A Novel Microbial Consortia Catalysis Strategy for the Production of Hydroxytyrosol from Tyrosine
Abstract
:1. Introduction
2. Results
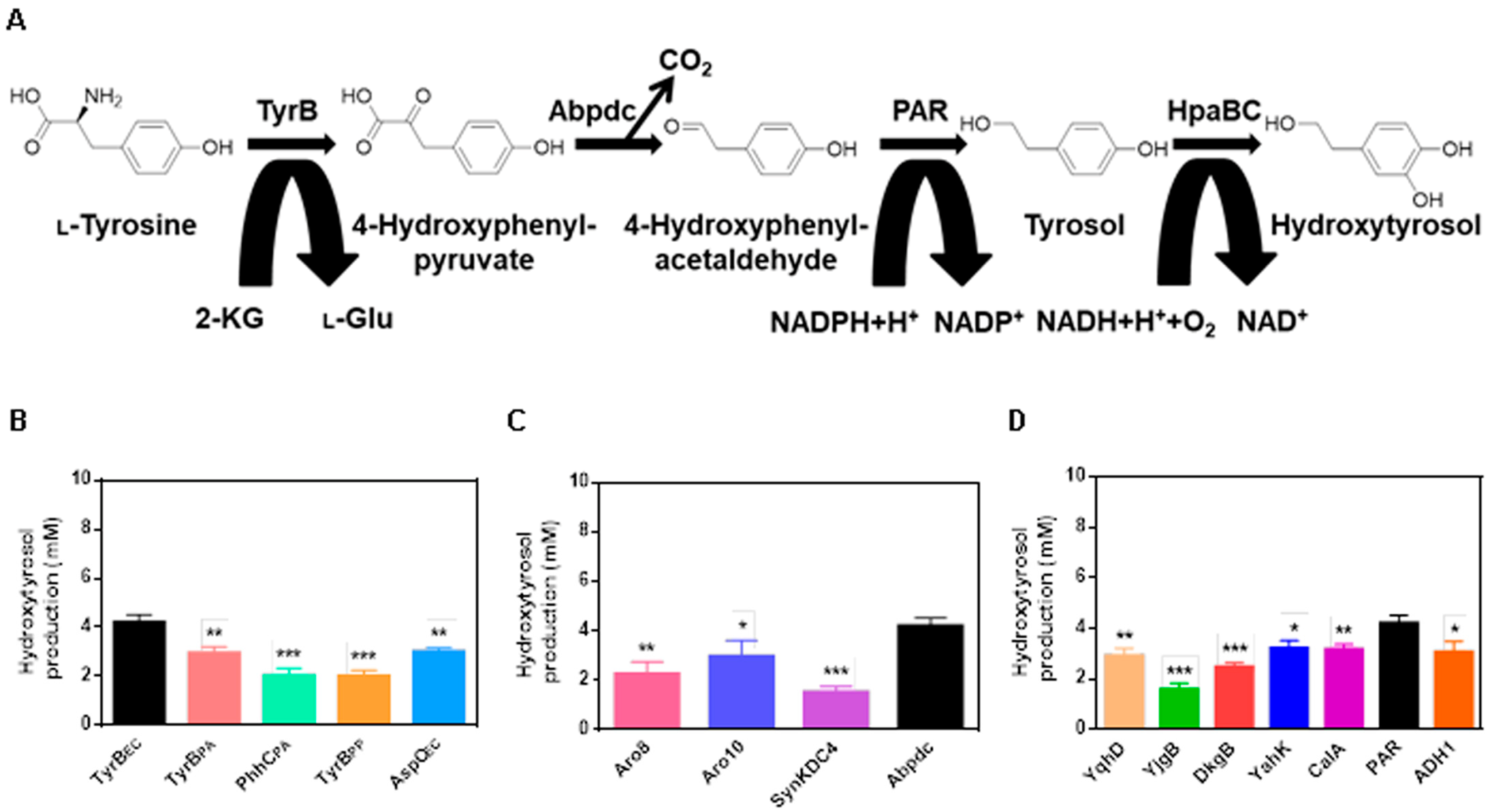
2.1. Design of the Hydroxytyrosol Biosynthetic Pathway
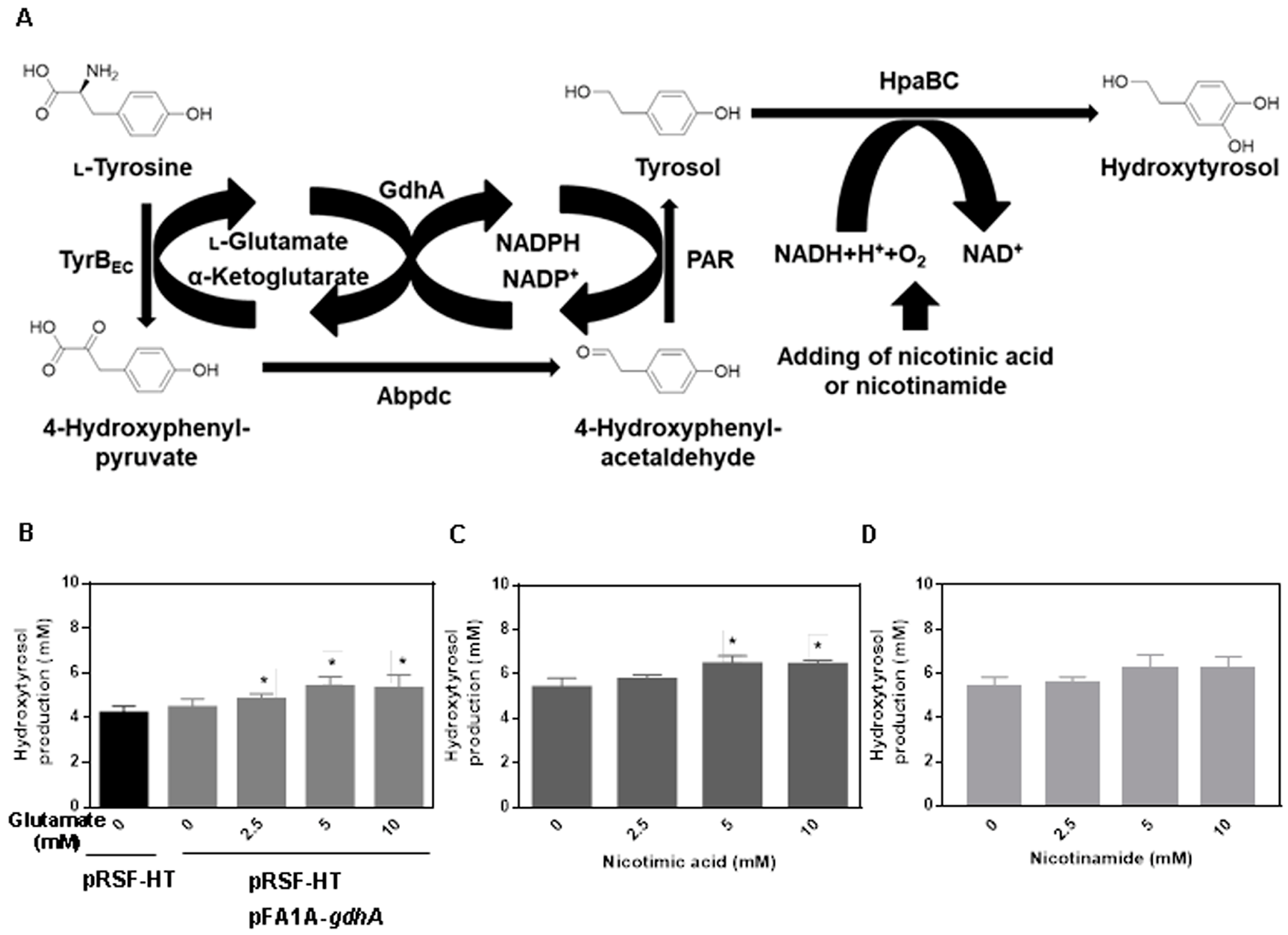
2.2. Cofactor Optimization
2.3. Design of a Microbial Consortia Catalysis Strategy
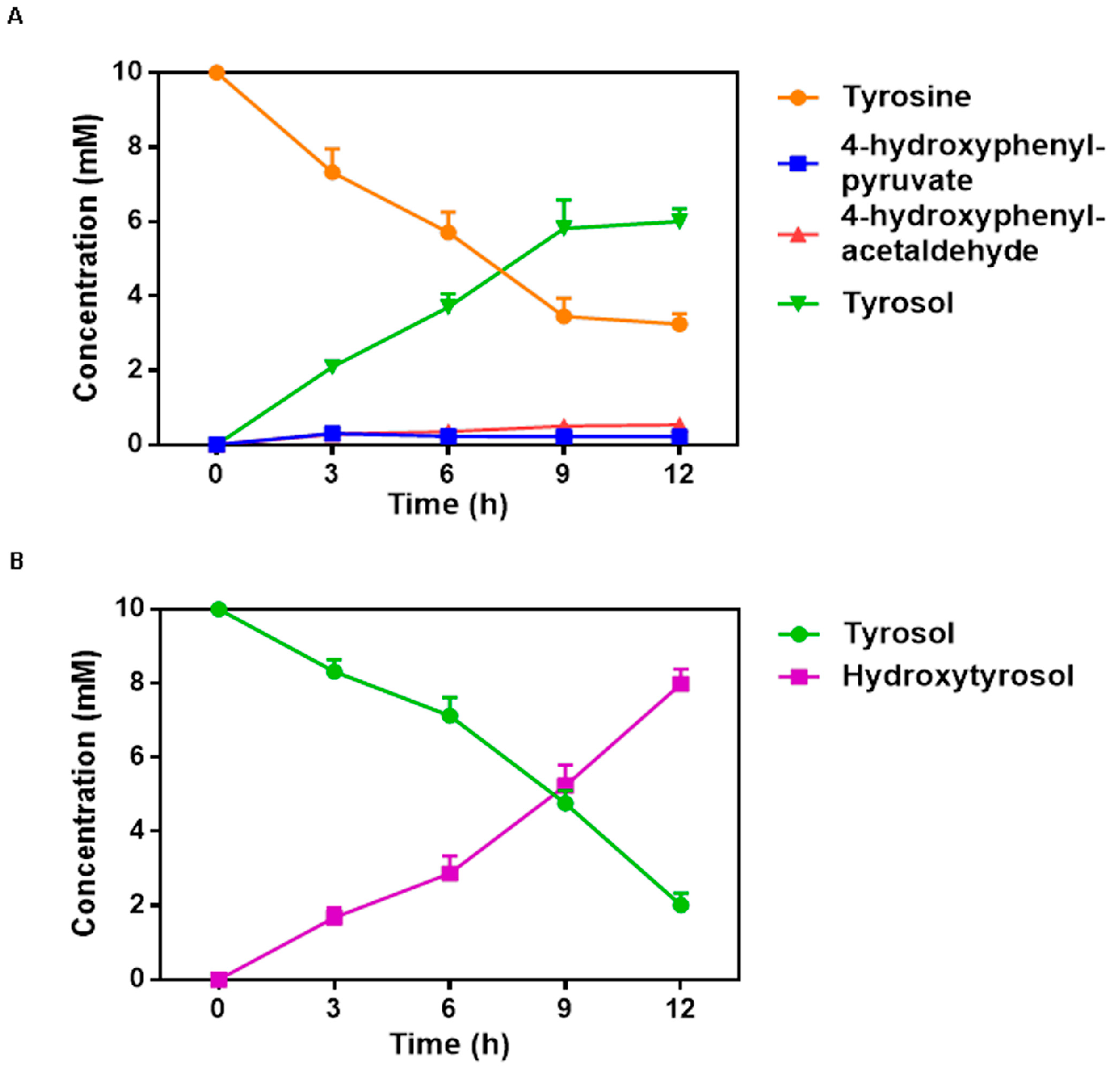
2.4. Fermentation Time Course of Two Individual Strains
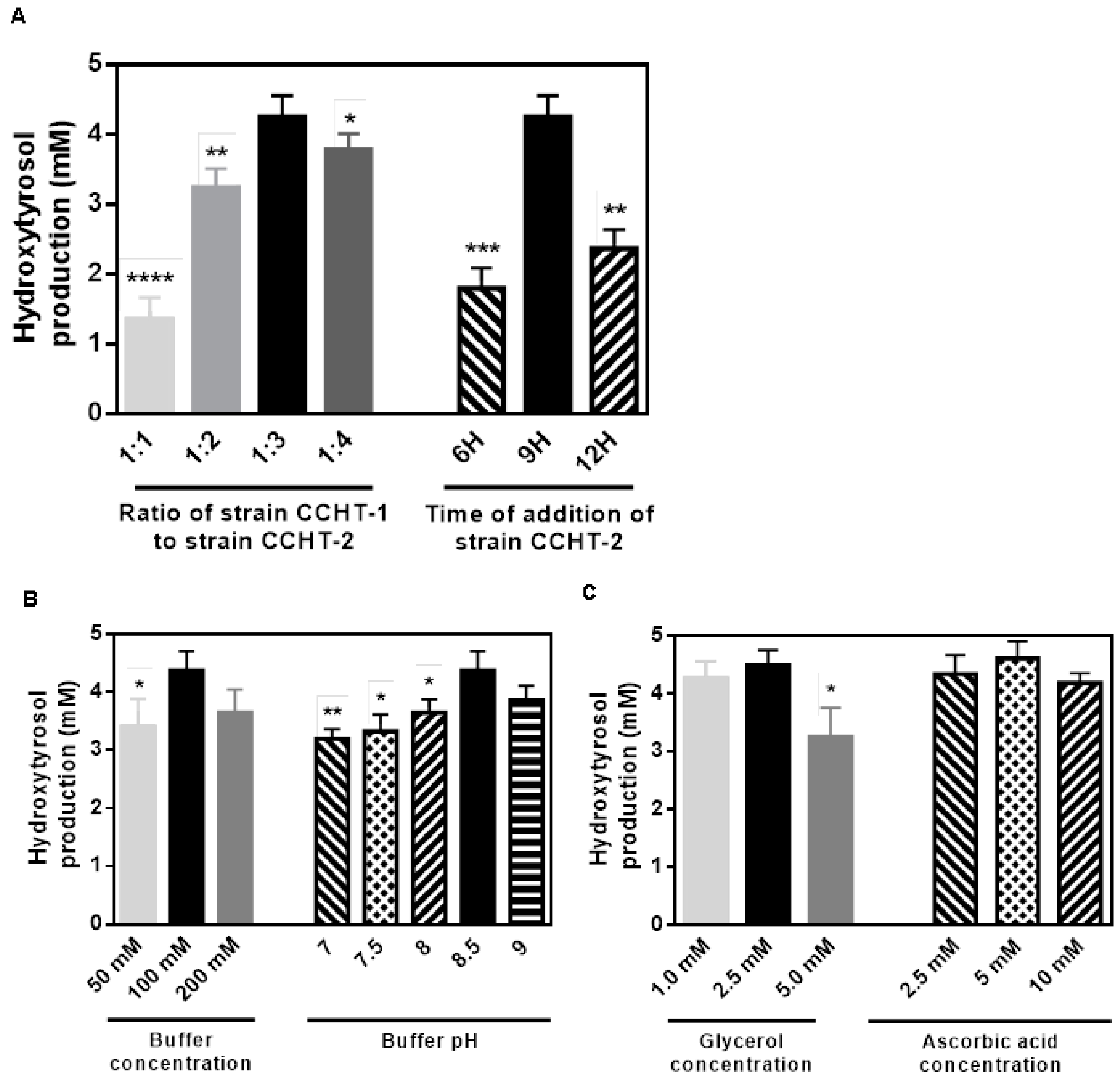
2.5. Optimization of the Microbial Consortia Catalysis
3. Discussion
4. Materials and Methods
4.1. General
4.2. Strain and Plasmid Construction
- (1)
- pRSF plasmids with different aminotransferases.
- (2)
- pRSF plasmids with different 4-hydroxyphenylpyruvate decarboxylases
- (3)
- pRSF plasmids with different reductases
- (4)
- Plasmids for co-culture strains
- (5)
- Plasmids for protein expression and purification
4.3. High-Performance Liquid Chromatography (HPLC) Quantification
4.4. Protein Expression and Purification
4.5. Activity Assay
4.6. Co-Culture Conditions
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, J.; Hansen, L.G.; Gudich, O.; Viehrig, K.; Lassen, L.M.M.; Schrubbers, L.; Adhikari, K.B.; Rubaszka, P.; Carrasquer-Alvarez, E.; Chen, L.; et al. A microbial supply chain for production of the anti-cancer drug vinblastine. Nature 2022, 609, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.H.; Li, B.Z.; Yuan, J.S.; Yuan, Y.J. Creative biological lignin conversion routes toward lignin valorization. Trends Biotechnol. 2022, 40, 1550–1566. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Qiao, K.; Edgar, S.; Stephanopoulos, G. Distributing a metabolic pathway among a microbial consortium enhances production of natural products. Nat. Biotechnol. 2015, 33, 377–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Pereira, B.; Li, Z.; Stephanopoulos, G. Engineering Escherichia coli coculture systems for the production of biochemical products. Proc. Natl. Acad. Sci. USA 2015, 112, 8266–8271. [Google Scholar] [CrossRef] [Green Version]
- Thuan, N.H.; Trung, N.T.; Cuong, N.X.; Van Cuong, D.; Van Quyen, D.; Malla, S. Escherichia coli modular coculture system for resveratrol glucosides production. World J. Microbiol. Biotechnol. 2018, 34, 75. [Google Scholar] [CrossRef]
- Wu, S.; Xue, Y.; Yang, S.; Xu, C.; Liu, C.; Liu, X.; Liu, J.; Zhu, H.; Zhao, G.R.; Yang, A.; et al. Combinational quorum sensing devices for dynamic control in cross-feeding cocultivation. Metab. Eng. 2021, 67, 186–197. [Google Scholar] [CrossRef]
- Aditya, C.; Bertaux, F.; Batt, G.; Ruess, J. A light tunable differentiation system for the creation and control of consortia in yeast. Nat. Commun. 2021, 12, 5829. [Google Scholar] [CrossRef]
- Chappell, T.C.; Nair, N.U. Co-utilization of hexoses by a microconsortium of sugar-specific E. coli strains. Biotechnol. Bioeng. 2017, 114, 2309–2318. [Google Scholar] [CrossRef]
- Li, X.; Zhou, Z.; Li, W.; Yan, Y.; Shen, X.; Wang, J.; Sun, X.; Yuan, Q. Design of stable and self-regulated microbial consortia for chemical synthesis. Nat. Commun. 2022, 13, 1554. [Google Scholar] [CrossRef]
- Coats, E.R.; Loge, F.J.; Smith, W.A.; Thompson, D.N.; Wolcott, M.P. Functional stability of a mixed microbial consortium producing PHA from waste carbon sources. Appl. Biochem. Biotechnol. 2007, 137–140, 909–925. [Google Scholar] [CrossRef] [Green Version]
- Thommes, M.; Wang, T.; Zhao, Q.; Paschalidis, I.C.; Segre, D. Designing Metabolic Division of Labor in Microbial Communities. mSystems 2019, 4, e00263-18. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Li, L.; Zhao, G.R. Systems Metabolic Engineering of Escherichia coli Coculture for De Novo Production of Genistein. Acs. Synth. Biol. 2022, 11, 1746–1757. [Google Scholar] [CrossRef]
- Pastor, R.; Bouzas, C.; Tur, J.A. Beneficial effects of dietary supplementation with olive oil, oleic acid, or hydroxytyrosol in metabolic syndrome: Systematic review and meta-analysis. Free Radic. Biol. Med. 2021, 172, 372–385. [Google Scholar] [CrossRef]
- Bertelli, M.; Kiani, A.K.; Paolacci, S.; Manara, E.; Kurti, D.; Dhuli, K.; Bushati, V.; Miertus, J.; Pangallo, D.; Baglivo, M.; et al. Hydroxytyrosol: A natural compound with promising pharmacological activities. J. Biotechnol. 2020, 309, 29–33. [Google Scholar] [CrossRef]
- Vijakumaran, U.; Yazid, M.D.; Hj Idrus, R.B.; Abdul Rahman, M.R.; Sulaiman, N. Molecular Action of Hydroxytyrosol in Attenuation of Intimal Hyperplasia: A Scoping Review. Front. Pharmacol. 2021, 12, 663266. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, C.; Abdullah; Tian, W.; Qiu, Z.; Song, M.; Cao, Y.; Xiao, J. Hydroxytyrosol Alleviates Dextran Sulfate Sodium-Induced Colitis by Modulating Inflammatory Responses, Intestinal Barrier, and Microbiome. J. Agric. Food Chem. 2022, 70, 2241–2252. [Google Scholar] [CrossRef]
- Medina-Martinez, M.S.; Truchado, P.; Castro-Ibanez, I.; Allende, A. Antimicrobial activity of hydroxytyrosol: A current controversy. Biosci. Biotechnol. Biochem. 2016, 80, 801–810. [Google Scholar] [CrossRef] [Green Version]
- Parra-Perez, A.M.; Perez-Jimenez, A.; Gris-Cardenas, I.; Bonel-Perez, G.C.; Carrasco-Diaz, L.M.; Mokhtari, K.; Garcia-Salguero, L.; Lupianez, J.A.; Rufino-Palomares, E.E. Involvement of the PI3K/AKT Intracellular Signaling Pathway in the AntiCancer Activity of Hydroxytyrosol, a Polyphenol from Olea europaea, in Hematological Cells and Implication of HSP60 Levels in Its Anti-Inflammatory Activity. Int. J. Mol. Sci. 2022, 23, 7053. [Google Scholar] [CrossRef]
- Bernini, R.; Merendino, N.; Romani, A.; Velotti, F. Naturally occurring hydroxytyrosol: Synthesis and anticancer potential. Curr. Med. Chem. 2013, 20, 655–670. [Google Scholar] [CrossRef]
- Satoh, Y.; Tajima, K.; Munekata, M.; Keasling, J.D.; Lee, T.S. Engineering of L-tyrosine oxidation in Escherichia coli and microbial production of hydroxytyrosol. Metab. Eng. 2012, 14, 603–610. [Google Scholar] [CrossRef]
- Chen, Z.; Sun, X.; Li, Y.; Yan, Y.; Yuan, Q. Metabolic engineering of Escherichia coli for microbial synthesis of monolignols. Metab. Eng. 2017, 39, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Li, X.L.; Chen, Z.Y.; Wu, Y.F.; Yan, Y.J.; Sun, X.X.; Yuan, Q.P. Establishing an Artificial Pathway for Efficient Biosynthesis of Hydroxytyrosol. ACS Synth. Biol. 2018, 7, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Bisquert, R.; Planells-Carcel, A.; Valera-Garcia, E.; Guillamon, J.M.; Muniz-Calvo, S. Metabolic engineering of Saccharomyces cerevisiae for hydroxytyrosol overproduction directly from glucose. Microb. Biotechnol. 2022, 15, 1499–1510. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Liu, H.; Hu, H.T.; Ng, K.R.; Yang, R.J.; Lyu, X.M. De Novo Production of Hydroxytyrosol by Metabolic Engineering of Saccharomyces cerevisiae. J. Agric. Food Chem. 2022, 70, 7490–7499. [Google Scholar] [CrossRef]
- Muniz-Calvo, S.; Bisquert, R.; Puig, S.; Guillamon, J.M. Overproduction of hydroxytyrosol in Saccharomyces cerevisiae by heterologous overexpression of the Escherichia coli 4-hydroxyphenylacetate 3-monooxygenase. Food Chem. 2020, 308, 125646. [Google Scholar] [CrossRef]
- Zeng, B.Y.; Lai, Y.M.; Liu, L.J.; Cheng, J.; Zhang, Y.; Yuan, J.F. Engineering Escherichia coli for High-Yielding Hydroxytyrosol Synthesis from Biobased L-Tyrosine. J. Agric. Food Chem. 2020, 68, 7691–7696. [Google Scholar] [CrossRef]
- Ren, Q.; Henes, B.; Fairhead, M.; Thony-Meyer, L. High level production of tyrosinase in recombinant Escherichia coli. BMC Biotechnol. 2013, 13, 18. [Google Scholar] [CrossRef] [Green Version]
- Liebgott, P.P.; Amouric, A.; Comte, A.; Tholozan, J.L.; Lorquin, J. Hydroxytyrosol from tyrosol using hydroxyphenylacetic acid-induced bacterial cultures and evidence of the role of 4-HPA 3-hydroxylase. Res. Microbiol. 2009, 160, 757–766. [Google Scholar] [CrossRef]
- Jiang, J.; Yin, H.; Wang, S.; Zhuang, Y.; Liu, S.; Liu, T.; Ma, Y. Metabolic Engineering of Saccharomyces cerevisiae for High-Level Production of Salidroside from Glucose. J. Agric. Food Chem. 2018, 66, 4431–4438. [Google Scholar] [CrossRef]
- Wang, P.; Yang, X.; Lin, B.; Huang, J.; Tao, Y. Cofactor self-sufficient whole-cell biocatalysts for the production of 2-phenylethanol. Metab. Eng. 2017, 44, 143–149. [Google Scholar] [CrossRef]
- Huang, Q.; Lin, Y.; Yan, Y. Caffeic acid production enhancement by engineering a phenylalanine over-producing Escherichia coli strain. Biotechnol. Bioeng. 2013, 110, 3188–3196. [Google Scholar] [CrossRef]
- Chen, W.; Yao, J.; Meng, J.; Han, W.; Tao, Y.; Chen, Y.; Guo, Y.; Shi, G.; He, Y.; Jin, J.M.; et al. Promiscuous enzymatic activity-aided multiple-pathway network design for metabolic flux rearrangement in hydroxytyrosol biosynthesis. Nat. Commun. 2019, 10, 960. [Google Scholar] [CrossRef] [Green Version]
- Rosenberg, J.; Commichau, F.M. Harnessing Underground Metabolism for Pathway Development. Trends Biotechnol. 2019, 37, 29–37. [Google Scholar] [CrossRef]
- Jiang, L.L.; Zhou, J.J.; Quan, C.S.; Xiu, Z.L. Advances in industrial microbiome based on microbial consortium for biorefinery. Bioresour. Bioprocess. 2017, 4, 11. [Google Scholar] [CrossRef] [Green Version]
- Yao, J.; He, Y.; Su, N.; Bharath, S.R.; Tao, Y.; Jin, J.M.; Chen, W.; Song, H.; Tang, S.Y. Developing a highly efficient hydroxytyrosol whole-cell catalyst by de-bottlenecking rate-limiting steps. Nat. Commun. 2020, 11, 1515. [Google Scholar] [CrossRef] [Green Version]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gong, P.; Tang, J.; Wang, J.; Wang, C.; Chen, W. A Novel Microbial Consortia Catalysis Strategy for the Production of Hydroxytyrosol from Tyrosine. Int. J. Mol. Sci. 2023, 24, 6944. https://doi.org/10.3390/ijms24086944
Gong P, Tang J, Wang J, Wang C, Chen W. A Novel Microbial Consortia Catalysis Strategy for the Production of Hydroxytyrosol from Tyrosine. International Journal of Molecular Sciences. 2023; 24(8):6944. https://doi.org/10.3390/ijms24086944
Chicago/Turabian StyleGong, Pengfei, Jiali Tang, Jiaying Wang, Chengtao Wang, and Wei Chen. 2023. "A Novel Microbial Consortia Catalysis Strategy for the Production of Hydroxytyrosol from Tyrosine" International Journal of Molecular Sciences 24, no. 8: 6944. https://doi.org/10.3390/ijms24086944




