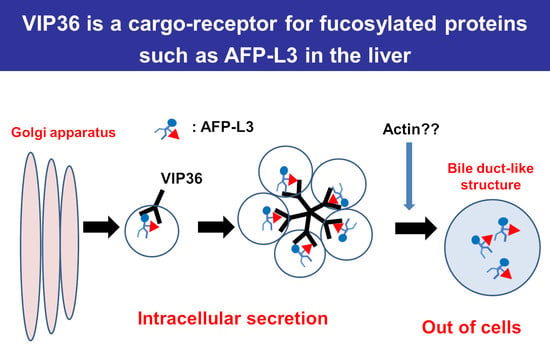
Vesicular Integral-Membrane Protein 36 Is Involved in the Selective Secretion of Fucosylated Proteins into Bile Duct-like Structures in HepG2 Cells
Abstract
:1. Introduction
2. Results
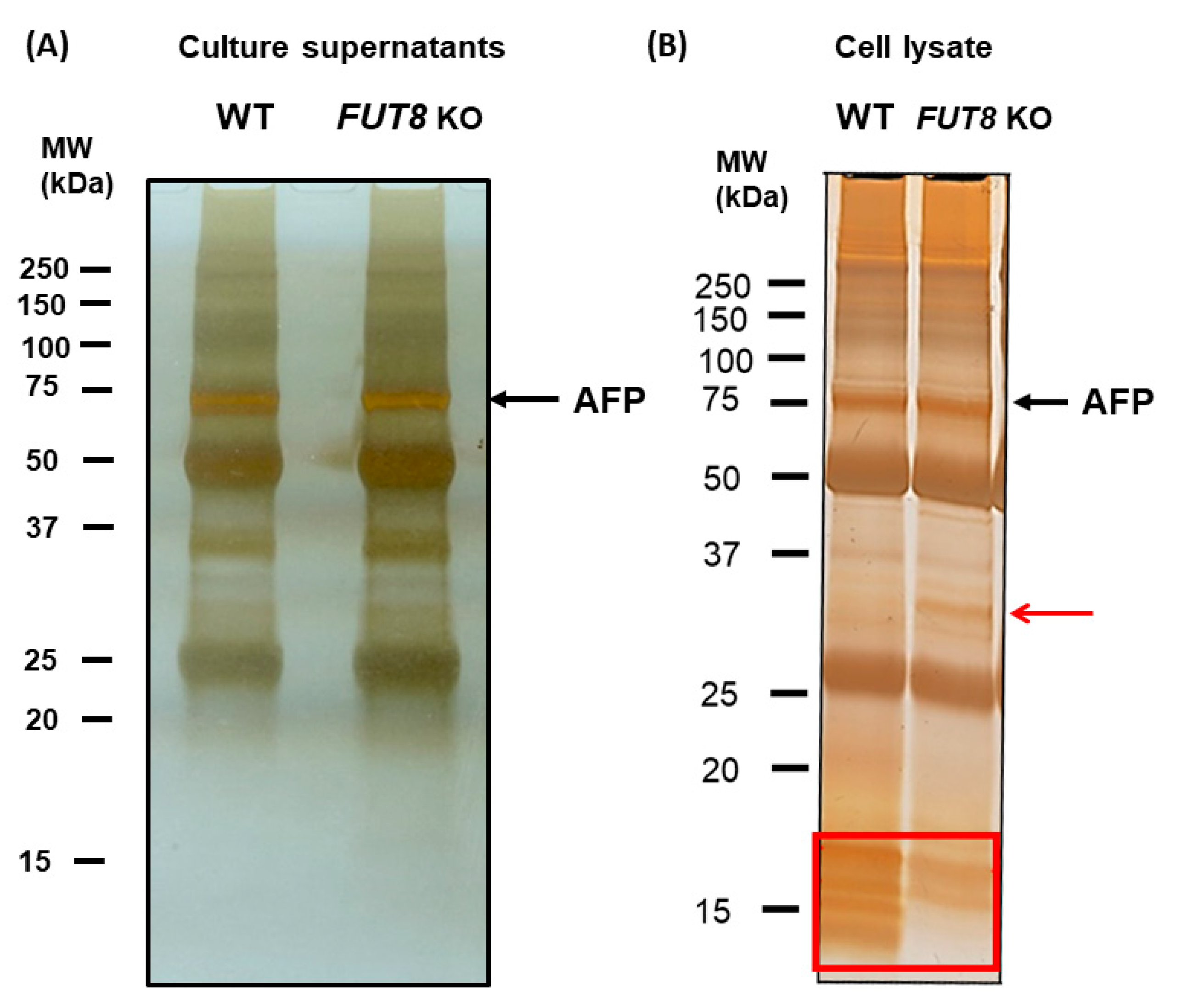
2.1. Immunostaining of AFP-L3 Was Not Observed in FUT8-Knockout HepG2 Cells
2.2. Identification of Cargo Receptors for AFP-L3 by Immunoprecipitation
2.3. Proteomics Analysis of AFP Using the Strep-Tag System
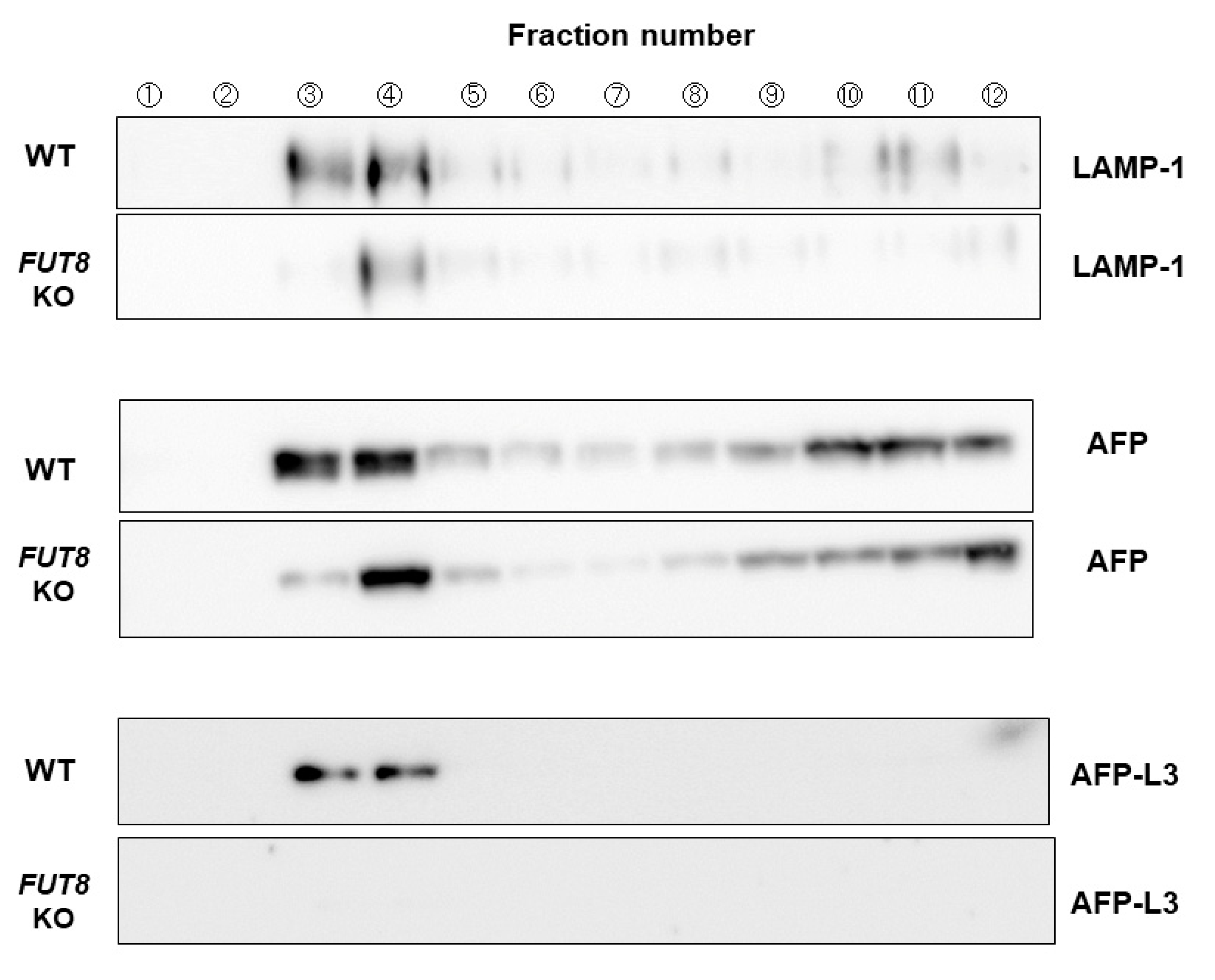
2.4. VIP36 Knockout Affects the Transport of AFP into the Bile Duct-like Structures of HepG2 Cells
2.5. Effect of VIP36 Knockout Affects the Alpha-1 Antitrypsin Localization in Bile Duct-like Structures of HepG2 Cells
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. Immunohistochemistry
4.3. Immunoprecipitation
4.4. Preparation of Endosome Fraction
4.5. The Proteomic Strep-Tag System
4.6. Mass Spectrometry and Immunocytechemistry
4.7. Production of VIP36 Knockout Vector
4.8. Transfection of AFP Gene, VIP36 KO Vector and Immunoblotting of VIP36
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| FUT8 | Alpha-1-6-fucosyltransferase |
| AFP | Alpha-fetoprotein |
| BC | Bile canaliculus |
| AFP-L3 | Fucosylated alpha-fetoprotein |
| FasMab | Fucosylated alpha-fetoprotein-specific monoclonal antibody |
| GDP | Guanosine diphosphate |
| HCC | Hepatocellular carcinoma |
| KO | Knockout |
| LAMP-1 | Lysosomal-associated membrane protein-1 |
| M2BP | Mac-2 binding protein |
| MVP | Major vault protein |
| MRP2 | Multidrug resistance-associated protein 2 |
| PCR | Polymerase chain reaction |
| PBS | Phosphate-buffered saline |
| PBS (−) | Phosphate-buffered saline without calcium and magnesium |
| SDS | Sodium dodecyl sulfate |
| VIP36 | Vesicular integral-membrane protein 36 |
| WT | Wild-type |
References
- Rademacher, T.W.; Parekh, R.B.; Dwek, R.A. Glycobiology. Annu. Rev. Biochem. 1988, 57, 785–838. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, E.; Kamada, Y.; Suzuki, T. Functional glycomics: Application to medical science and hepatology. Hepatol. Res. 2020, 50, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, E.; Moriwaki, K.; Nakagawa, T. Biological function of fucosylation in cancer biology. J. Biochem. 2008, 143, 725–729. [Google Scholar] [CrossRef]
- Verhelst, X.; Dias, A.M.; Colombel, J.F.; Vermeire, S.; Van Vlierberghe, H.; Callewaert, N.; Pinho, S.S. Protein Glycosylation as a Diagnostic and Prognostic Marker of Chronic Inflammatory Gastrointestinal and Liver Diseases. Gastroenterology 2020, 158, 95–110. [Google Scholar] [CrossRef] [Green Version]
- Kim, C.Y.; Kim, B.R.; Lee, S.S.; Jeon, D.H.; Lee, C.M.; Kim, W.S.; Cho, H.C.; Kim, J.J.; Lee, J.M.; Kim, H.J.; et al. Clinical features of hepatitis B and C virus infections, with high α-fetoprotein levels but not hepatocellular carcinoma. Medicine 2017, 96, e5844. [Google Scholar] [CrossRef]
- Taketa, K. Alpha-fetoprotein: Reevaluation in hepatology. Hepatology 1990, 12, 1420–1432. [Google Scholar] [CrossRef]
- Aoyagi, Y. Carbohydrate-based measurements on alpha-fetoprotein in the early diagnosis of hepatocellular carcinoma. Glycoconj. J. 1995, 12, 194–199. [Google Scholar] [CrossRef]
- Kagebayashi, C.; Yamaguchi, I.; Akinaga, A.; Kitano, H.; Yokoyama, K.; Satomura, M.; Kurosawa, T.; Watanabe, M.; Kawabata, T.; Chang, W.; et al. Automated immunoassay system for AFP-L3% using on-chip electrokinetic reaction and separation by affinity electrophoresis. Anal. Biochem. 2009, 388, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Force, M.; Park, G.; Chalikonda, D.; Roth, C.; Cohen, M.; Halegoua-DeMarzio, D.; Hann, H.W. Alpha-Fetoprotein (AFP) and AFP-L3 Is Most Useful in Detection of Recurrence of Hepatocellular Carcinoma in Patients after Tumor Ablation and with Low AFP Level. Viruses 2022, 14, 775. [Google Scholar] [CrossRef]
- Yi, X.; Yu, S.; Bao, Y. Alpha-fetoprotein-L3 in hepatocellular carcinoma: A meta-analysis. Clin. Chim. Acta 2013, 425, 212–220. [Google Scholar] [CrossRef]
- Cheng, J.; Wang, W.; Zhang, Y.; Liu, X.; Li, M.; Wu, Z.; Liu, Z.; Lv, Y.; Wang, B. Prognostic role of pre-treatment serum AFP-L3% in hepatocellular carcinoma: Systematic review and meta-analysis. PLoS ONE 2014, 9, e87011. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.M.; Wang, T.; Zhang, K.H. AFP-L3 for the diagnosis of early hepatocellular carcinoma: A meta-analysis. Medicine 2021, 100, e27673. [Google Scholar] [CrossRef]
- Liu, A.; Li, Y.; Shen, L.; Shen, L.; Li, Z. Clinical utility of serum fucosylated fraction of alpha-fetoprotein in the diagnostic of hepatocellular carcinoma: A comprehensive analysis with large sample size. Aging 2022, 14, 2645–2664. [Google Scholar] [CrossRef] [PubMed]
- Uozumi, N.; Yanagidani, S.; Miyoshi, E.; Ihara, Y.; Sakuma, T.; Gao, C.X.; Teshima, T.; Fujii, S.; Shiba, T.; Taniguchi, N. Purification and cDNA cloning of porcine brain GDP-L-Fuc:N-acetyl-beta-D-glucosaminide alpha1-->6fucosyltransferase. J. Biol. Chem. 1996, 271, 27810–27817. [Google Scholar] [CrossRef] [Green Version]
- Yanagidani, S.; Uozumi, N.; Ihara, Y.; Miyoshi, E.; Yamaguchi, N.; Taniguchi, N. Purification and cDNA cloning of GDP-L-Fuc:N-acetyl-beta-D-glucosaminide:alpha1-6 fucosyltransferase (alpha1-6 FucT) from human gastric cancer MKN45 cells. J. Biochem. 1997, 121, 626–632. [Google Scholar] [CrossRef]
- Noda, K.; Miyoshi, E.; Uozumi, N.; Yanagidani, S.; Ikeda, Y.; Gao, C.; Suzuki, K.; Yoshihara, H.; Yoshikawa, K.; Kawano, K.; et al. Gene expression of alpha1-6 fucosyltransferase in human hepatoma tissues: A possible implication for increased fucosylation of alpha-fetoprotein. Hepatology 1998, 28, 944–952. [Google Scholar] [CrossRef]
- Moriwaki, K.; Noda, K.; Nakagawa, T.; Asahi, M.; Yoshihara, H.; Taniguchi, N.; Hayashi, N.; Miyoshi, E. A high expression of GDP-fucose transporter in hepatocellular carcinoma is a key factor for increases in fucosylation. Glycobiology 2007, 17, 1311–1320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noda, K.; Miyoshi, E.; Gu, J.; Gao, C.X.; Nakahara, S.; Kitada, T.; Honke, K.; Suzuki, K.; Yoshihara, H.; Yoshikawa, K.; et al. Relationship between elevated FX expression and increased production of GDP-L-fucose, a common donor substrate for fucosylation in human hepatocellular carcinoma and hepatoma cell lines. Cancer Res. 2003, 63, 6282–6289. [Google Scholar]
- Nakagawa, T.; Uozumi, N.; Nakano, M.; Mizuno-Horikawa, Y.; Okuyama, N.; Taguchi, T.; Gu, J.; Kondo, A.; Taniguchi, N.; Miyoshi, E. Fucosylation of N-glycans regulates the secretion of hepatic glycoproteins into bile ducts. J. Biol. Chem. 2006, 281, 29797–29806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakagawa, T.; Takeishi, S.; Kameyama, A.; Yagi, H.; Yoshioka, T.; Moriwaki, K.; Masuda, T.; Matsumoto, H.; Kato, K.; Narimatsu, H.; et al. Glycomic analyses of glycoproteins in bile and serum during rat hepatocarcinogenesis. J. Proteome Res. 2010, 9, 4888–4896. [Google Scholar] [CrossRef]
- Nakagawa, T.; Moriwaki, K.; Terao, N.; Nakagawa, T.; Miyamoto, Y.; Kamada, Y.; Miyoshi, E. Analysis of polarized secretion of fucosylated alpha-fetoprotein in HepG2 cells. J. Proteome Res. 2012, 11, 2798–2806. [Google Scholar] [CrossRef] [PubMed]
- Egashira, Y.; Suganuma, M.; Kataoka, Y.; Higa, Y.; Ide, N.; Morishita, K.; Kamada, Y.; Gu, J.; Fukagawa, K.; Miyoshi, E. Establishment and characterization of a fucosylated α-fetoprotein-specific monoclonal antibody: A potential application for clinical research. Sci. Rep. 2019, 9, 12359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiedler, K. VIP36 preferentially binds to core-fucosylated N-glycans: A molecular docking study. bioRxiv 2016, 092460. [Google Scholar] [CrossRef] [Green Version]
- Hara-Kuge, S.; Seko, A.; Shimada, O.; Tosaka-Shimada, H.; Yamashita, K. The binding of VIP36 and alpha-amylase in the secretory vesicles via high-mannose type glycans. Glycobiology 2004, 14, 739–744. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Fukuda, T.; Isaji, T.; Lu, J.; Im, S.; Hang, Q.; Gu, W.; Hou, S.; Ohtsubo, K.; Gu, J. Loss of α1,6-fucosyltransferase inhibits chemical-induced hepatocellular carcinoma and tumorigenesis by down-regulating several cell signaling pathways. FASEB J.: Off. Publ. Fed. Am. Soc. Exp. Biol. 2015, 29, 3217–3227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Araújo, M.E.; Lamberti, G.; Huber, L.A. Isolation of Early and Late Endosomes by Density Gradient Centrifugation. Cold Spring Harb. Protoc. 2015, 2015, 1013–1016. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muranaka, M.; Takamatsu, S.; Ouchida, T.; Kanazawa, Y.; Kondo, J.; Nakagawa, T.; Egashira, Y.; Fukagawa, K.; Gu, J.; Okamoto, T.; et al. Vesicular Integral-Membrane Protein 36 Is Involved in the Selective Secretion of Fucosylated Proteins into Bile Duct-like Structures in HepG2 Cells. Int. J. Mol. Sci. 2023, 24, 7037. https://doi.org/10.3390/ijms24087037
Muranaka M, Takamatsu S, Ouchida T, Kanazawa Y, Kondo J, Nakagawa T, Egashira Y, Fukagawa K, Gu J, Okamoto T, et al. Vesicular Integral-Membrane Protein 36 Is Involved in the Selective Secretion of Fucosylated Proteins into Bile Duct-like Structures in HepG2 Cells. International Journal of Molecular Sciences. 2023; 24(8):7037. https://doi.org/10.3390/ijms24087037
Chicago/Turabian StyleMuranaka, Mizuki, Shinji Takamatsu, Tsunenori Ouchida, Yuri Kanazawa, Jumpei Kondo, Tsutomu Nakagawa, Yuriko Egashira, Koji Fukagawa, Jianguo Gu, Toru Okamoto, and et al. 2023. "Vesicular Integral-Membrane Protein 36 Is Involved in the Selective Secretion of Fucosylated Proteins into Bile Duct-like Structures in HepG2 Cells" International Journal of Molecular Sciences 24, no. 8: 7037. https://doi.org/10.3390/ijms24087037