Oriented Insertion of ESR-Containing Hybrid Proteins in Proteoliposomes
Abstract
:1. Introduction
2. Results
2.1. Recombinant Fusion Protein Construction and Expression
2.2. Characterization of the Fusion Proteins in Micelles
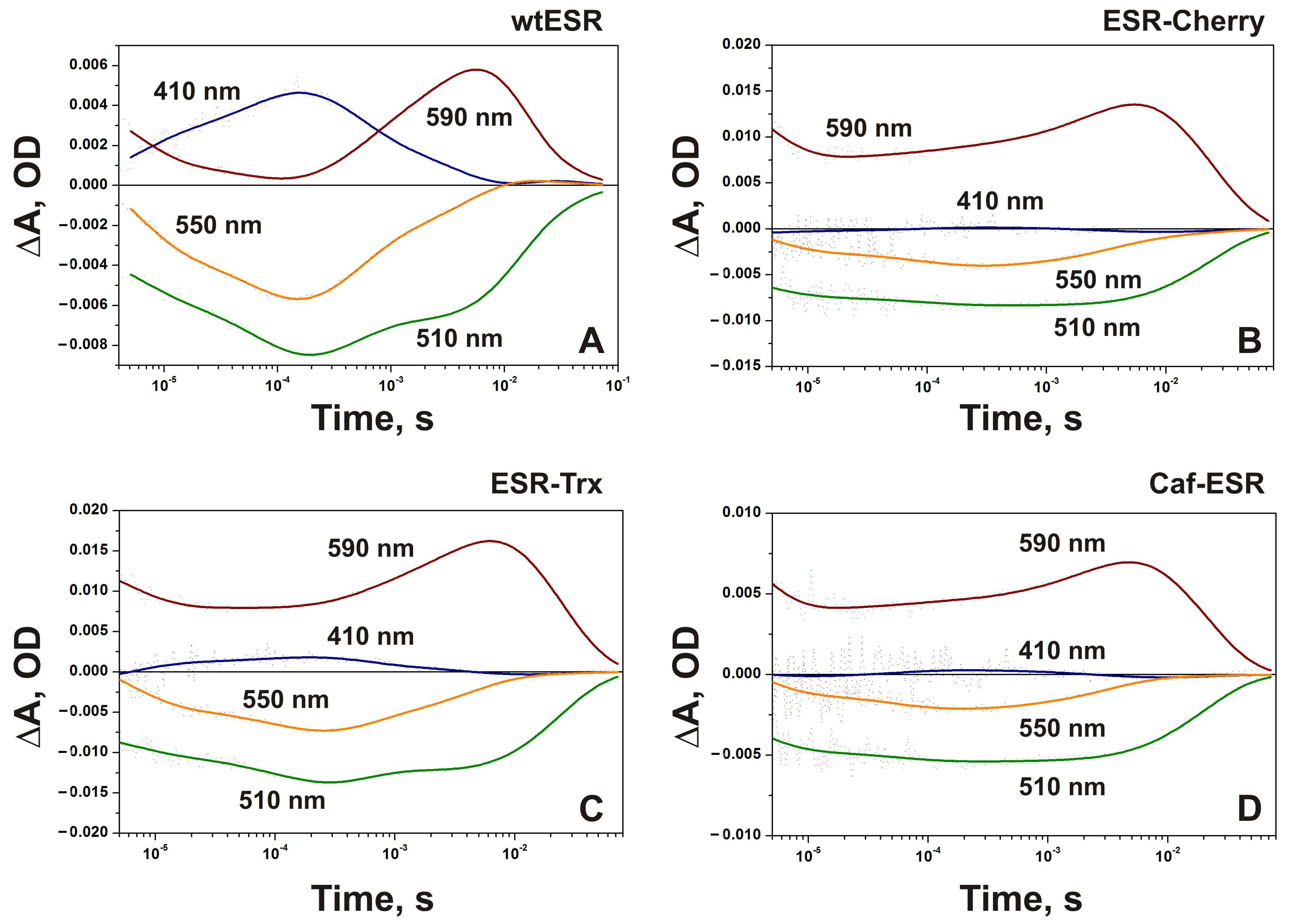
2.3. Photocycle of the Hybrid Proteins in Proteoliposomes
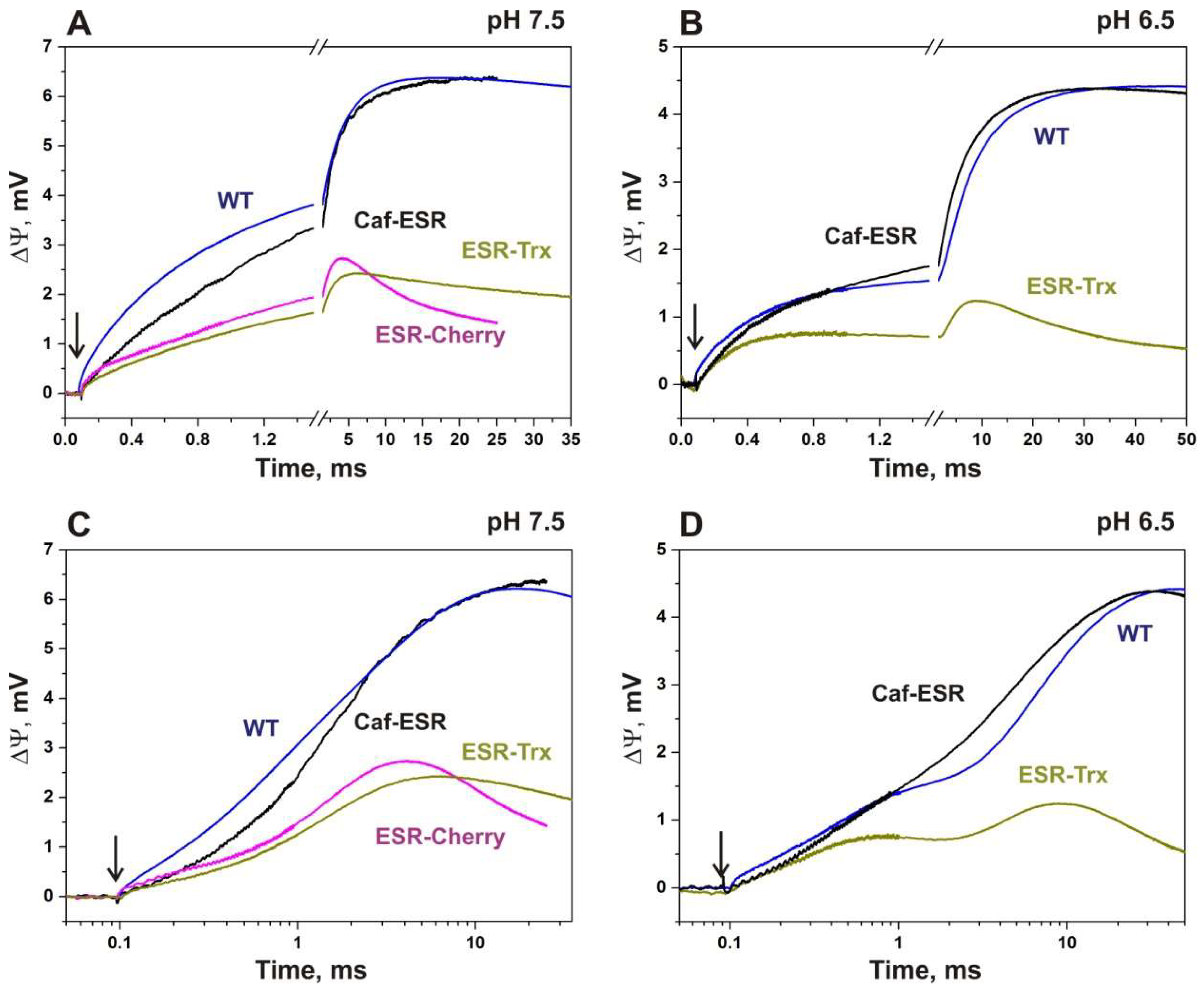
2.4. Electrogenic Response of the Fusion Proteins Incorporated into Proteoliposomes
2.4.1. ESR-Cherry
2.4.2. ESR-Trx
2.4.3. Caf-ESR
3. Discussion
4. Materials and Methods
4.1. Recombinant Gene Construction and Expression
4.2. Hybrid Protein Purification
4.3. Protein Electrophoresis and Western Blot
4.4. Spectroscopic Characterization
4.5. Electrometric Time-Resolved Measurements of the Membrane Potential ΔΨ Generation
4.6. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Ernst, O.P.; Lodowski, D.T.; Elstner, M.; Hegemann, P.; Brown, L.S.; Kandori, H. Microbial and animal rhodopsins: Structures, functions, and molecular mechanisms. Chem. Rev. 2014, 114, 126–163. [Google Scholar] [CrossRef]
- Govorunova, E.G.; Sineshchekov, O.A.; Li, H.; Spudich, J.L. Microbial Rhodopsins: Diversity, Mechanisms, and Optogenetic Applications. Annu. Rev. Biochem. 2017, 86, 845–872. [Google Scholar] [CrossRef]
- Gushchin, I.; Gordeliy, V. Microbial rhodopsins. In Membrane Protein Complexes: Structure and Function; Harris, J.R., Boekema, E.J., Eds.; Springer: Singapore, 2018; pp. 19–56. [Google Scholar] [CrossRef]
- Skulachev, V.P.; Bogachev, A.V.; Kasparinsky, F.O. Bacteriorhodopsin. In Principles of Bioenergetics; Springer: Berlin/Heidelberg, Germany, 2013; pp. 139–156. [Google Scholar] [CrossRef]
- Bamann, C.; Bamberg, E.; Wachtveitl, J.; Glaubitz, C. Proteorhodopsin. Biochim. Biophys. Acta (BBA)-Bioenerg. 2014, 1837, 614–625. [Google Scholar] [CrossRef]
- Pushkarev, A.; Béjà, O. Functional metagenomic screen reveals new and diverse microbial rhodopsins. ISME J. 2016, 10, 2331–2335. [Google Scholar] [CrossRef] [PubMed]
- Chazan, A.; Das, I.; Fujiwara, T.; Murakoshi, S.; Rozenberg, A.; Molina-Marquez, A.; Sano, F.K.; Tanaka, T.; Gomez-Villegas, P.; Larom, S.; et al. Phototrophy by antenna-containing rhodopsin pumps in aquatic environments. Nature 2023, 615, 525–540. [Google Scholar] [CrossRef]
- Shenkarev, Z.O.; Lyukmanova, E.N.; Butenko, I.O.; Petrovskaya, L.E.; Paramonov, A.S.; Shulepko, M.A.; Nekrasova, O.V.; Kirpichnikov, M.P.; Arseniev, A.S. Lipid-protein nanodiscs promote in vitro folding of transmembrane domains of multi-helical and multimeric membrane proteins. Biochim. Biophys. Acta 2013, 1828, 776–784. [Google Scholar] [CrossRef] [PubMed]
- Amati, A.M.; Graf, S.; Deutschmann, S.; Dolder, N.; von Ballmoos, C. Current problems and future avenues in proteoliposome research. Biochem. Soc. Trans. 2020, 48, 1473–1492. [Google Scholar] [CrossRef] [PubMed]
- Dürr, U.H.; Gildenberg, M.; Ramamoorthy, A. The magic of bicelles lights up membrane protein structure. Chem. Rev. 2012, 112, 6054–6074. [Google Scholar] [CrossRef] [PubMed]
- Denisov, I.G.; Sligar, S.G. Nanodiscs for structural and functional studies of membrane proteins. Nat. Struct. Mol. Biol. 2016, 23, 481–486. [Google Scholar] [CrossRef]
- Wang, Z.; Bai, J.; Xu, Y. The effect of charged lipids on bacteriorhodopsin membrane reconstitution and its photochemical activities. Biochem. Biophys. Res. Commun. 2008, 371, 814–817. [Google Scholar] [CrossRef] [PubMed]
- Corin, K.; Bowie, J.U. How bilayer properties influence membrane protein folding. Protein Sci. 2020, 29, 2348–2362. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.-S.; Bayley, H.; Khorana, H.G. Delipidation of bacteriorhodopsin and reconstitution with exogenous phospholipid. Proc. Nat. Acad. Sci. USA 1980, 77, 323–327. [Google Scholar] [CrossRef] [PubMed]
- Dioumaev, A.K.; Wang, J.M.; Bálint, Z.; Váró, G.; Lanyi, J.K. Proton transport by proteorhodopsin requires that the retinal Schiff base counterion Asp-97 be anionic. Biochemistry 2003, 42, 6582–6587. [Google Scholar] [CrossRef] [PubMed]
- Kandori, H. Biophysics of rhodopsins and optogenetics. Biophys. Rev. 2020, 12, 355–361. [Google Scholar] [CrossRef]
- Bräuchle, C.; Hampp, N.; Oesterhelt, D. Optical applications of bacteriorhodopsin and its mutated variants. Adv. Mater. 1991, 3, 420–428. [Google Scholar] [CrossRef]
- Choi, H.-J.; Montemagno, C.D. Artificial organelle: ATP synthesis from cellular mimetic polymersomes. Nano Lett. 2005, 5, 2538–2542. [Google Scholar] [CrossRef]
- Lee, K.Y.; Park, S.-J.; Lee, K.A.; Kim, S.-H.; Kim, H.; Meroz, Y.; Mahadevan, L.; Jung, K.-H.; Ahn, T.K.; Parker, K.K. Photosynthetic artificial organelles sustain and control ATP-dependent reactions in a protocellular system. Nat. Biotechnol. 2018, 36, 530–535. [Google Scholar] [CrossRef]
- Shen, H.H.; Lithgow, T.; Martin, L. Reconstitution of membrane proteins into model membranes: Seeking better ways to retain protein activities. Int. J. Mol. Sci. 2013, 14, 1589–1607. [Google Scholar] [CrossRef] [PubMed]
- Rigaud, J.-L.; Pitard, B.; Levy, D. Reconstitution of membrane proteins into liposomes: Application to energy-transducing membrane proteins. Biochim. Biophys. Acta (BBA)-Bioenerg. 1995, 1231, 223–246. [Google Scholar] [CrossRef]
- Bogdanov, M.; Dowhan, W.; Vitrac, H. Lipids and topological rules governing membrane protein assembly. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2014, 1843, 1475–1488. [Google Scholar] [CrossRef]
- Cymer, F.; Von Heijne, G.; White, S.H. Mechanisms of integral membrane protein insertion and folding. J. Mol. Biol. 2015, 427, 999–1022. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, A.; Koch, H.-G.; Dalbey, R.E.; Lovett, S.T.; Bernstein, H.D. Targeting and insertion of membrane proteins. EcoSal Plus 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Seigneuret, M.; Rigaud, J.-L. Use of the fluorescent pH probe pyranine to detect heterogeneous directions of proton movement in bacteriorhodopsin reconstituted large liposomes. FEBS Lett. 1985, 188, 101–106. [Google Scholar] [CrossRef]
- Pfleger, N.; Wörner, A.C.; Yang, J.; Shastri, S.; Hellmich, U.A.; Aslimovska, L.; Maier, M.S.; Glaubitz, C. Solid-state NMR and functional studies on proteorhodopsin. Biochim. Biophys. Acta (BBA)-Bioenerg. 2009, 1787, 697–705. [Google Scholar] [CrossRef] [PubMed]
- Tunuguntla, R.; Bangar, M.; Kim, K.; Stroeve, P.; Ajo-Franklin, C.M.; Noy, A. Lipid bilayer composition can influence the orientation of proteorhodopsin in artificial membranes. Biophys. J. 2013, 105, 1388–1396. [Google Scholar] [CrossRef]
- Lee, H.; Kim, H. Membrane topology of transmembrane proteins: Determinants and experimental tools. Biochem. Biophys. Res. Commun. 2014, 453, 268–276. [Google Scholar] [CrossRef]
- Denzer, A.J.; Nabholz, C.E.; Spiess, M. Transmembrane orientation of signal-anchor proteins is affected by the folding state but not the size of the N-terminal domain. EMBO J. 1995, 14, 6311–6317. [Google Scholar] [CrossRef] [PubMed]
- Ritzmann, N.; Thoma, J.; Hirschi, S.; Kalbermatter, D.; Fotiadis, D.; Muller, D.J. Fusion domains guide the oriented insertion of light-driven proton pumps into liposomes. Biophys. J. 2017, 113, 1181–1186. [Google Scholar] [CrossRef] [PubMed]
- Petrovskaya, L.E.; Lukashev, E.P.; Chupin, V.V.; Sychev, S.V.; Lyukmanova, E.N.; Kryukova, E.A.; Ziganshin, R.H.; Spirina, E.V.; Rivkina, E.M.; Khatypov, R.A.; et al. Predicted bacteriorhodopsin from Exiguobacterium sibiricum is a functional proton pump. FEBS Lett. 2010, 584, 4193–4196. [Google Scholar] [CrossRef]
- Balashov, S.P.; Petrovskaya, L.E.; Lukashev, E.P.; Imasheva, E.S.; Dioumaev, A.K.; Wang, J.M.; Sychev, S.V.; Dolgikh, D.A.; Rubin, A.B.; Kirpichnikov, M.P.; et al. Aspartate-histidine interaction in the retinal Schiff base counterion of the light-driven proton pump of Exiguobacterium sibiricum. Biochemistry 2012, 51, 5748–5762. [Google Scholar] [CrossRef]
- Gushchin, I.; Chervakov, P.; Kuzmichev, P.; Popov, A.N.; Round, E.; Borshchevskiy, V.; Ishchenko, A.; Petrovskaya, L.; Chupin, V.; Dolgikh, D.A.; et al. Structural insights into the proton pumping by unusual proteorhodopsin from nonmarine bacteria. Proc. Natl. Acad. Sci. USA 2013, 110, 12631–12636. [Google Scholar] [CrossRef] [PubMed]
- Balashov, S.P.; Petrovskaya, L.E.; Imasheva, E.S.; Lukashev, E.P.; Dioumaev, A.K.; Wang, J.M.; Sychev, S.V.; Dolgikh, D.A.; Rubin, A.B.; Kirpichnikov, M.P.; et al. Breaking the carboxyl rule: Lysine 96 facilitates reprotonation of the Schiff base in the photocycle of a retinal protein from Exiguobacterium sibiricum. J. Biol. Chem. 2013, 288, 21254–21265. [Google Scholar] [CrossRef] [PubMed]
- Siletsky, S.A.; Mamedov, M.D.; Lukashev, E.P.; Balashov, S.P.; Dolgikh, D.A.; Rubin, A.B.; Kirpichnikov, M.P.; Petrovskaya, L.E. Electrogenic steps of light-driven proton transport in ESR, a retinal protein from Exiguobacterium sibiricum. Biochim. Biophys. Acta (BBA)-Bioenerg. 2016, 1857, 1741–1750. [Google Scholar] [CrossRef] [PubMed]
- Siletsky, S.A.; Mamedov, M.D.; Lukashev, E.P.; Balashov, S.P.; Dolgikh, D.A.; Rubin, A.B.; Kirpichnikov, M.P.; Petrovskaya, L.E. Elimination of proton donor strongly affects directionality and efficiency of proton transport in ESR, a light-driven proton pump from Exiguobacterium sibiricum. Biochim. Biophys. Acta (BBA)-Bioenerg. 2019, 1860, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Siletsky, S.A.; Lukashev, E.P.; Mamedov, M.D.; Borisov, V.B.; Balashov, S.P.; Dolgikh, D.A.; Rubin, A.B.; Kirpichnikov, M.P.; Petrovskaya, L.E. His57 controls the efficiency of ESR, a light-driven proton pump from Exiguobacterium sibiricum at low and high pH. Biochim. Biophys. Acta (BBA)-Bioenerg. 2021, 1862, 148328. [Google Scholar] [CrossRef]
- Petrovskaya, L.E.; Lukashev, E.P.; Siletsky, S.A.; Imasheva, E.S.; Wang, J.M.; Mamedov, M.D.; Kryukova, E.A.; Dolgikh, D.A.; Rubin, A.B.; Kirpichnikov, M.P.; et al. Proton transfer reactions in donor site mutants of ESR, a retinal protein from Exiguobacterium sibiricum. J. Photochem. Photobiol. B Biol. 2022, 234, 112529. [Google Scholar] [CrossRef]
- Shaner, N.C.; Campbell, R.E.; Steinbach, P.A.; Giepmans, B.N.; Palmer, A.E.; Tsien, R.Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004, 22, 1567–1572. [Google Scholar] [CrossRef]
- Lavallie, E.R.; DiBlasio, E.A.; Kovacic, S.; Grant, K.L.; Schendel, P.F.; McCoy, J.M. A thioredoxin gene fusion expression system that circumvents inclusion body formation in the E. coli cytoplasm. Nat. Biotechnol. 1993, 11, 187–193. [Google Scholar] [CrossRef]
- Zavialov, A.V.; Batchikova, N.V.; Korpela, T.; Petrovskaya, L.E.; Korobko, V.G.; Kersley, J.; MacIntyre, S.; Zav’yalov, V.P. Secretion of recombinant proteins via the chaperone/usher pathway in Escherichia coli. Appl. Environ. Microbiol. 2001, 67, 1805–1814. [Google Scholar] [CrossRef]
- Dioumaev, A.K.; Petrovskaya, L.E.; Wang, J.M.; Balashov, S.P.; Dolgikh, D.A.; Kirpichnikov, M.P.; Lanyi, J.K. Photocycle of Exiguobacterium sibiricum rhodopsin characterized by low-temperature trapping in the IR and time-resolved studies in the visible. J. Phys. Chem. B 2013, 117, 7235–7253. [Google Scholar] [CrossRef] [PubMed]
- Drachev, L.A.; Kaulen, A.D.; Khitrina, L.V.; Skulachev, V.P. Fast stages of photoelectric processes in biological membranes. I. Bacteriorhodopsin. Eur. J. Biochem. 1981, 117, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Kalaidzidis, I.; Kaulen, A.; Radionov, A.; Khitrina, L. Photoelectrochemical cycle of bacteriorhodopsin. Biochemistry 2001, 66, 1220–1233. [Google Scholar] [CrossRef]
- Chun, E.; Thompson, A.A.; Liu, W.; Roth, C.B.; Griffith, M.T.; Katritch, V.; Kunken, J.; Xu, F.; Cherezov, V.; Hanson, M.A.; et al. Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors. Structure 2012, 20, 967–976. [Google Scholar] [CrossRef]
- Petrovskaya, L.; Shulga, A.; Bocharova, O.; Ermolyuk, Y.S.; Kryukova, E.; Chupin, V.; Blommers, M.; Arseniev, A.; Kirpichnikov, M. Expression of G-protein coupled receptors in Escherichia coli for structural studies. Biochemistry (Moscow) 2010, 75, 881–891. [Google Scholar] [CrossRef]
- Schlegel, S.; Hjelm, A.; Baumgarten, T.; Vikström, D.; de Gier, J.-W. Bacterial-based membrane protein production. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2014, 1843, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Kobe, B.; Ve, T.; Williams, S.J. Fusion-protein-assisted protein crystallization. Acta Crystallogr. F Struct. Biol. Commun. 2015, 71, 861–869. [Google Scholar] [CrossRef]
- Kiefer, H.; Krieger, J.; Olszewski, J.D.; von Heijne, G.; Prestwich, G.D.; Breer, H. Expression of an olfactory receptor in Escherichia coli: Purification, reconstitution, and ligand binding. Biochemistry 1996, 35, 16077–16084. [Google Scholar] [CrossRef]
- Banères, J.-L.; Mesnier, D.; Martin, A.; Joubert, L.; Dumuis, A.; Bockaert, J. Molecular Characterization of a Purified 5-HT4 Receptor: A structural basis for drug efficacy. J. Biol. Chem. 2005, 280, 20253–20260. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Qin, H.; Gao, F.P.; Cross, T.A. A systematic assessment of mature MBP in membrane protein production: Overexpression, membrane targeting and purification. Protein Expr. Purif. 2011, 80, 34–40. [Google Scholar] [CrossRef]
- Ishihara, G.; Goto, M.; Saeki, M.; Ito, K.; Hori, T.; Kigawa, T.; Shirouzu, M.; Yokoyama, S. Expression of G protein coupled receptors in a cell-free translational system using detergents and thioredoxin-fusion vectors. Protein Expr. Purif. 2005, 41, 27–37. [Google Scholar] [CrossRef]
- Chen, G.Q.; Gouaux, J.E. Overexpression of bacterio-opsin in Escherichia coli as a water-soluble fusion to maltose binding protein: Efficient regeneration of the fusion protein and selective cleavage with trypsin. Protein Sci. 1996, 5, 456–467. [Google Scholar] [CrossRef] [PubMed]
- Nekrasova, O.V.; Wulfson, A.N.; Tikhonov, R.V.; Yakimov, S.A.; Simonova, T.N.; Tagvey, A.I.; Dolgikh, D.A.; Ostrovsky, M.A.; Kirpichnikov, M.P. A new hybrid protein for production of recombinant bacteriorhodopsin in Escherichia coli. J. Biotechnol. 2010, 147, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Drew, D.; Lerch, M.; Kunji, E.; Slotboom, D.-J.; de Gier, J.-W. Optimization of membrane protein overexpression and purification using GFP fusions. Nat. Methods 2006, 3, 303. [Google Scholar] [CrossRef]
- Duellman, T.; Burnett, J.; Yang, J. Quantitation of secreted proteins using mCherry fusion constructs and a fluorescent microplate reader. Anal. Biochem. 2015, 473, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Kano, K.; Fendler, J.H. Pyranine as a sensitive pH probe for liposome interiors and surfaces. pH gradients across phospholipid vesicles. Biochim. Biophys. Acta (BBA)-Biomembr. 1978, 509, 289–299. [Google Scholar] [CrossRef]
- Kyrychenko, A.; Vasquez-Montes, V.; Ulmschneider, M.B.; Ladokhin, A.S. Lipid Headgroups Modulate Membrane Insertion of pHLIP Peptide. Biophys. J. 2015, 108, 791–794. [Google Scholar] [CrossRef]
- Kalaidzidis, Y.L.; Gavrilov, A.; Zaitsev, P.; Kalaidzidis, A.; Korolev, E. PLUK-an environment for software development. Program. Comp. Softw. 1997, 23, 206–211. [Google Scholar]
- Kaulen, A.D. Electrogenic processes and protein conformational changes accompanying the bacteriorhodopsin photocycle. Biochim. Biophys. Acta (BBA)-Bioenerg. 2000, 1460, 204–219. [Google Scholar] [CrossRef]
- Mamedov, M.D.; Tyunyatkina, A.A.; Siletsky, S.A.; Semenov, A.Y. Voltage changes involving photosystem II quinone-iron complex turnover. Eur. Biophys. J. 2006, 35, 647–654. [Google Scholar] [CrossRef]
- Siletsky, S.A.; Zhu, J.; Gennis, R.B.; Konstantinov, A.A. Partial steps of charge translocation in the nonpumping N139L mutant of Rhodobacter sphaeroides cytochrome c oxidase with a blocked D-channel. Biochemistry 2010, 49, 3060–3073. [Google Scholar] [CrossRef]
- Siletsky, S.A.; Mamedov, M.D.; Lukashev, E.P.; Balashov, S.P.; Petrovskaya, L.E. Application of direct electrometry in studies of microbial rhodopsins reconstituted in proteoliposomes. Biophys. Rev. 2022, 14, 771–778. [Google Scholar] [CrossRef] [PubMed]
- Siletsky, S.A.; Han, D.; Brand, S.; Morgan, J.E.; Fabian, M.; Geren, L.; Millett, F.; Durham, B.; Konstantinov, A.A.; Gennis, R.B. Single-electron photoreduction of the P M intermediate of cytochrome c oxidase. Biochim. Biophys. Acta (BBA)-Bioenerg. 2006, 1757, 1122–1132. [Google Scholar] [CrossRef] [PubMed]
- Siletsky, S.A. Steps of the coupled charge translocation in the catalytic cycle of cytochrome c oxidase. Front. Biosci. (Landmark Ed.) 2013, 18, 36–57. [Google Scholar] [CrossRef] [PubMed]
- Siletsky, S.A.; Borisov, V.B.; Mamedov, M.D. Photosystem II and terminal respiratory oxidases: Molecular machines operating in opposite directions. Front. Biosci. (Landmark Ed.) 2017, 22, 1379–1426. [Google Scholar] [CrossRef]
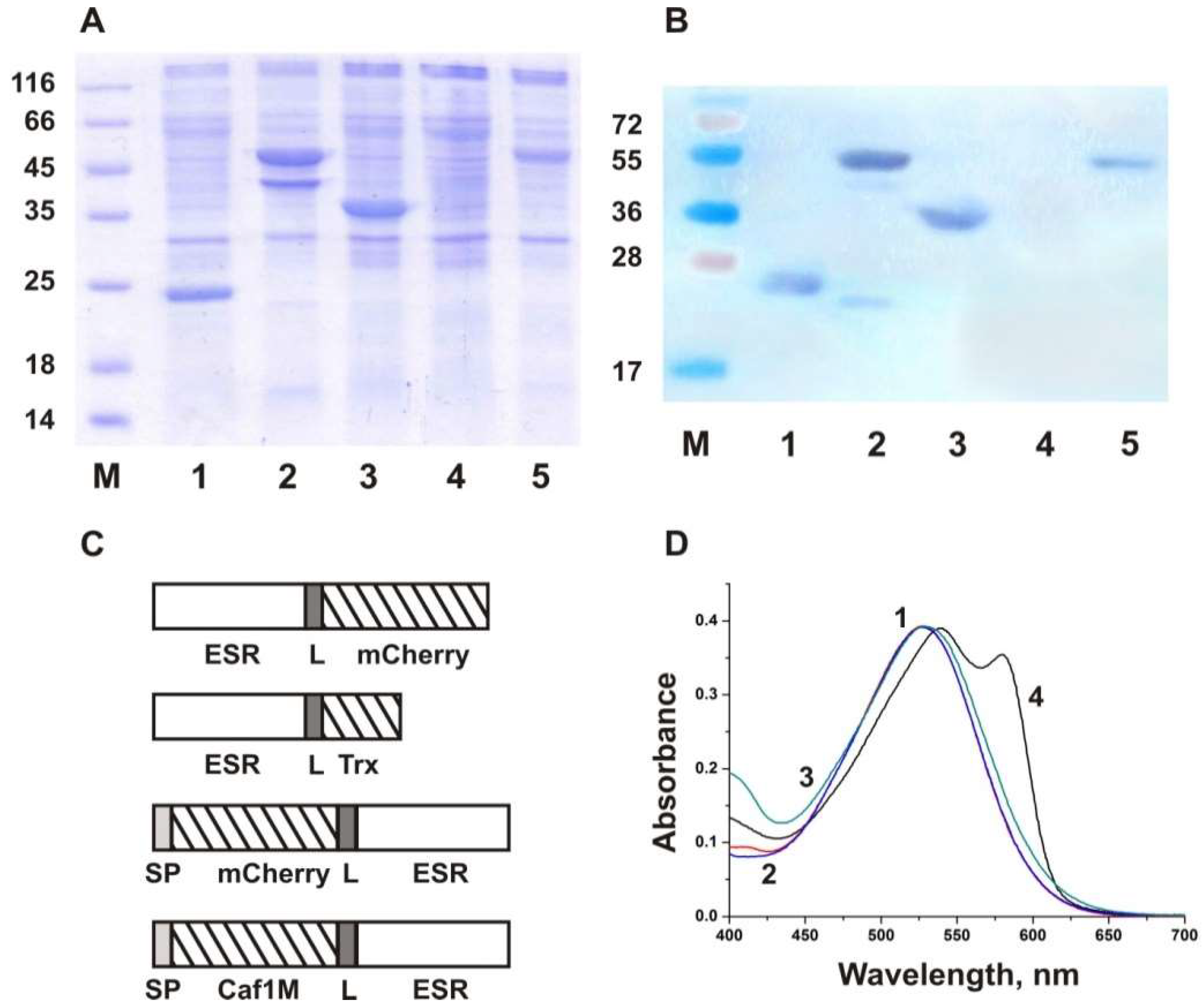



| Protein | Molecular Weight, kDa | pI ** | Reference |
|---|---|---|---|
| mCherry | 26.6 | 5.63 | [39] |
| Trx | 11.7 | 4.67 | [40] |
| Caf1M * | 26.8 | 7.9 | [41] |
| Protein | τ1 | τ2 | τ3 | τ4 | τ5 | τ6 |
|---|---|---|---|---|---|---|
| wt ESR | 0.0044 ± 0.0002 | 0.13 ± 0.008 | 2.24 ± 0.13 | 12.1 ± 0.38 | 82.6 ± 3.6 | 310 ± 14 |
| ESR-Cherry | 0.0092 ± 0.0011 | 0.18 ± 0.013 | 2.9 ± 0.27 | 10.0 ± 0.77 | 73.6 ± 5.2 | 293 ± 21 |
| ESR-Trx | 0.008 ± 0.0006 | 0.15 ± 0.02 | 2.28 ± 0.14 | 13.3 ± 0.52 | 79.6 ± 5.4 | 319 ± 20 |
| Caf-ESR | 0.004 ± 0.0003 | 0.08 ± 0.01 | 2.23 ± 0.11 | 14.2 ± 0.66 | 60.0 ± 3.9 | 322 ± 29 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petrovskaya, L.E.; Lukashev, E.P.; Mamedov, M.D.; Kryukova, E.A.; Balashov, S.P.; Dolgikh, D.A.; Rubin, A.B.; Kirpichnikov, M.P.; Siletsky, S.A. Oriented Insertion of ESR-Containing Hybrid Proteins in Proteoliposomes. Int. J. Mol. Sci. 2023, 24, 7369. https://doi.org/10.3390/ijms24087369
Petrovskaya LE, Lukashev EP, Mamedov MD, Kryukova EA, Balashov SP, Dolgikh DA, Rubin AB, Kirpichnikov MP, Siletsky SA. Oriented Insertion of ESR-Containing Hybrid Proteins in Proteoliposomes. International Journal of Molecular Sciences. 2023; 24(8):7369. https://doi.org/10.3390/ijms24087369
Chicago/Turabian StylePetrovskaya, Lada E., Evgeniy P. Lukashev, Mahir D. Mamedov, Elena A. Kryukova, Sergei P. Balashov, Dmitry A. Dolgikh, Andrei B. Rubin, Mikhail P. Kirpichnikov, and Sergey A. Siletsky. 2023. "Oriented Insertion of ESR-Containing Hybrid Proteins in Proteoliposomes" International Journal of Molecular Sciences 24, no. 8: 7369. https://doi.org/10.3390/ijms24087369







