The Effect of Cholesterol in MCF7 Human Breast Cancer Cells
Abstract
:1. Introduction
1.1. Breast Cancer
1.2. Cholesterol in Cancer
1.3. Cholesterol–Sphingomyelin Relationship in Cancer
2. Results
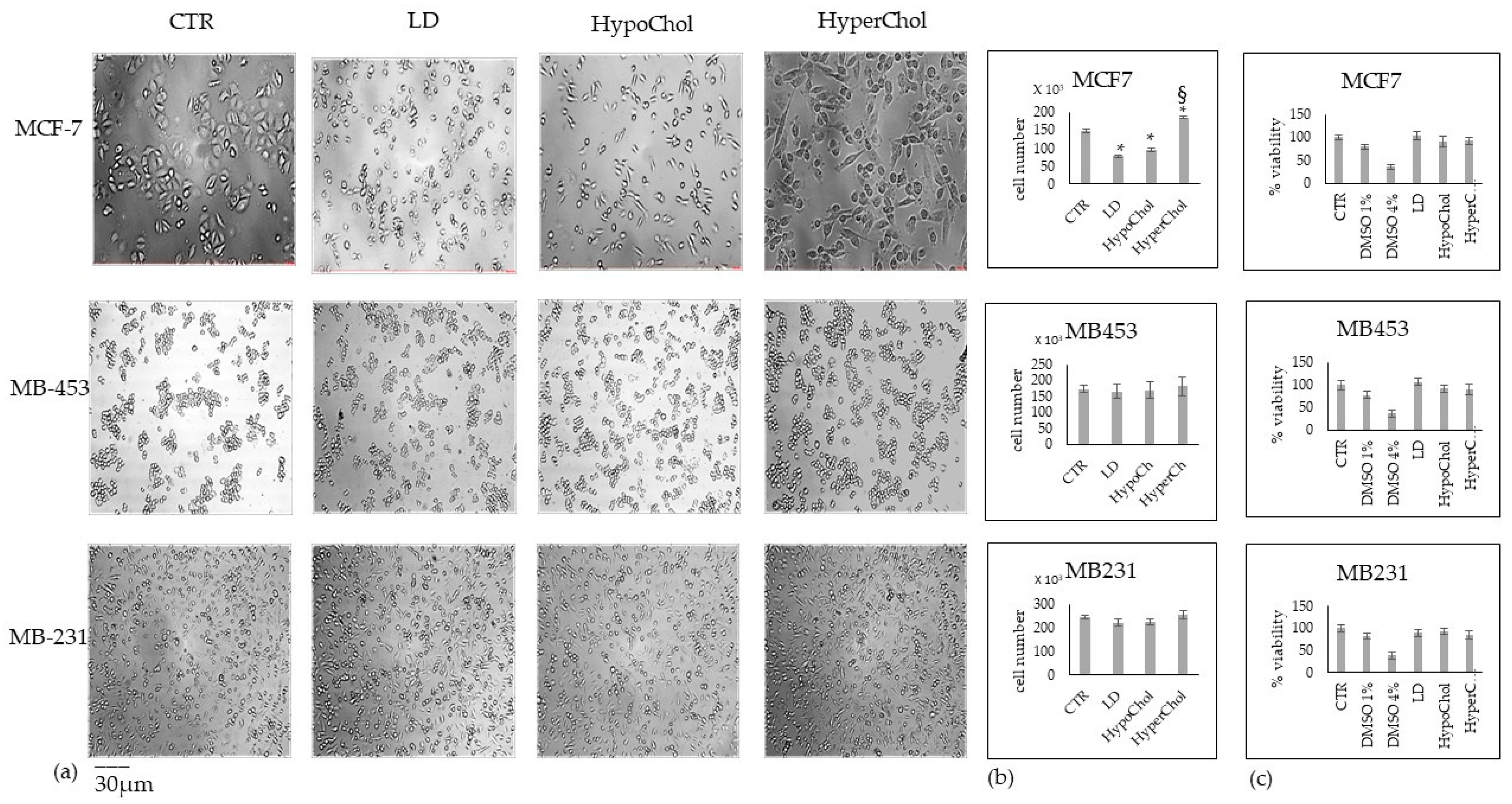
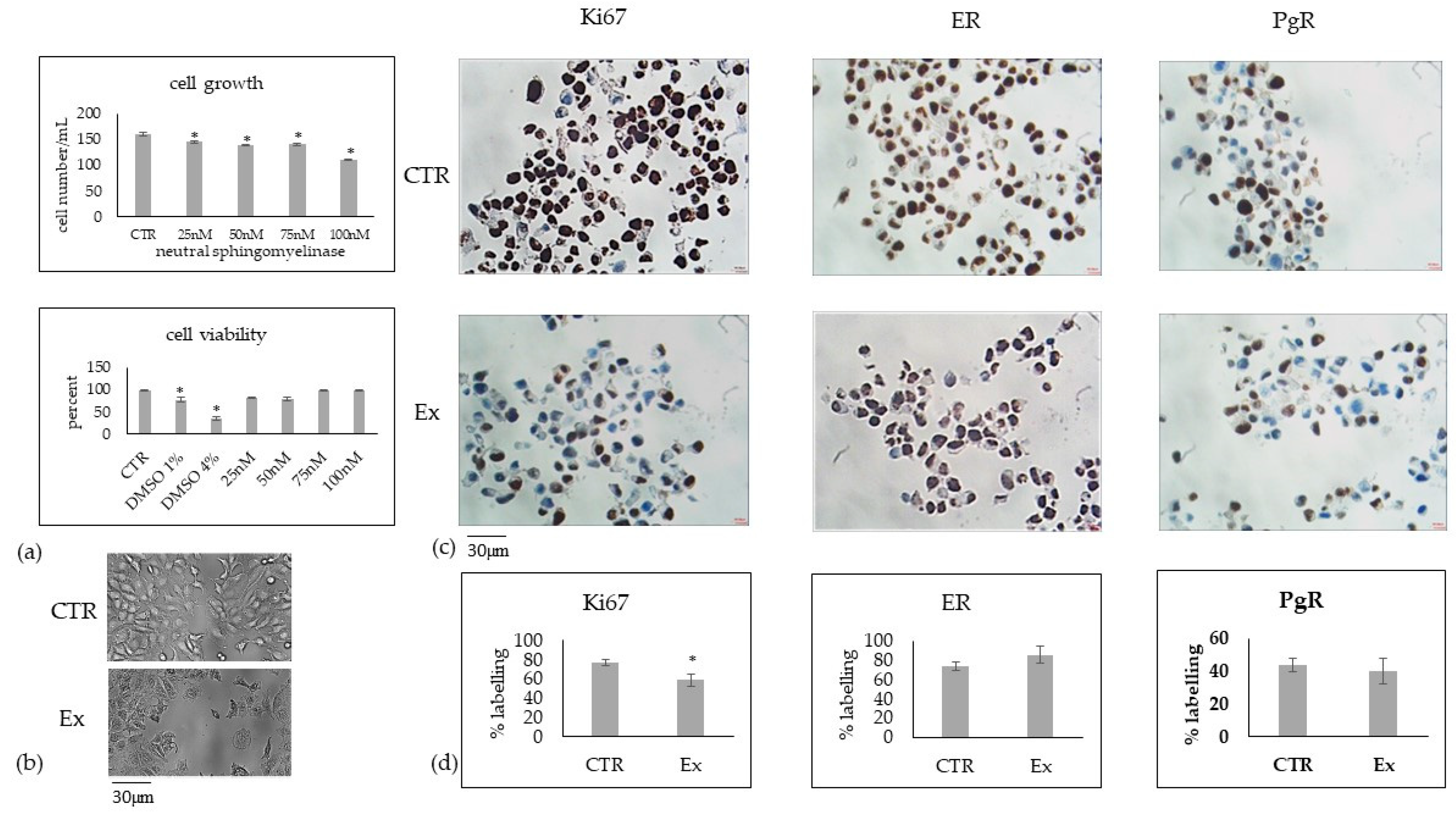
2.1. Cholesterol Does Not Affect MCF7, MB453 and MB231 Breast Cancer Cells in the Same Way
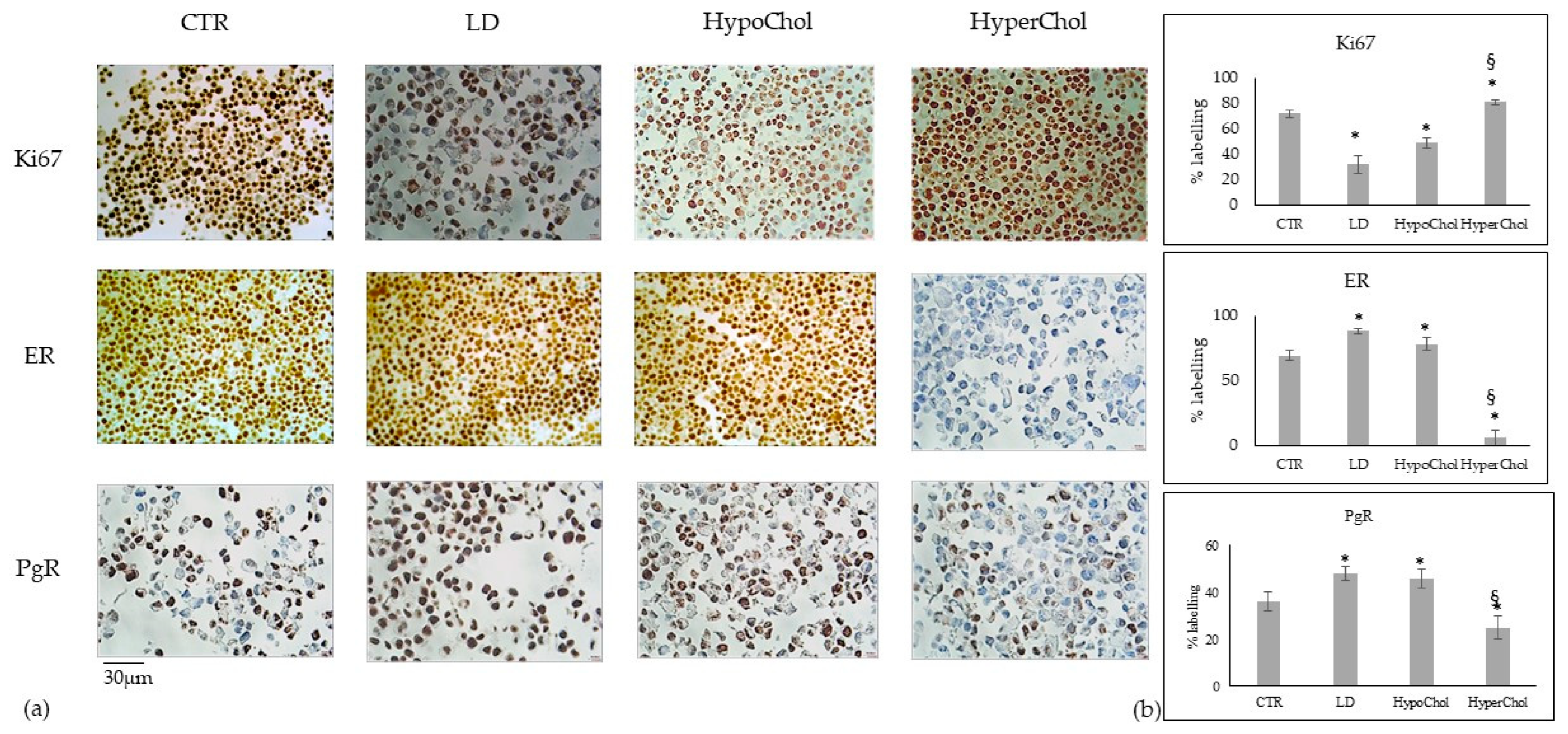
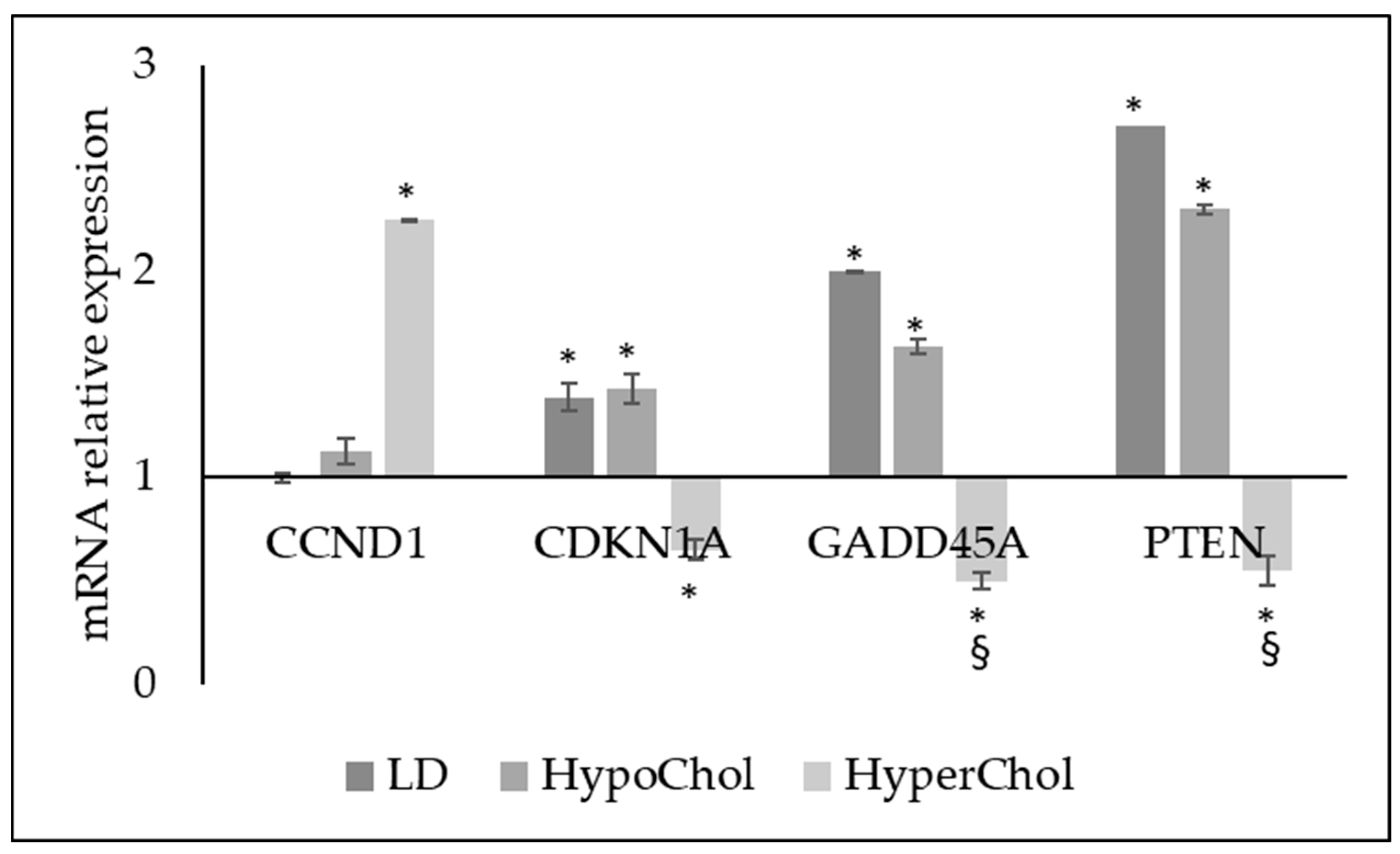
2.2. Involvement of Cholesterol in Several Biological Phenomena of MCF7 Cells
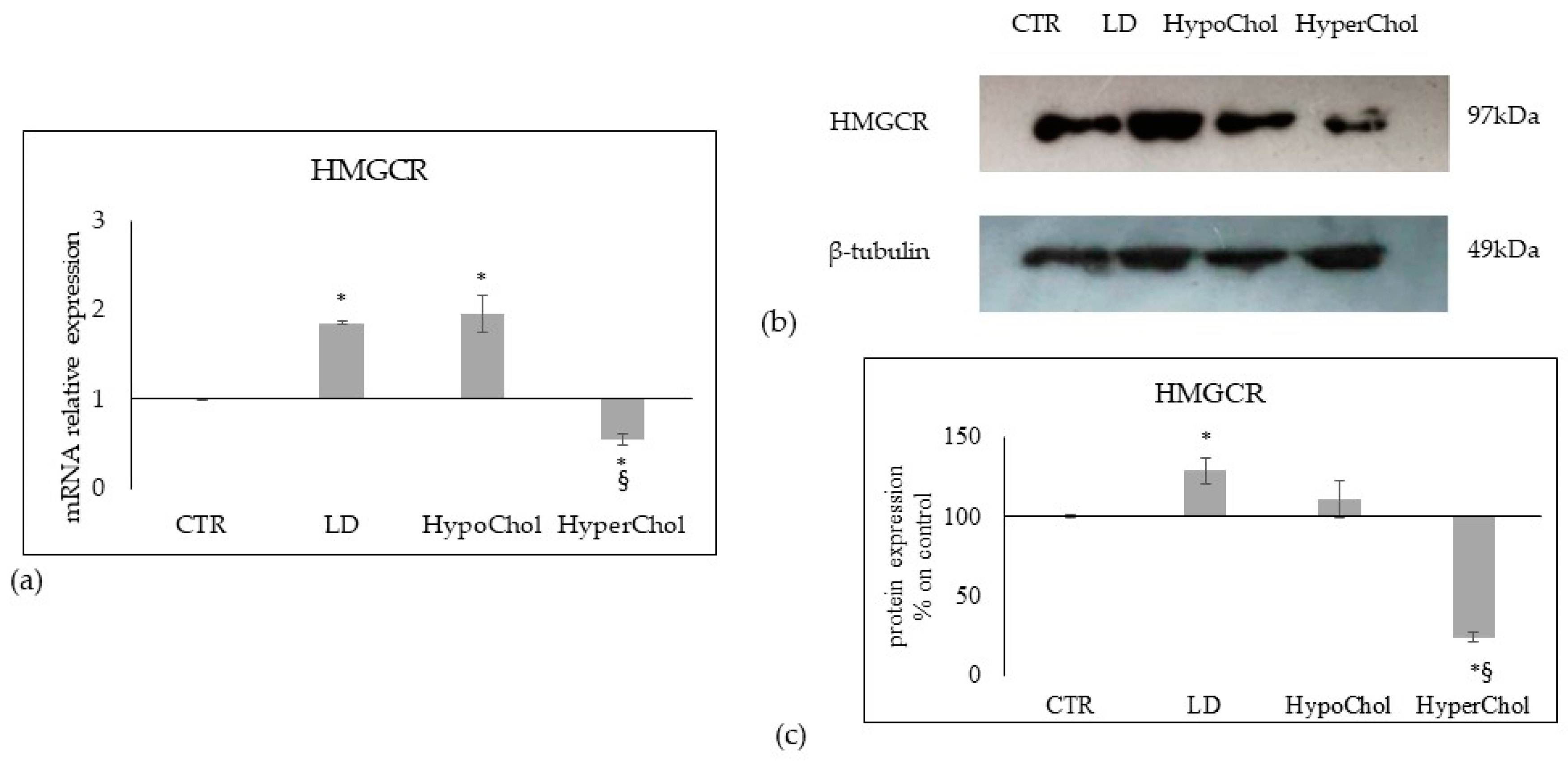
2.3. Relationship between Chol Level and Sphingomyelin Metabolism in MCF7 Cells
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Cell Culture and Treatments
4.3. Cell Viability
4.4. Cell Morphology
4.5. Cholesterol Analysis
4.6. Immunocytochemistry
4.7. Reverse Transcription Quantitative PCR (RTqPCR)
4.8. Protein Concentration and Western Blotting
4.9. Neutral Sphingomyelinase Activity Assay
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Anastasiadi, Z.; Lianos, G.D.; Ignatiadou, E.; Harissis, H.V.; Mitsis, M. Breast cancer in young women: An overview. Updates Surg. 2017, 69, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Veronesi, U.; Boyle, P.; Goldhirsch, A.; Orecchia, R.; Viale, G. Breast cancer. Lancet 2005, 365, 1727–1741. [Google Scholar] [CrossRef] [PubMed]
- Kolak, A.; Kamińska, M.; Sygit, K.; Budny, A.; Surdyka, D.; Kukiełka-Budny, B.; Burdan, F. Primary and secondary prevention of breast cancer. Ann. Agric. Environ. Med. 2017, 24, 549–553. [Google Scholar] [CrossRef] [PubMed]
- Odle, T.G. Precision Medicine in Breast Cancer. Radiol. Technol. 2017, 88, 401M–421M. [Google Scholar]
- Yeo, S.K.; Guan, J.L. Breast Cancer: Multiple Subtypes within a Tumor? Trends Cancer 2017, 3, 753–760. [Google Scholar] [CrossRef]
- Kondov, B.; Milenkovikj, Z.; Kondov, G.; Petrushevska, G.; Basheska, N.; Bogdanovska-Todorovska, M.; Tolevska, N.; Ivkovski, L. Presentation of the Molecular Subtypes of Breast Cancer Detected By Immunohistochemistry in Surgically Treated Patients. Open Access Maced. J. Med. Sci. 2018, 6, 961–967. [Google Scholar] [CrossRef] [Green Version]
- Blows, F.M.; Driver, K.E.; Schmidt, M.K.; Broeks, A.; van Leeuwen, F.E.; Wesseling, J.; Cheang, M.C.; Gelmon, K.; Nielsen, T.O.; Blomqvist, C.; et al. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: A collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med. 2010, 25, e1000279. [Google Scholar] [CrossRef]
- Holliday, D.L.; Speirs, V. Choosing the right cell line for breast cancer research. Breast Cancer Res. 2011, 13, 215. [Google Scholar] [CrossRef] [Green Version]
- Gizzi, G.; Cataldi, S.; Mazzeschi, C.; Delvecchio, E.; Ceccarini, M.R.; Codini, M.; Albi, E. Hypercholesterolemia in Cancer and in Anorexia Nervosa: A Hypothesis for a Crosstalk. Int. J. Mol. Sci. 2022, 23, 7466. [Google Scholar] [CrossRef]
- Baba, A.I.; Câtoi, C. Comparative Oncology; Publishing House of the Romanian Academy: Bucharest, Romania, 2007. [Google Scholar]
- Goldstein, J.L.; Brown, M.S. Regulation of low-density lipoprotein receptors: Implications for pathogenesis and therapy of hypercholesterolemia and atherosclerosis. Circulation 2021, 76, 504–507. [Google Scholar] [CrossRef] [Green Version]
- Xu, F.; Yan, J.; Peng, Z.; Liu, J.; Li, Z. Comprehensive analysis of a glycolysis and cholesterol synthesis-related genes signature for predicting prognosis and immune landscape in osteosarcoma. Front. Immunol. 2022, 13, 1096009. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.; Yuan, W.; Chen, T.; Zhou, C.; Liu, C.; Huang, Y.; Han, D.; Huang, Q. HMGCR positively regulated the growth and migration of glioblastoma cells. Gene 2016, 576, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Ashida, S.; Kawada, C.; Inoue, K. Stromal regulation of prostate cancer cell growth by mevalonate pathway enzymes HMGCS1 and HMGCR. Oncol. Lett. 2017, 14, 6533–6542. [Google Scholar] [CrossRef] [Green Version]
- Murai, T. Cholesterol lowering: Role in cancer prevention and treatment. Biol. Chem. 2015, 396, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Codini, M.; Garcia-Gil, M.; Albi, E. Cholesterol and sphingolipid enriched lipid rafts as therapeutic targets in cancer. Int. J. Mol. Sci. 2021, 22, 726. [Google Scholar] [CrossRef]
- Codini, M.; Conte, C.; Cataldi, S.; Arcuri, C.; Lazzarini, A.; Ceccarini, M.R.; Patria, F.; Floridi, A.; Mecca, C.; Ambesi-Impiombato, F.S.; et al. Nuclear Lipid Microdomains Regulate Daunorubicin Resistance in Hepatoma Cells. Int. J. Mol. Sci. 2018, 19, 3424. [Google Scholar] [CrossRef] [Green Version]
- Pugliese, L.; Bernardini, I.; Viola-Magni, M.P.; Albi, E. Low levels of serum cholesterol/phospholipids are associated with the antiphospholipid antibodies in monoclonal gammopathy. Int. J. Immunopathol. Pharmacol. 2006, 19, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Yue, S.; Li, J.; Lee, S.Y.; Lee, H.J.; Shao, T.; Song, B.; Cheng, L.; Masterson, T.A.; Liu, X.; Ratliff, T.L.; et al. Cholesteryl ester accumulation induced by PTEN loss and PI3K/AKT activation underlies human prostate cancer aggressiveness. Cell Metab. 2014, 19, 393–406. [Google Scholar] [CrossRef] [Green Version]
- Xi, Y.; Yani, Z.; Jing, M.; Yinhang, W.; Xiaohui, H.; Jing, Z.; Quan, Q.; Shuwen, H. Mechanisms of induction of tumors by cholesterol and potential therapeutic prospects. Biomed. Pharmacother. 2021, 144, 112277. [Google Scholar] [CrossRef]
- Pugliese, L.; Bernardini, I.; Pacifico, N.; Peverini, M.; Damaskopoulou, E.; Cataldi, S.; Albi, E. Severe hypocholesterolaemia is often neglected in haematological malignancies. Eur. J. Cancer 2010, 46, 1735–1743. [Google Scholar] [CrossRef]
- Olsson, A.G.; Angelin, B.; Assmann, G.; Binder, C.J.; Björkhem, I.; Cedazo-Minguez, A.; Cohen, J.; von Eckardstein, A.; Farinaro, E.; Müller-Wieland, D.; et al. Can LDL cholesterol be too low? Possible risks of extremely low levels. J. Intern. Med. 2017, 281, 534–553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pugliese, L.; Bernardini, I.; Pacifico, E.; Viola-Magni, M.; Albi, E. Antiphospholipid antibodies in patients with cancer. Int. J. Immunopathol. Pharmacol. 2006, 19, 879–888. [Google Scholar] [CrossRef] [PubMed]
- Codini, M.; Cataldi, S.; Lazzarini, A.; Tasegian, A.; Ceccarini, M.R.; Floridi, A.; Lazzarini, R.; Ambesi-Impiombato, F.S.; Curcio, F.; Beccari, T.; et al. Why high cholesterol levels help hematological malignancies: Role of nuclear lipid microdomains. Lipids Health Dis. 2016, 15, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuzmen, S.; Hostetter, G.; Watanabe, A.; Ekmekçi, C.; Carrigan, E.P.; Shechter, I.; Kallioniemi, O.; Miller, L.J.; Mousses, S. Characterization of farnesyl diphosphate farnesyl transferase 1 (FDFT1) expression in cancer. Pers. Med. 2019, 16, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Goossens, P.; Rodriguez-Vita, J.; Etzerodt, A.; Masse, M.; Rastoin, O.; Gouirand, V.; Ulas, T.; Papantonopoulou, O.; Van Eck, M.; Auphan-Anezin, N.; et al. Membrane Cholesterol Efflux Drives Tumor-Associated Macrophage Reprogramming and Tumor Progression. Cell Metab. 2019, 29, 1376–1389.e4. [Google Scholar] [CrossRef] [Green Version]
- Horváth, Á.; Erostyák, J.; Szőke, É. Effect of Lipid Raft Disruptors on Cell Membrane Fluidity Studied by Fluorescence Spectroscopy. Int. J. Mol. Sci. 2022, 23, 13729. [Google Scholar] [CrossRef]
- Pitson, S.M. Regulation of sphingosine kinase and sphingolipid signaling. Trends Biochem. Sci. 2011, 36, 97–107. [Google Scholar] [CrossRef]
- Pulkoski-Gross, M.J.; Obeid, L.M. Molecular mechanisms of regulation of sphingosine kinase 1. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 2018, 1863, 1413–1422. [Google Scholar] [CrossRef]
- Kar, A.; Medatwal, N.; Rajput, K.; Mandal, S.; Pani, T.; Khan, A.; Sharma, P.; Oberoi, A.S.; Vishwakarma, G.; Deo, S.; et al. Unique Sphingolipid Signature Identifies Luminal and Triple-Negative Breast Cancer Subtypes. Int. J. Cancer 2023, 5, 1–14. [Google Scholar] [CrossRef]
- Corcoran, C.A.; He, Q.; Ponnusamy, S.; Ogretmen, B.; Huang, Y.; Sheikh, M.S. Neutral sphingomyelinase-3 is a DNA damage and nongenotoxic stress-regulated gene that is deregulated in human malignancies. Mol. Cancer Res. 2008, 6, 795–807. [Google Scholar] [CrossRef] [Green Version]
- Comşa, Ş.; Cîmpean, A.M.; Raica, M. The Story of MCF-7 Breast Cancer Cell Line: 40 years of Experience in Research. Anticancer Res. 2015, 35, 3147–3154. [Google Scholar]
- Chen, H.W. Role of cholesterol metabolism in cell growth. Fed. Proc. 1984, 43, 126–130. [Google Scholar]
- Beckwitt, C.H.; Brufsky, A.; Oltvai, Z.N.; Wells, A. Statin drugs to reduce breast cancer recurrence and mortality. Breast Cancer Res. 2018, 20, 144. [Google Scholar] [CrossRef] [PubMed]
- Ahern, T.P.; Lash, T.L.; Damkier, P.; Christiansen, P.M.; Cronin-Fenton, D.P. Statins and breast cancer prognosis: Evidence and opportunities. Lancet Oncol. 2014, 15, e461–e468. [Google Scholar] [CrossRef] [Green Version]
- Amerizadeh, A.; Vaseghi, G.; Farajzadegan, Z.; Asgary, S. An Updated Systematic Review and Meta-Analysis on Association of Serum Lipid Profile With Risk of Breast Cancer Incidence. Int. J. Prev. Med. 2022, 13, 142. [Google Scholar] [CrossRef]
- Sun, X.B.; Liu, W.W.; Wang, B.; Yang, Z.P.; Tang, H.Z.; Lu, S.; Wang, Y.Y.; Qu, J.X.; Rao, B.Q. Correlations between serum lipid and Ki-67 levels in different breast cancer molecular subcategories. Oncol. Lett. 2022, 25, 53. [Google Scholar] [CrossRef]
- Baek, A.E.; Nelson, E.R. The Contribution of Cholesterol and Its Metabolites to the Pathophysiology of Breast Cancer. Horm. Cancer 2016, 7, 219–228. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.Y.; Bae, S.J.; Choi, J.W.; Han, S.; Bae, S.H.; Cheong, J.H.; Jang, H. Cholesterol Synthesis Is Important for Breast Cancer Cell Tumor Sphere Formation and Invasion. Biomedicines 2022, 10, 1908. [Google Scholar] [CrossRef]
- Maja, M.; Mohammed, D.; Dumitru, A.C.; Verstraeten, S.; Lingurski, M.; Mingeot-Leclercq, M.P.; Alsteens, D.; Tyteca, D. Surface cholesterol-enriched domains specifically promote invasion of breast cancer cell lines by controlling invadopodia and extracellular matrix degradation. Cell Mol. Life Sci. 2022, 79, 417. [Google Scholar] [CrossRef]
- Shi, Y.; Ye, Z.; Lu, G.; Yang, N.; Zhang, J.; Wang, L.; Cui, J.; Del Pozo, M.A.; Wu, Y.; Xia, D.; et al. Cholesterol-enriched membrane micro-domaindeficiency induces doxorubicin resistancevia promoting autophagy in breast cancer. Mol. Ther. Oncolytics 2021, 23, 311–329. [Google Scholar] [CrossRef]
- Shammout, O.D.A.; Ashmawy, N.S.; Shakartalla, S.B.; Altaie, A.M.; Semreen, M.H.; Omar, H.A.; Soliman, S.S.M. Comparative sphingolipidomic analysis reveals significant differences between doxorubicin-sensitive and -resistance MCF-7 cells. PLoS ONE 2021, 16, e0258363. [Google Scholar] [CrossRef]
- Pal, P.; Atilla-Gokcumen, G.E.; Frasor, J. Emerging Roles of Ceramides in Breast Cancer Biology and Therapy. Int. J. Mol. Sci. 2022, 23, 11178. [Google Scholar] [CrossRef]
- Haylett, A.K.; Moore, J.V. Comparative analysis of foetal calf and human low density lipoprotein: Relevance for pharmacodynamics of photosensitizers. J. Photochem. Photobiol. B 2002, 66, 171–178. [Google Scholar] [CrossRef]
- Patria, F.F.; Ceccarini, M.R.; Codini, M.; Conte, C.; Perioli, L.; Beccari, T.; Albi, E. A Role for Neutral Sphingomyelinase in Wound Healing Induced by Keratinocyte Proliferation upon 1α, 25-Dihydroxyvitamin D3 Treatment. Int. J. Mol. Sci. 2019, 20, 3634. [Google Scholar] [CrossRef] [Green Version]
- Ben Trivedi, A.; Kitabatake, N.; Doi, E. Toxicity of dimethyl sulfoxide as a solvent in bioassay system with HeLa cells evaluated colorimetrically with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide. Agric. Biol. Chem. 1990, 54, 2961. [Google Scholar] [CrossRef]
- Oz, E.S.; Aydemir, E.; Fışkın, K. DMSO exhibits similar cytotoxicity effects to thalidomide in mouse breast cancer cells. Oncol. Lett. 2012, 3, 927. [Google Scholar] [CrossRef] [Green Version]
- Galvao, J.; Davis, B.; Tilley, M.; Normando, E.; Duchen, M.R.; Cordeiro, M.F. Unexpected low-dose toxicity of the universal solvent DMSO. FASEB J. 2014, 28, 1317. [Google Scholar] [CrossRef]
- Albi, E.; Arcuri, C.; Kobayashi, T.; Tomishige, N.; Cas, M.D.; Paroni, R.; Signorelli, P.; Cerquiglini, L.; Troiani, S.; Gizzi, C.; et al. Sphingomyelin in Human Breast Milk might be Essential for the Hippocampus Maturation. Front. Biosci. 2022, 27, 247. [Google Scholar] [CrossRef]
- Rossi, G.; Magni, M.V.; Albi, E. Sphingomyelin-cholesterol and double stranded RNA relationship in the intranuclear complex. Arch. Biochem. Biophys. 2007, 459, 27–32. [Google Scholar] [CrossRef]
- Albi, E.; Viola Magni, P. The presence and the role of chromatin cholesterol in rat liver regeneration. J. Hepatol. 2002, 36, 395–400. [Google Scholar] [CrossRef]
- Rudel, L.L.; Morris, M.D. Determination of cholesterol using o-phthalaldehyde. J. Lipid Res. 1973, 14, 364–366. [Google Scholar] [CrossRef]
- Cataldi, S.; Ceccarini, M.R.; Patria, F.; Beccari, T.; Mandarano, M.; Ferri, I.; Lazzarini, A.; Curcio, F.; Albi, E. The Effect of Vitamin D3 and Silver Nanoparticles on HaCaT Cell Viability. Int. J. Mol. Sci. 2022, 23, 1410. [Google Scholar] [CrossRef]
- Albi, E.; Cataldi, S.; Ferri, I.; Sidoni, A.; Traina, G.; Fettucciari, K.; Ambesi-Impiombato, F.S.; Lazzarini, A.; Curcio, F.; Ceccarini, M.R.; et al. VDR independent induction of acid-sphingomyelinase by 1,23(OH)2 D3 in gastric cancer cells: Impact on apoptosis and cell morphology. Biochimie 2018, 146, 35–42. [Google Scholar] [CrossRef]
- Marini, F.; Bartoccini, E.; Cascianelli, G.; Voccoli, V.; Baviglia, M.G.; Magni, M.V.; Garcia-Gil, M.; Albi, E. Effect of 1alpha,25-dihydroxyvitamin D3 in embryonic hippocampal cells. Hippocampus 2010, 20, 696–705. [Google Scholar] [CrossRef]
- Cataldi, S.; Borrelli, A.; Ceccarini, M.R.; Nakashidze, I.; Codini, M.; Belov, O.; Ivanov, A.; Krasavin, E.; Ferri, I.; Conte, C.; et al. Acid and Neutral Sphingomyelinase Behavior in Radiation-Induced Liver Pyroptosis and in the Protective/Preventive Role of rMnSOD. Int. J. Mol. Sci. 2020, 21, 3281. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Albi, E.; Mandarano, M.; Cataldi, S.; Ceccarini, M.R.; Fiorani, F.; Beccari, T.; Sidoni, A.; Codini, M. The Effect of Cholesterol in MCF7 Human Breast Cancer Cells. Int. J. Mol. Sci. 2023, 24, 5935. https://doi.org/10.3390/ijms24065935
Albi E, Mandarano M, Cataldi S, Ceccarini MR, Fiorani F, Beccari T, Sidoni A, Codini M. The Effect of Cholesterol in MCF7 Human Breast Cancer Cells. International Journal of Molecular Sciences. 2023; 24(6):5935. https://doi.org/10.3390/ijms24065935
Chicago/Turabian StyleAlbi, Elisabetta, Martina Mandarano, Samuela Cataldi, Maria Rachele Ceccarini, Federico Fiorani, Tommaso Beccari, Angelo Sidoni, and Michela Codini. 2023. "The Effect of Cholesterol in MCF7 Human Breast Cancer Cells" International Journal of Molecular Sciences 24, no. 6: 5935. https://doi.org/10.3390/ijms24065935




