadella, I.J.; Bimczok, D.; Smythies, L.E.; Harris, P.R.; Ravichandran, K.S.; et al. Brain angiogenesis inhibitor 1 is expressed by gastric phagocytes during infection with Helicobacter pylori and mediates the recognition and engulfment of human apoptotic gastric epithelial cells. FASEB J. 2014, 28, 2214–2224. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.S.; Penberthy, K.K.; Wheeler, K.M.; Juncadella, I.J.; Vandenabeele, P.; Lysiak, J.J.; Ravichandran, K.S. Boosting Apoptotic Cell Clearance by Colonic Epithelial Cells Attenuates Inflammation In Vivo. Immunity 2016, 44, 807–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fond, A.M.; Lee, C.S.; Schulman, I.G.; Kiss, R.S.; Ravichandran, K.S. Apoptotic cells trigger a membrane-initiated pathway to increase ABCA1. J. Clin. Investig. 2015, 125, 2748–2758. [Google Scholar] [CrossRef] [PubMed]
- Emerging Risk Factors, C.; Di Angelantonio, E.; Sarwar, N.; Perry, P.; Kaptoge, S.; Ray, K.K.; Thompson, A.; Wood, A.M.; Lewington, S.; Sattar, N.; et al. Major lipids. apolipoproteins, and risk of vascular disease. JAMA 2009, 302, 1993–2000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazaheri, F.; Breus, O.; Durdu, S.; Haas, P.; Wittbrodt, J.; Gilmour, D.; Peri, F. Distinct roles for BAI1 and TIM-4 in the engulfment of dying neurons by microglia. Nat. Commun. 2014, 5, 4046. [Google Scholar] [CrossRef] [Green Version]
- Billings, E.A.; Lee, C.S.; Owen, K.A.; D’Souza, R.S.; Ravichandran, K.S.; Casanova, J.E. The adhesion GPCR BAI1 mediates macrophage ROS production and microbicidal activity against Gram-negative bacteria. Sci. Signal. 2016, 9, ra14. [Google Scholar] [CrossRef] [Green Version]
- Bolyard, C.; Meisen, W.H.; Banasavadi-Siddegowda, Y.; Hardcastle, J.; Yoo, J.Y.; Wohleb, E.S.; Wojton, J.; Yu, J.G.; Dubin, S.; Khosla, M.; et al. BAI1 Orchestrates Macrophage Inflammatory Response to HSV Infection-Implications for Oncolytic Viral Therapy. Clin. Cancer Res. 2017, 23, 1809–1819. [Google Scholar] [CrossRef] [Green Version]
- Yuan, S.; Wang, X.; Hou, S.; Guo, T.; Lan, Y.; Yang, S.; Zhao, F.; Gao, J.; Wang, Y.; Chu, Y.; et al. PHF6 and JAK3 mutations cooperate to drive T-cell acute lymphoblastic leukemia progression. Leukemia 2022, 36, 370–382. [Google Scholar] [CrossRef]
- Zhu, D.; Osuka, S.; Zhang, Z.; Reichert, Z.R.; Yang, L.; Kanemura, Y.; Jiang, Y.; You, S.; Zhang, H.; Devi, N.S.; et al. BAI1 Suppresses Medulloblastoma Formation by Protecting p53 from Mdm2-Mediated Degradation. Cancer Cell. 2018, 33, 1004–1016.e5. [Google Scholar] [CrossRef] [Green Version]
- Hsiao, C.C.; van der Poel, M.; van Ham, T.J.; Hamann, J. Macrophages Do Not Express the Phagocytic Receptor BAI1/ADGRB1. Front. Immunol. 2019, 10, 962. [Google Scholar] [CrossRef] [Green Version]
- Austyn, J.M.; Gordon, S. F4/80. a monoclonal antibody directed specifically against the mouse macrophage. Eur. J. Immunol. 1981, 11, 805–815. [Google Scholar] [CrossRef]
- Lin, H.H.; Stacey, M.; Stein-Streilein, J.; Gordon, S. F4/80: The macrophage-specific adhesion-GPCR and its role in immunoregulation. Adv. Exp. Med. Biol. 2010, 706, 149–156. [Google Scholar] [PubMed]
- Gordon, S.; Hamann, J.; Lin, H.H.; Stacey, M. F4/80 and the related adhesion-GPCRs. Eur. J. Immunol. 2011, 41, 2472–2476. [Google Scholar] [CrossRef] [PubMed]
- McGarry, M.P.; Stewart, C.C. Murine eosinophil granulocytes bind the murine macrophage-monocyte specific monoclonal antibody F4/80. J. Leukoc Biol. 1991, 50, 471–478. [Google Scholar] [CrossRef]
- Taylor, P.R.; Martinez-Pomares, L.; Stacey, M.; Lin, H.H.; Brown, G.D.; Gordon, S. Macrophage receptors and immune recognition. Annu. Rev. Immunol. 2005, 23, 901–944. [Google Scholar] [CrossRef]
- Qian, B.Z.; Pollard, J.W. Macrophage diversity enhances tumor progression and metastasis. Cell 2010, 141, 39–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKnight, A.J.; Gordon, S. EGF-TM7: A novel subfamily of seven-transmembrane-region leukocyte cell-surface molecules. Immunol. Today 1996, 17, 283–287. [Google Scholar] [CrossRef]
- McKnight, A.J.; Gordon, S. The EGF-TM7 family: Unusual structures at the leukocyte surface. J. Leukoc. Biol. 1998, 63, 271–280. [Google Scholar] [CrossRef]
- Baud, V.; Chissoe, S.L.; Viegas-Pequignot, E.; Diriong, S.; N’Guyen, V.C.; Roe, B.A.; Lipinski, M. EMR1. an unusual member in the family of hormone receptors with seven transmembrane segments, Genomics 1995, 26, 334–344. [Google Scholar]
- McKnight, A.J.; Macfarlane, A.J.; Dri, P.; Turley, L.; Willis, A.C.; Gordon, S. Molecular cloning of F4/80. a murine macrophage-restricted cell surface glycoprotein with homology to the G-protein-linked transmembrane 7 hormone receptor family. J. Biol. Chem. 1996, 271, 486–489. [Google Scholar] [CrossRef] [Green Version]
- Lin, H.H.; Stubbs, L.J.; Mucenski, M.L. Identification and characterization of a seven transmembrane hormone receptor using differential display. Genomics 1997, 41, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Warschkau, H.; Kiderlen, A.F. A monoclonal antibody directed against the murine macrophage surface molecule F4/80 modulates natural immune response to Listeria monocytogenes. J. Immunol. 1999, 163, 3409–3416. [Google Scholar] [CrossRef] [PubMed]
- Schaller, E.; Macfarlane, A.J.; Rupec, R.A.; Gordon, S.; McKnight, A.J.; Pfeffer, K. Inactivation of the F4/80 glycoprotein in the mouse germ line. Mol. Cell. Biol. 2002, 22, 8035–8043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, H.H.; Faunce, D.E.; Stacey, M.; Terajewicz, A.; Nakamura, T.; Zhang-Hoover, J.; Kerley, M.; Mucenski, M.L.; Gordon, S.; Stein-Streilein, J. The macrophage F4/80 receptor is required for the induction of antigen-specific efferent regulatory T cells in peripheral tolerance. J. Exp. Med. 2005, 201, 1615–1625. [Google Scholar] [CrossRef] [Green Version]
- Hamann, J.; Koning, N.; Pouwels, W.; Ulfman, L.H.; van Eijk, M.; Stacey, M.; Lin, H.H.; Gordon, S.; Kwakkenbos, M.J. EMR1. the human homolog of F4/80, is an eosinophil-specific receptor. Eur. J. Immunol. 2007, 37, 2797–2802. [Google Scholar] [CrossRef]
- Legrand, F.; Tomasevic, N.; Simakova, O.; Lee, C.C.; Wang, Z.; Raffeld, M.; Makiya, M.A.; Palath, V.; Leung, J.; Baer, M.; et al. The eosinophil surface receptor epidermal growth factor-like module containing mucin-like hormone receptor 1 (EMR1): A novel therapeutic target for eosinophilic disorders. J. Allergy Clin. Immunol. 2014, 133, 1439–1447.e8. [Google Scholar] [CrossRef] [Green Version]
- Waddell, L.A.; Lefevre, L.; Bush, S.J.; Raper, A.; Young, R.; Lisowski, Z.M.; McCulloch, M.E.B.; Muriuki, C.; Sauter, K.A.; Clark, E.L.; et al. ADGRE1 (EMR1. F4/80) Is a Rapidly-Evolving Gene Expressed in Mammalian Monocyte-Macrophages. Front. Immunol. 2018, 9, 2246. [Google Scholar] [CrossRef] [Green Version]
- Lin, H.H.; Stacey, M.; Hamann, J.; Gordon, S.; McKnight, A.J. Human EMR2. a novel EGF-TM7 molecule on chromosome 19p13.1, is closely related to CD97. Genomics 2000, 67, 188–200. [Google Scholar] [CrossRef]
- Kwakkenbos, M.J.; Kop, E.N.; Stacey, M.; Matmati, M.; Gordon, S.; Lin, H.H.; Hamann, J. The EGF-TM7 family: A postgenomic view. Immunogenetics 2004, 55, 655–666. [Google Scholar]
- Stacey, M.; Lin, H.H.; Hilyard, K.L.; Gordon, S.; McKnight, A.J. Human epidermal growth factor (EGF) module-containing mucin-like hormone receptor 3 is a new member of the EGF-TM7 family that recognizes a ligand on human macrophages and activated neutrophils. J. Biol. Chem. 2001, 276, 18863–18870. [Google Scholar] [CrossRef] [Green Version]
- Kwakkenbos, M.J.; Matmati, M.; Madsen, O.; Pouwels, W.; Wang, Y.; Bontrop, R.E.; Heidt, P.J.; Hoek, R.M.; Hamann, J. An unusual mode of concerted evolution of the EGF-TM7 receptor chimera EMR2. FASEB J. 2006, 20, 2582–2584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stacey, M.; Chang, G.W.; Davies, J.Q.; Kwakkenbos, M.J.; Sanderson, R.D.; Hamann, J.; Gordon, S.; Lin, H.H. The epidermal growth factor-like domains of the human EMR2 receptor mediate cell attachment through chondroitin sulfate glycosaminoglycans. Blood 2003, 102, 2916–2924. [Google Scholar] [CrossRef]
- Lin, H.H.; Stacey, M.; Saxby, C.; Knott, V.; Chaudhry, Y.; Evans, D.; Gordon, S.; McKnight, A.J.; Handford, P.; Lea, S. Molecular analysis of the epidermal growth factor-like short consensus repeat domain-mediated protein-protein interactions: Dissection of the CD97-CD55 complex. J. Biol. Chem. 2001, 276, 24160–24169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwakkenbos, M.J.; Chang, G.W.; Lin, H.H.; Pouwels, W.; de Jong, E.C.; van Lier, R.A.; Gordon, S.; Hamann, J. The human EGF-TM7 family member EMR2 is a heterodimeric receptor expressed on myeloid cells. J. Leukoc. Biol. 2002, 71, 854–862. [Google Scholar] [CrossRef]
- Chang, G.W.; Davies, J.Q.; Stacey, M.; Yona, S.; Bowdish, D.M.; Hamann, J.; Chen, T.C.; Lin, C.Y.; Gordon, S.; Lin, H.H. CD312. the human adhesion-GPCR EMR2, is differentially expressed during differentiation, maturation, and activation of myeloid cells. Biochem. Biophys. Res. Commun. 2007, 353, 133–138. [Google Scholar] [CrossRef]
- Van Eijk, M.; Aust, G.; Brouwer, M.S.; van Meurs, M.; Voerman, J.S.; Dijke, I.E.; Pouwels, W.; Sandig, I.; Wandel, E.; Aerts, J.M.; et al. Differential expression of the EGF-TM7 family members CD97 and EMR2 in lipid-laden macrophages in atherosclerosis. multiple sclerosis and Gaucher disease. Immunol. Lett. 2010, 129, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Yona, S.; Lin, H.H.; Dri, P.; Davies, J.Q.; Hayhoe, R.P.; Lewis, S.M.; Heinsbroek, S.E.; Brown, K.A.; Perretti, M.; Hamann, J.; et al. Ligation of the adhesion-GPCR EMR2 regulates human neutrophil function. FASEB J. 2008, 22, 741–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, T.Y.; Hwang, T.L.; Lin, C.Y.; Lin, T.N.; Lai, H.Y.; Tsai, W.P.; Lin, H.H. EMR2 receptor ligation modulates cytokine secretion profiles and cell survival of lipopolysaccharide-treated neutrophils. Chang Gung Med. J. 2011, 34, 468–477. [Google Scholar]
- Lewis, S.M.; Treacher, D.F.; Edgeworth, J.; Mahalingam, G.; Brown, C.S.; Mare, T.A.; Stacey, M.; Beale, R.; Brown, K.A. Expression of CD11c and EMR2 on neutrophils: Potential diagnostic biomarkers for sepsis and systemic inflammation. Clin. Exp. Immunol. 2015, 182, 184–194. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.H.; Jeng, W.J.; Ho, Y.P.; Teng, W.; Hsieh, Y.C.; Chen, W.T.; Chen, Y.C.; Lin, H.H.; Sheen, I.S.; Lin, C.Y. Increased EMR2 expression on neutrophils correlates with disease severity and predicts overall mortality in cirrhotic patients. Sci. Rep. 2016, 6, 38250. [Google Scholar] [CrossRef] [Green Version]
- Kop, E.N.; Kwakkenbos, M.J.; Teske, G.J.; Kraan, M.C.; Smeets, T.J.; Stacey, M.; Lin, H.H.; Tak, P.P.; Hamann, J. Identification of the epidermal growth factor-TM7 receptor EMR2 and its ligand dermatan sulfate in rheumatoid synovial tissue. Arthritis Rheumatol. 2005, 52, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Boyden, S.E.; Desai, A.; Cruse, G.; Young, M.L.; Bolan, H.C.; Scott, L.M.; Eisch, A.R.; Long, R.D.; Lee, C.C.; Satorius, C.L.; et al. Vibratory Urticaria Associated with a Missense Variant in ADGRE2. N. Engl. J. Med. 2016, 374, 656–663. [Google Scholar] [CrossRef]
- Naranjo, A.N.; Bandara, G.; Bai, Y.; Smelkinson, M.G.; Tobio, A.; Komarow, H.D.; Boyden, S.E.; Kastner, D.L.; Metcalfe, D.D.; Olivera, A. Critical Signaling Events in the Mechanoactivation of Human Mast Cells through p.C492Y-ADGRE2. J. Investig. Dermatol. 2020, 140, 2210–2220.e5. [Google Scholar] [CrossRef]
- Le, Q.T.; Lyons, J.J.; Naranjo, A.N.; Olivera, A.; Lazarus, R.A.; Metcalfe, D.D.; Milner, J.D.; Schwartz, L.B. Impact of naturally forming human alpha/beta-tryptase heterotetramers in the pathogenesis of hereditary alpha-tryptasemia. J. Exp. Med. 2019, 216, 2348–2361. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.S.; Chiang, N.Y.; Hu, C.H.; Hsiao, C.C.; Cheng, K.F.; Tsai, W.P.; Yona, S.; Stacey, M.; Gordon, S.; Chang, G.W.; et al. Activation of myeloid cell-specific adhesion class G protein-coupled receptor EMR2 via ligation-induced translocation and interaction of receptor subunits in lipid raft microdomains. Mol. Cell. Biol. 2012, 32, 1408–1420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.S.; Hu, C.H.; Tseng, W.Y.; Cheng, C.H.; Stacey, M.; Gordon, S.; Chang, G.W.; Lin, H.H. Activation of Adhesion GPCR EMR2/ADGRE2 Induces Macrophage Differentiation and Inflammatory Responses via Galpha(16)/Akt/MAPK/NF-kappaB Signaling Pathways. Front Immunol. 2017, 8, 373. [Google Scholar]
- Tseng, W.Y.; Wang, W.C.; Gordon, S.; Ng, K.F.; Lin, H.H. Stimulation of Vibratory Urticaria-Associated Adhesion-GPCR. EMR2/ADGRE2, Triggers the NLRP3 Inflammasome Activation Signal in Human Monocytes. Front. Immunol. 2020, 11, 602016. [Google Scholar]
- Irmscher, S.; Brix, S.R.; Zipfel, S.L.H.; Halder, L.D.; Mutluturk, S.; Wulf, S.; Girdauskas, E.; Reichenspurner, H.; Stahl, R.A.K.; Jungnickel, B.; et al. Serum FHR1 binding to necrotic-type cells activates monocytic inflammasome and marks necrotic sites in vasculopathies. Nat. Commun. 2019, 10, 2961. [Google Scholar] [CrossRef] [Green Version]
- Gray, J.X.; Haino, M.; Roth, M.J.; Maguire, J.E.; Jensen, P.N.; Yarme, A.; Stetler-Stevenson, M.A.; Siebenlist, U.; Kelly, K. CD97 is a processed. seven-transmembrane, heterodimeric receptor associated with inflammation. J. Immunol. 1996, 157, 5438–5447. [Google Scholar] [CrossRef]
- Hamann, J.; Vogel, B.; van Schijndel, G.M.; van Lier, R.A. The seven-span transmembrane receptor CD97 has a cellular ligand (CD55. DAF). J. Exp. Med. 1996, 184, 1185–1189. [Google Scholar] [CrossRef]
- Wang, T.; Ward, Y.; Tian, L.; Lake, R.; Guedez, L.; Stetler-Stevenson, W.G.; Kelly, K. CD97. an adhesion receptor on inflammatory cells, stimulates angiogenesis through binding integrin counterreceptors on endothelial cells. Blood 2005, 105, 2836–2844. [Google Scholar] [CrossRef] [PubMed]
- Wandel, E.; Saalbach, A.; Sittig, D.; Gebhardt, C.; Aust, G. Thy-1 (CD90) is an interacting partner for CD97 on activated endothelial cells. J. Immunol. 2012, 188, 1442–1450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, Y.; Lake, R.; Yin, J.J.; Heger, C.D.; Raffeld, M.; Goldsmith, P.K.; Merino, M.; Kelly, K. LPA receptor heterodimerizes with CD97 to amplify LPA-initiated RHO-dependent signaling and invasion in prostate cancer cells. Cancer Res. 2011, 71, 7301–7311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamann, J.; Eichler, W.; Hamann, D.; Kerstens, H.M.; Poddighe, P.J.; Hoovers, J.M.; Hartmann, E.; Strauss, M.; van Lier, R.A. Expression cloning and chromosomal mapping of the leukocyte activation antigen CD97. a new seven-span transmembrane molecule of the secretion receptor superfamily with an unusual extracellular domain. J. Immunol. 1995, 155, 1942–1950. [Google Scholar] [CrossRef] [PubMed]
- Eichler, W.; Aust, G.; Hamann, D. Characterization of an early activation-dependent antigen on lymphocytes defined by the monoclonal antibody BL-Ac(F2). Scand J. Immunol. 1994, 39, 111–115. [Google Scholar] [CrossRef]
- Van Pel, M.; Hagoort, H.; Hamann, J.; Fibbe, W.E. CD97 is differentially expressed on murine hematopoietic stem-and progenitor-cells. Haematologica 2008, 93, 1137–1144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kop, E.N.; Matmati, M.; Pouwels, W.; Leclercq, G.; Tak, P.P.; Hamann, J. Differential expression of CD97 on human lymphocyte subsets and limited effect of CD97 antibodies on allogeneic T-cell stimulation. Immunol. Lett. 2009, 123, 160–168. [Google Scholar] [CrossRef]
- Jaspars, L.H.; Vos, W.; Aust, G.; Van Lier, R.A.; Hamann, J. Tissue distribution of the human CD97 EGF-TM7 receptor. Tissue Antigens 2001, 57, 325–331. [Google Scholar] [CrossRef]
- Aust, G.; Wandel, E.; Boltze, C.; Sittig, D.; Schutz, A.; Horn, L.C.; Wobus, M. Diversity of CD97 in smooth muscle cells. Cell Tissue Res. 2006, 324, 139–147. [Google Scholar] [CrossRef]
- Aust, G.; Zhu, D.; Van Meir, E.G.; Xu, L. Adhesion GPCRs in Tumorigenesis. Handb. Exp. Pharmacol. 2016, 234, 369–396. [Google Scholar]
- Kwakkenbos, M.J.; Pouwels, W.; Matmati, M.; Stacey, M.; Lin, H.H.; Gordon, S.; van Lier, R.A.; Hamann, J. Expression of the largest CD97 and EMR2 isoforms on leukocytes facilitates a specific interaction with chondroitin sulfate on B cells. J. Leukoc. Biol. 2005, 77, 112–119. [Google Scholar] [CrossRef] [Green Version]
- Hamann, J.; Wishaupt, J.O.; van Lier, R.A.; Smeets, T.J.; Breedveld, F.C.; Tak, P.P. Expression of the activation antigen CD97 and its ligand CD55 in rheumatoid synovial tissue. Arthritis Rheumatol. 1999, 42, 650–658. [Google Scholar] [CrossRef]
- Visser, L.; de Vos, A.F.; Hamann, J.; Melief, M.J.; van Meurs, M.; van Lier, R.A.; Laman, J.D.; Hintzen, R.Q. Expression of the EGF-TM7 receptor CD97 and its ligand CD55 (DAF) in multiple sclerosis. J. Neuroimmunol. 2002, 132, 156–163. [Google Scholar] [CrossRef]
- Leemans, J.C.; te Velde, A.A.; Florquin, S.; Bennink, R.J.; de Bruin, K.; van Lier, R.A.; van der Poll, T.; Hamann, J. The epidermal growth factor-seven transmembrane (EGF-TM7) receptor CD97 is required for neutrophil migration and host defense. J. Immunol. 2004, 172, 1125–1131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kop, E.N.; Adriaansen, J.; Smeets, T.J.; Vervoordeldonk, M.J.; van Lier, R.A.; Hamann, J.; Tak, P.P. CD97 neutralisation increases resistance to collagen-induced arthritis in mice. Arthritis Res. Ther. 2006, 8, R155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Pel, M.; Hagoort, H.; Kwakkenbos, M.J.; Hamann, J.; Fibbe, W.E. Differential role of CD97 in interleukin-8-induced and granulocyte-colony stimulating factor-induced hematopoietic stem and progenitor cell mobilization. Haematologica 2008, 93, 601–604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamann, J.; Veninga, H.; de Groot, D.M.; Visser, L.; Hofstra, C.L.; Tak, P.P.; Laman, J.D.; Boots, A.M.; van Eenennaam, H. CD97 in leukocyte trafficking. Adv. Exp. Med. Biol. 2010, 706, 128–137. [Google Scholar]
- Veninga, H.; de Groot, D.M.; McCloskey, N.; Owens, B.M.; Dessing, M.C.; Verbeek, J.S.; Nourshargh, S.; van Eenennaam, H.; Boots, A.M.; Hamann, J. CD97 antibody depletes granulocytes in mice under conditions of acute inflammation via a Fc receptor-dependent mechanism. J. Leukoc. Biol. 2011, 89, 413–421. [Google Scholar] [CrossRef]
- Veninga, H.; Becker, S.; Hoek, R.M.; Wobus, M.; Wandel, E.; van der Kaa, J.; van der Valk, M.; de Vos, A.F.; Haase, H.; Owens, B.; et al. Analysis of CD97 expression and manipulation: Antibody treatment but not gene targeting curtails granulocyte migration. J. Immunol. 2008, 181, 6574–6583. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Tian, L.; Haino, M.; Gao, J.L.; Lake, R.; Ward, Y.; Wang, H.; Siebenlist, U.; Murphy, P.M.; Kelly, K. Improved antibacterial host defense and altered peripheral granulocyte homeostasis in mice lacking the adhesion class G protein receptor CD97. Infect. Immun. 2007, 75, 1144–1153. [Google Scholar] [CrossRef] [Green Version]
- Veninga, H.; Hoek, R.M.; de Vos, A.F.; de Bruin, A.M.; An, F.Q.; van der Poll, T.; van Lier, R.A.; Medof, M.E.; Hamann, J. A novel role for CD55 in granulocyte homeostasis and anti-bacterial host defense. PLoS ONE 2011, 6, e24431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoek, R.M.; de Launay, D.; Kop, E.N.; Yilmaz-Elis, A.S.; Lin, F.; Reedquist, K.A.; Verbeek, J.S.; Medof, M.E.; Tak, P.P.; Hamann, J. Deletion of either CD55 or CD97 ameliorates arthritis in mouse models. Arthritis Rheumatol. 2010, 62, 1036–1042. [Google Scholar] [CrossRef] [PubMed]
- Capasso, M.; Durrant, L.G.; Stacey, M.; Gordon, S.; Ramage, J.; Spendlove, I. Costimulation via CD55 on human CD4+ T cells mediated by CD97. J. Immunol. 2006, 177, 1070–1077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sutavani, R.V.; Bradley, R.G.; Ramage, J.M.; Jackson, A.M.; Durrant, L.G.; Spendlove, I. CD55 costimulation induces differentiation of a discrete T regulatory type 1 cell population with a stable phenotype. J. Immunol. 2013, 191, 5895–5903. [Google Scholar] [CrossRef] [Green Version]
- Karpus, O.N.; Veninga, H.; Hoek, R.M.; Flierman, D.; van Buul, J.D.; Vandenakker, C.C.; vanBavel, E.; Medof, M.E.; van Lier, R.A.; Reedquist, K.A.; et al. Shear stress-dependent downregulation of the adhesion-G protein-coupled receptor CD97 on circulating leukocytes upon contact with its ligand CD55. J. Immunol. 2013, 190, 3740–3748. [Google Scholar] [CrossRef] [Green Version]
- Niu, M.; Xu, S.; Yang, J.; Yao, D.; Li, N.; Yan, J.; Zhong, G.; Song, G. Structural basis for CD97 recognition of the decay-accelerating factor CD55 suggests mechanosensitive activation of adhesion GPCRs. J. Biol. Chem. 2021, 296, 100776. [Google Scholar] [CrossRef]
- Liu, D.; Duan, L.; Rodda, L.B.; Lu, E.; Xu, Y.; An, J.; Qiu, L.; Liu, F.; Looney, M.R.; Yang, Z.; et al. CD97 promotes spleen dendritic cell homeostasis through the mechanosensing of red blood cells. Science 2022, 375, eabi5965. [Google Scholar] [CrossRef]
- Cerny, O.; Godlee, C.; Tocci, R.; Cross, N.E.; Shi, H.; Williamson, J.C.; Alix, E.; Lehner, P.J.; Holden, D.W. CD97 stabilises the immunological synapse between dendritic cells and T cells and is targeted for degradation by the Salmonella effector SteD. PLoS Pathog. 2021, 17, e1009771. [Google Scholar] [CrossRef]
- Xu, L.; Wang, X.; Wang, W.; Sun, M.; Choi, W.J.; Kim, J.Y.; Hao, C.; Li, S.; Qu, A.; Lu, M.; et al. Enantiomer-dependent immunological response to chiral nanoparticles. Nature 2022, 601, 366–373. [Google Scholar] [CrossRef]
- Wobus, M.; Bornhauser, M.; Jacobi, A.; Krater, M.; Otto, O.; Ortlepp, C.; Guck, J.; Ehninger, G.; Thiede, C.; Oelschlagel, U. Association of the EGF-TM7 receptor CD97 expression with FLT3-ITD in acute myeloid leukemia. Oncotarget 2015, 6, 38804–38815. [Google Scholar] [CrossRef] [Green Version]
- Maiga, A.; Lemieux, S.; Pabst, C.; Lavallee, V.P.; Bouvier, M.; Sauvageau, G.; Hebert, J. Transcriptome analysis of G protein-coupled receptors in distinct genetic subgroups of acute myeloid leukemia: Identification of potential disease-specific targets. Blood Cancer J. 2016, 6, e431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleo, K.; Dimitrova, L.; Oker, E.; Tomaszewski, N.; Berg, E.; Taruttis, F.; Engelmann, J.C.; Schwarzfischer, P.; Reinders, J.; Spang, R.; et al. Identification of ADGRE5 as discriminating MYC target between Burkitt lymphoma and diffuse large B-cell lymphoma. BMC Cancer 2019, 19, 322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaikari, V.P.; Yang, J.; Wu, S.; Alachkar, H. CD97 expression is associated with poor overall survival in acute myeloid leukemia. Exp. Hematol. 2019, 75, 64–73.e4. [Google Scholar] [CrossRef]
- Martin, G.H.; Roy, N.; Chakraborty, S.; Desrichard, A.; Chung, S.S.; Woolthuis, C.M.; Hu, W.; Berezniuk, I.; Garrett-Bakelman, F.E.; Hamann, J.; et al. CD97 is a critical regulator of acute myeloid leukemia stem cell function. J. Exp. Med. 2019, 216, 2362–2377. [Google Scholar] [CrossRef] [Green Version]
- Piao, X.; Hill, R.S.; Bodell, A.; Chang, B.S.; Basel-Vanagaite, L.; Straussberg, R.; Dobyns, W.B.; Qasrawi, B.; Winter, R.M.; Innes, A.M.; et al. G protein-coupled receptor-dependent development of human frontal cortex. Science 2004, 303, 2033–2036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salzman, G.S.; Ackerman, S.D.; Ding, C.; Koide, A.; Leon, K.; Luo, R.; Stoveken, H.M.; Fernandez, C.G.; Tall, G.G.; Piao, X.; et al. Structural Basis for Regulation of GPR56/ADGRG1 by Its Alternatively Spliced Extracellular Domains. Neuron 2016, 91, 1292–1304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Little, K.D.; Hemler, M.E.; Stipp, C.S. Dynamic regulation of a GPCR-tetraspanin-G protein complex on intact cells: Central role of CD81 in facilitating GPR56-Galpha q/11 association. Mol. Biol. Cell 2004, 15, 2375–2387. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Begum, S.; Hearn, J.D.; Hynes, R.O. GPR56. an atypical G protein-coupled receptor, binds tissue transglutaminase, TG2, and inhibits melanoma tumor growth and metastasis. Proc. Natl. Acad. Sci. USA 2006, 103, 9023–9028. [Google Scholar] [CrossRef] [Green Version]
- Luo, R.; Jeong, S.J.; Jin, Z.; Strokes, N.; Li, S.; Piao, X. G protein-coupled receptor 56 and collagen III. a receptor-ligand pair, regulates cortical development and lamination. Proc. Natl. Acad. Sci. USA 2011, 108, 12925–12930. [Google Scholar] [CrossRef] [Green Version]
- Chiang, N.Y.; Chang, G.W.; Huang, Y.S.; Peng, Y.M.; Hsiao, C.C.; Kuo, M.L.; Lin, H.H. Heparin interacts with the adhesion GPCR GPR56. reduces receptor shedding, and promotes cell adhesion and motility. J. Cell Sci. 2016, 129, 2156–2169. [Google Scholar] [CrossRef] [Green Version]
- Giera, S.; Luo, R.; Ying, Y.; Ackerman, S.D.; Jeong, S.J.; Stoveken, H.M.; Folts, C.J.; Welsh, C.A.; Tall, G.G.; Stevens, B.; et al. Microglial transglutaminase-2 drives myelination and myelin repair via GPR56/ADGRG1 in oligodendrocyte precursor cells. Elife 2018, 7, e33385. [Google Scholar] [CrossRef] [PubMed]
- Jin, G.; Sakitani, K.; Wang, H.; Jin, Y.; Dubeykovskiy, A.; Worthley, D.L.; Tailor, Y.; Wang, T.C. The G-protein coupled receptor 56. expressed in colonic stem and cancer cells, binds progastrin to promote proliferation and carcinogenesis. Oncotarget 2017, 8, 40606–40619. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Nwe, P.K.; Yang, Y.; Rosen, C.E.; Bielecka, A.A.; Kuchroo, M.; Cline, G.W.; Kruse, A.C.; Ring, A.M.; Crawford, J.M.; et al. A Forward Chemical Genetic Screen Reveals Gut Microbiota Metabolites That Modulate Host Physiology. Cell 2019, 177, 1217–1231.e18. [Google Scholar] [CrossRef]
- Li, T.; Chiou, B.; Gilman, C.K.; Luo, R.; Koshi, T.; Yu, D.; Oak, H.C.; Giera, S.; Johnson-Venkatesh, E.; Muthukumar, A.K.; et al. A splicing isoform of GPR56 mediates microglial synaptic refinement via phosphatidylserine binding. EMBO J. 2020, 39, e104136. [Google Scholar] [CrossRef] [PubMed]
- Della Chiesa, M.; Falco, M.; Parolini, S.; Bellora, F.; Petretto, A.; Romeo, E.; Balsamo, M.; Gambarotti, M.; Scordamaglia, F.; Tabellini, G.; et al. GPR56 as a novel marker identifying the CD56dull CD16+ NK cell subset both in blood stream and in inflamed peripheral tissues. Int. Immunol. 2010, 22, 91–100. [Google Scholar] [CrossRef] [Green Version]
- Peng, Y.M.; van de Garde, M.D.; Cheng, K.F.; Baars, P.A.; Remmerswaal, E.B.; van Lier, R.A.; Mackay, C.R.; Lin, H.H.; Hamann, J. Specific expression of GPR56 by human cytotoxic lymphocytes. J. Leukoc. Biol. 2011, 90, 735–740. [Google Scholar] [CrossRef] [Green Version]
- Chang, G.W.; Hsiao, C.C.; Peng, Y.M.; Vieira Braga, F.A.; Kragten, N.A.; Remmerswaal, E.B.; van de Garde, M.D.; Straussberg, R.; Konig, G.M.; Kostenis, E.; et al. The Adhesion G Protein-Coupled Receptor GPR56/ADGRG1 Is an Inhibitory Receptor on Human NK Cells. Cell Rep. 2016, 15, 1757–1770. [Google Scholar] [CrossRef] [Green Version]
- Truong, K.L.; Schlickeiser, S.; Vogt, K.; Boes, D.; Stanko, K.; Appelt, C.; Streitz, M.; Grutz, G.; Stobutzki, N.; Meisel, C.; et al. Killer-like receptors and GPR56 progressive expression defines cytokine production of human CD4(+) memory T cells. Nat. Commun. 2019, 10, 2263. [Google Scholar] [CrossRef] [Green Version]
- Argyriou, A.; Wadsworth, M.H., 2nd; Lendvai, A.; Christensen, S.M.; Hensvold, A.H.; Gerstner, C.; van Vollenhoven, A.; Kravarik, K.; Winkler, A.; Malmstrom, V.; et al. Single cell sequencing identifies clonally expanded synovial CD4(+) T(PH) cells expressing GPR56 in rheumatoid arthritis. Nat. Commun. 2022, 13, 4046. [Google Scholar] [CrossRef]
- Lutter, L.; van der Wal, M.M.; Brand, E.C.; Maschmeyer, P.; Vastert, S.; Mashreghi, M.F.; van Loosdregt, J.; van Wijk, F. Human regulatory T cells locally differentiate and are functionally heterogeneous within the inflamed arthritic joint. Clin. Transl. Immunol. 2022, 11, e1420. [Google Scholar] [CrossRef]
- Tseng, W.Y.; Jan Wu, Y.J.; Yang, T.Y.; Chiang, N.Y.; Tsai, W.P.; Gordon, S.; Chang, G.W.; Kuo, C.F.; Luo, S.F.; Lin, H.H. High levels of soluble GPR56/ADGRG1 are associated with positive rheumatoid factor and elevated tumor necrosis factor in patients with rheumatoid arthritis. J. Microbiol. Immunol. Infect. 2018, 51, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Zheng, M.; Liu, T.; Bahabayi, A.; Song, S.; Alimu, X.; Kang, R.; Lu, S.; Song, Y.; Liu, C. Cytotoxic T Lymphocytes Expressing GPR56 are Up-regulated in the Peripheral Blood of Patients with Active Rheumatoid Arthritis and Reflect Disease Progression. Immunol. Investig. 2022, 51, 1804–1819. [Google Scholar] [CrossRef] [PubMed]
- Bilemjian, V.; Vlaming, M.R.; Alvarez Freile, J.; Huls, G.; De Bruyn, M.; Bremer, E. The Novel Immune Checkpoint GPR56 Is Expressed on Tumor-Infiltrating Lymphocytes and Selectively Upregulated upon TCR Signaling. Cancers 2022, 14, 3164. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Fass, J.N.; Shih, Y.P.; Gunderson, A.J.; Sanjuan Silva, N.; Huang, H.; Bernard, B.M.; Rajamanickam, V.; Slagel, J.; Bifulco, C.B.; et al. Transcriptomic profiles of neoantigen-reactive T cells in human gastrointestinal cancers. Cancer Cell 2022, 40, 410–423.e7. [Google Scholar] [CrossRef]
- Yeung, J.; Adili, R.; Stringham, E.N.; Luo, R.; Vizurraga, A.; Rosselli-Murai, L.K.; Stoveken, H.M.; Yu, M.; Piao, X.; Holinstat, M.; et al. GPR56/ADGRG1 is a platelet collagen-responsive GPCR and hemostatic sensor of shear force. Proc. Natl. Acad. Sci. USA 2020, 117, 28275–28286. [Google Scholar] [CrossRef]
- Rao, T.N.; Marks-Bluth, J.; Sullivan, J.; Gupta, M.K.; Chandrakanthan, V.; Fitch, S.R.; Ottersbach, K.; Jang, Y.C.; Piao, X.; Kulkarni, R.N.; et al. High-level Gpr56 expression is dispensable for the maintenance and function of hematopoietic stem and progenitor cells in mice. Stem Cell Res. 2015, 14, 307–322. [Google Scholar] [CrossRef] [Green Version]
- Tokoro, Y.; Yamada, Y.; Takayanagi, S.I.; Hagiwara, T. 57R2A. a newly established monoclonal antibody against mouse GPR56, marks long-term repopulating hematopoietic stem cells. Exp. Hematol. 2018, 59, 51–59.e1. [Google Scholar] [CrossRef]
- Solaimani Kartalaei, P.; Yamada-Inagawa, T.; Vink, C.S.; de Pater, E.; van der Linden, R.; Marks-Bluth, J.; van der Sloot, A.; van den Hout, M.; Yokomizo, T.; van Schaick-Solerno, M.L.; et al. Whole-transcriptome analysis of endothelial to hematopoietic stem cell transition reveals a requirement for Gpr56 in HSC generation. J. Exp. Med. 2015, 212, 93–106. [Google Scholar] [CrossRef] [Green Version]
- Maglitto, A.; Mariani, S.A.; de Pater, E.; Rodriguez-Seoane, C.; Vink, C.S.; Piao, X.; Lukke, M.L.; Dzierzak, E. Unexpected redundancy of Gpr56 and Gpr97 during hematopoietic cell development and differentiation. Blood Adv. 2021, 5, 829–842. [Google Scholar] [CrossRef]
- Saito, Y.; Kaneda, K.; Suekane, A.; Ichihara, E.; Nakahata, S.; Yamakawa, N.; Nagai, K.; Mizuno, N.; Kogawa, K.; Miura, I.; et al. Maintenance of the hematopoietic stem cell pool in bone marrow niches by EVI1-regulated GPR56. Leukemia 2013, 27, 1637–1649. [Google Scholar] [CrossRef] [Green Version]
- Daria, D.; Kirsten, N.; Muranyi, A.; Mulaw, M.; Ihme, S.; Kechter, A.; Hollnagel, M.; Bullinger, L.; Dohner, K.; Dohner, H.; et al. GPR56 contributes to the development of acute myeloid leukemia in mice. Leukemia 2016, 30, 1734–1741. [Google Scholar] [CrossRef] [PubMed]
- Pabst, C.; Bergeron, A.; Lavallee, V.P.; Yeh, J.; Gendron, P.; Norddahl, G.L.; Krosl, J.; Boivin, I.; Deneault, E.; Simard, J.; et al. GPR56 identifies primary human acute myeloid leukemia cells with high repopulating potential in vivo. Blood 2016, 127, 2018–2027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daga, S.; Rosenberger, A.; Quehenberger, F.; Krisper, N.; Prietl, B.; Reinisch, A.; Zebisch, A.; Sill, H.; Wolfler, A. High GPR56 surface expression correlates with a leukemic stem cell gene signature in CD34-positive AML. Cancer Med. 2019, 8, 1771–1778. [Google Scholar] [CrossRef]
- Von Palffy, S.; Thorsson, H.; Pena-Martinez, P.; Puente-Moncada, N.; Sanden, C.; Blom, A.M.; Henningsson, R.; Juliusson, G.; King, B.C.; Landberg, N.; et al. The complement receptor C3AR constitutes a novel therapeutic target in NPM1-mutated AML. Blood Adv. 2022. Online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Jentzsch, M.; Bill, M.; Grimm, J.; Schulz, J.; Schuhmann, L.; Brauer, D.; Goldmann, K.; Wilke, F.; Franke, G.N.; Behre, G.; et al. High expression of the stem cell marker GPR56 at diagnosis identifies acute myeloid leukemia patients at higher relapse risk after allogeneic stem cell transplantation in context with the CD34+/CD38- population. Haematologica 2020, 105, e507. [Google Scholar] [CrossRef] [Green Version]
- Sleckman, B.P.; Khan, W.N.; Xu, W.; Bassing, C.H.; Malynn, B.A.; Copeland, N.G.; Bardon, C.G.; Breit, T.M.; Davidson, L.; Oltz, E.M.; et al. Cloning and functional characterization of the early-lymphocyte-specific Pb99 gene. Mol. Cell. Biol. 2000, 20, 4405–4410. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.J.; Zhang, L.L.; Zhang, H.X.; Shen, C.L.; Lu, S.Y.; Kuang, Y.; Wan, Y.H.; Wang, W.G.; Yan, H.M.; Dang, S.Y.; et al. Gpr97 is essential for the follicular versus marginal zone B-lymphocyte fate decision. Cell Death Dis. 2013, 4, e853. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Zhang, X.; Wang, S.; Wang, J.; Du, B.; Wang, Z.; Liu, M.; Jiang, W.; Qian, M.; Ren, H. Gpr97 is dispensable for metabolic syndrome but is involved in macrophage inflammation in high-fat diet-induced obesity in mice. Sci. Rep. 2016, 6, 24649. [Google Scholar] [CrossRef] [Green Version]
- Fang, W.; Wang, Z.; Li, Q.; Wang, X.; Zhang, Y.; Sun, Y.; Tang, W.; Ma, C.; Sun, J.; Li, N.; et al. Gpr97 Exacerbates AKI by Mediating Sema3A Signaling. J. Am. Soc. Nephrol. 2018, 29, 1475–1489. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wang, X.; Chen, X.; Lu, S.; Kuang, Y.; Fei, J.; Wang, Z. Gpr97/Adgrg3 ameliorates experimental autoimmune encephalomyelitis by regulating cytokine expression. Acta Biochim. Biophys. Sin. 2018, 50, 666–675. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.P.; Li, X.N.; Zhang, X.Y.; Du, B.; Jiang, W.Z.; Liu, M.Y.; Wang, J.J.; Wang, Z.G.; Ren, H.; Qian, M. Gpr97 Is Dispensable for Inflammation in OVA-Induced Asthmatic Mice. PLoS ONE 2015, 10, e0131461. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, C.C.; Chu, T.Y.; Wu, C.J.; van den Biggelaar, M.; Pabst, C.; Hebert, J.; Kuijpers, T.W.; Scicluna, B.P.; I, K.-Y.; Chen, T.C.; et al. The Adhesion G Protein-Coupled Receptor GPR97/ADGRG3 Is Expressed in Human Granulocytes and Triggers Antimicrobial Effector Functions. Front. Immunol. 2018, 9, 2830. [Google Scholar] [CrossRef] [PubMed]
- Chu, T.Y.; Zheng-Gerard, C.; Huang, K.Y.; Chang, Y.C.; Chen, Y.W.; I, K.-Y.; Lo, Y.L.; Chiang, N.Y.; Chen, H.Y.; Stacey, M.; et al. GPR97 triggers inflammatory processes in human neutrophils via a macromolecular complex upstream of PAR2 activation. Nat. Commun. 2022, 13, 6385. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.; Qiu, N.; Wang, M.; Zhang, B.; Du, J.; Zhong, Z.; Xu, W.; Chu, X.; Ma, L.; Yi, C.; et al. Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1. Nature 2022, 604, 779–785. [Google Scholar] [CrossRef] [PubMed]
- Ping, Y.Q.; Xiao, P.; Yang, F.; Zhao, R.J.; Guo, S.C.; Yan, X.; Wu, X.; Zhang, C.; Lu, Y.; Zhao, F.; et al. Structural basis for the tethered peptide activation of adhesion GPCRs. Nature 2022, 604, 763–770. [Google Scholar] [CrossRef]
- Xiao, P.; Guo, S.; Wen, X.; He, Q.T.; Lin, H.; Huang, S.M.; Gou, L.; Zhang, C.; Yang, Z.; Zhong, Y.N.; et al. Tethered peptide activation mechanism of the adhesion GPCRs ADGRG2 and ADGRG4. Nature 2022, 604, 771–778. [Google Scholar] [CrossRef]
- Barros-Alvarez, X.; Nwokonko, R.M.; Vizurraga, A.; Matzov, D.; He, F.; Papasergi-Scott, M.M.; Robertson, M.J.; Panova, O.; Yardeni, E.H.; Seven, A.B.; et al. The tethered peptide activation mechanism of adhesion GPCRs. Nature 2022, 604, 757–762. [Google Scholar] [CrossRef]

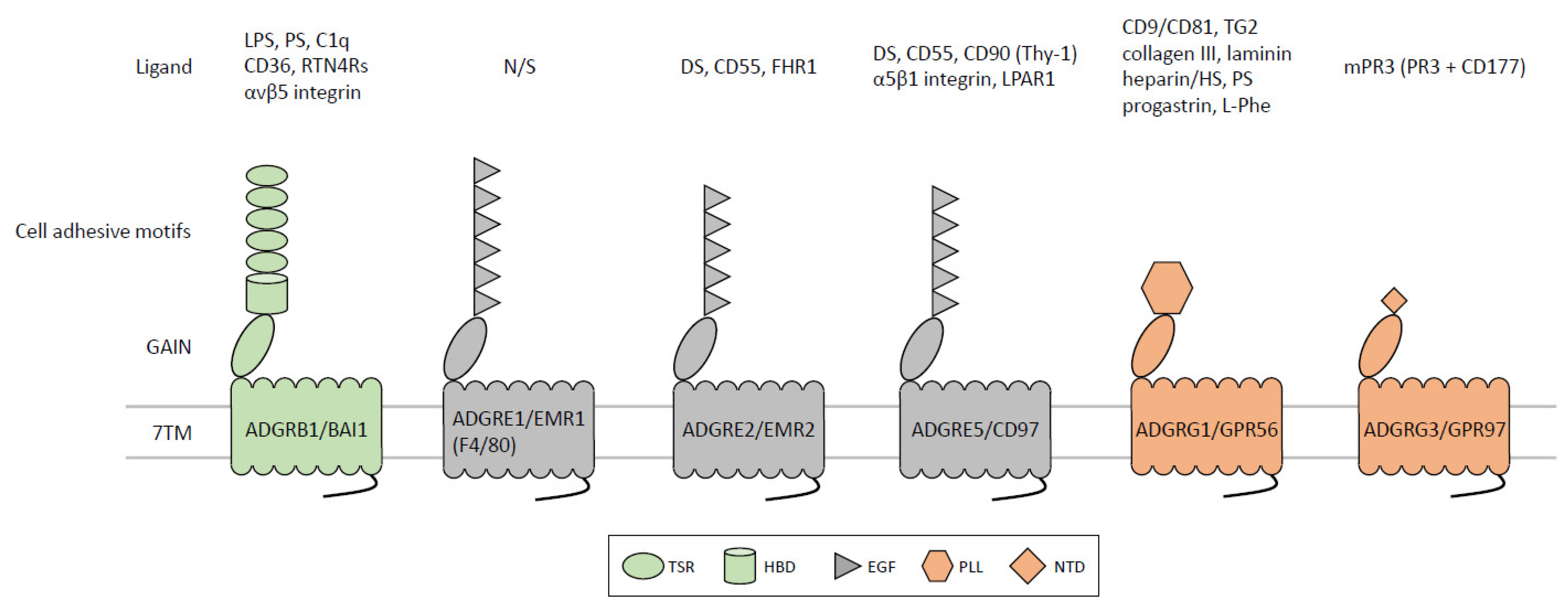
| Receptor | Expression in Immune Cells | Ligand/Interacting Partner | Physiopathological Functions in the Immune System |
|---|---|---|---|
| ADGRB1/ BAI1 | Macrophage, gastric phagocyte, microglia, T-ALL * | LPS, PS, C1q, CD36, αvβ5 integrin, RTN4Rs | Apoptotic cell clearance, colonic inflammatory disorders, HDL biogenesis, microglial neuronal removal, bacteria phagocytosis, anti-viral response, T-ALL pathogenesis. |
| ADGRE1/ EMR1 (Emr1, F4/80) | Monocyte, macrophage, eosinophil, and DC | N/S | Emr1 (F4/80): Anti-intracellular bacterial infection, induction of Ag-specific peripheral immune tolerance. EMR1: homeostatic control of human eosinophils. |
| ADGRE2/ EMR2 | Monocyte, macrophage, granulocyte, mast cell, and DC | DS, CD55, FHR1 | Neutrophil activation, differentiation and inflammatory activation of macrophage, NLRP3 inflammasome activation, vibration-induced mast cell activation, and degranulation (VU, hereditary α-tryptasemia). |
| ADGRE5/ CD97 | T and B lymphocytes, monocyte, macrophage, granulocyte, DC, HSPCs, AML, BL, LSC | DS, CD55, CD90 (Thy-1), α5β1 integrin, LPAR1 | Leukocyte adhesion/trafficking, homeostasis and anti-bacterial response of granulocytes, activation, and expansion of T cell subsets, homeostasis and Agpresenting functions of splenic cDC2s, IS stability, regulation of hematopoiesis and LSC maintenance. |
| ADGRG1/ GPR56 | NK, cytotoxic T cells, TPH and Treg cells, TILs, platelet, HSPCs, AML, LSC | CD9/CD81, TG2, collagen III, laminin, heparin/HS, progastrin, L-Phe, PS | NK inhibitory receptor, up-regulated expression in T cell subsets of RA, JIA, and various cancer types, platelet activation in response to exposed collagen and blood flow, hematopoietic development, tumorigenesis of AML. |
| ADGRG3/ GPR97 | Gpr97: pre-B cells and thymocytes. GPR97: neutrophil and eosinophil | N/A mPR3 (PR3 + CD177) | Mouse: B-lymphocyte fate decision, obesity-associated macrophage inflammation, pathogenic modulation of AKI and EAE. Human: inflammatory activation of neutrophilic granulocytes. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tseng, W.-Y.; Stacey, M.; Lin, H.-H. Role of Adhesion G Protein-Coupled Receptors in Immune Dysfunction and Disorder. Int. J. Mol. Sci. 2023, 24, 5499. https://doi.org/10.3390/ijms24065499
Tseng W-Y, Stacey M, Lin H-H. Role of Adhesion G Protein-Coupled Receptors in Immune Dysfunction and Disorder. International Journal of Molecular Sciences. 2023; 24(6):5499. https://doi.org/10.3390/ijms24065499
Chicago/Turabian StyleTseng, Wen-Yi, Martin Stacey, and Hsi-Hsien Lin. 2023. "Role of Adhesion G Protein-Coupled Receptors in Immune Dysfunction and Disorder" International Journal of Molecular Sciences 24, no. 6: 5499. https://doi.org/10.3390/ijms24065499






