Proteomic Exploration of Paraoxonase 1 Function in Health and Disease
Abstract
:1. Introduction
2. PON1, Lipid Oxidation, and CVD
3. Mechanistic Bases of PON1 Involvement in CVD
3.1. PON1 Controls NO Synthesis
3.2. PON1 Is Not a Redox Protein
3.3. PON1 Interacts with Redox-Related Proteins
3.4. HHcy Diet Exacerbates Pro-Oxidative and Pro-Atherogenic Changes in Mouse Proteome
4. PON1, Lipid Oxidation, and Alzheimer’s Disease
4.1. PON1 and Cognition
4.2. Mechanistic Bases of PON1 Involvement in AD
4.2.1. Pon1 Depletion Induces Pro-Neurodegenerative Changes in Mouse Brain Proteome
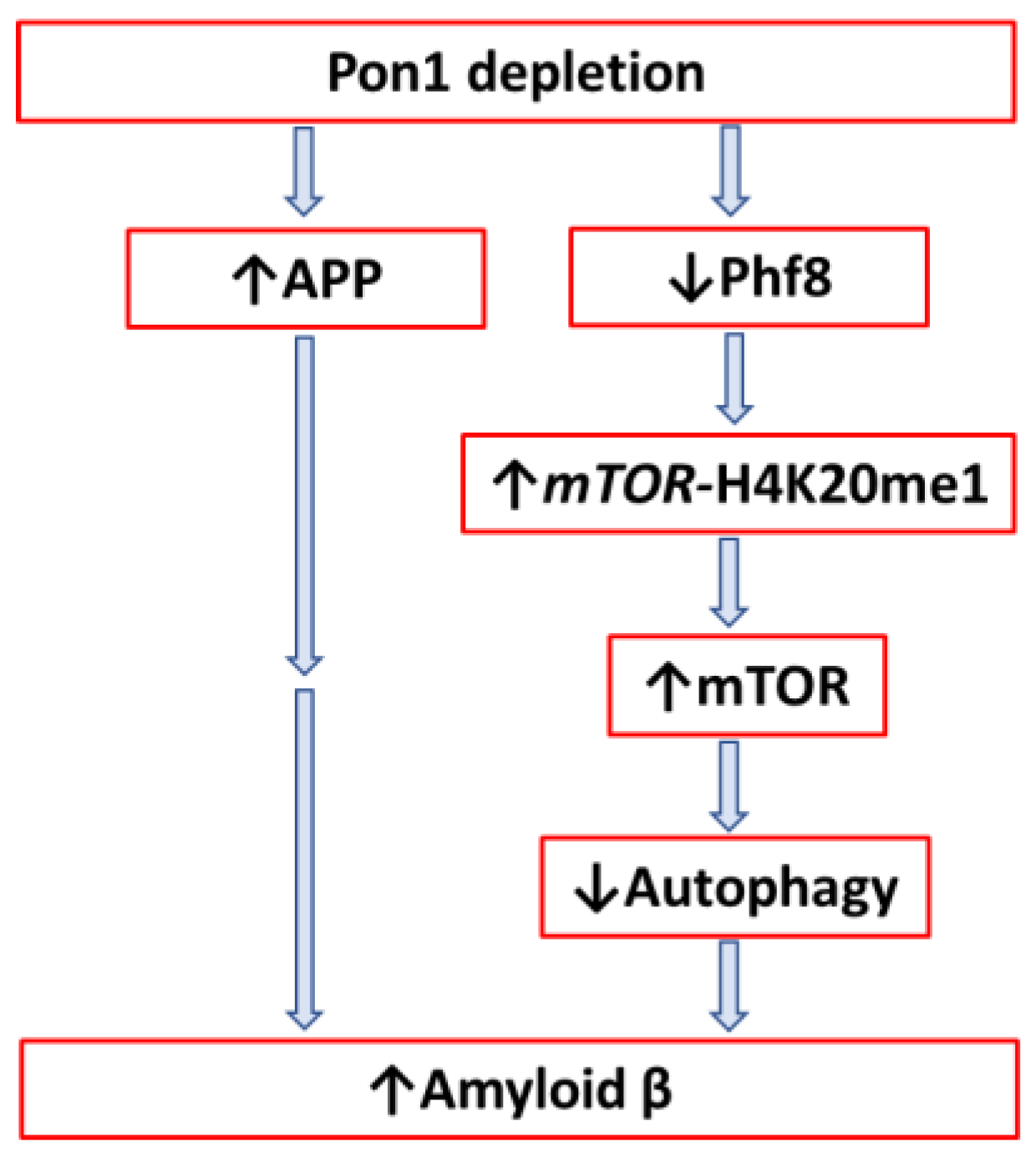
4.2.2. Pon1 Depletion Induces Accumulation of Amyloid β in Mouse Brain
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mackness, B.; Beltran-Debon, R.; Aragones, G.; Joven, J.; Camps, J.; Mackness, M. Human tissue distribution of paraoxonases 1 and 2 mRNA. IUBMB Life 2010, 62, 480–482. [Google Scholar] [CrossRef]
- Leduc, V.; Legault, V.; Dea, D.; Poirier, J. Normalization of gene expression using SYBR green qPCR: A case for paraoxonase 1 and 2 in Alzheimer’s disease brains. J. Neurosci. Methods 2011, 200, 14–19. [Google Scholar] [CrossRef]
- Witucki, L.; Jakubowski, H. Depletion of Paraoxonase 1 (Pon1) Dysregulates mTOR, Autophagy, and Accelerates Amyloid Beta Accumulation in Mice. Cells 2023, 12, 746. [Google Scholar] [CrossRef] [PubMed]
- Marsillach, J.; Mackness, B.; Mackness, M.; Riu, F.; Beltran, R.; Joven, J.; Camps, J. Immunohistochemical analysis of paraoxonases-1, 2, and 3 expression in normal mouse tissues. Free. Radic. Biol. Med. 2008, 45, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Moren, X.; Lhomme, M.; Bulla, A.; Sanchez, J.-C.; Kontush, A.; James, R.W. Proteomic and lipidomic analyses of paraoxonase defined high density lipoprotein particles: Association of paraoxonase with the anti-coagulant, protein S. Proteom. Clin. Appl. 2016, 10, 230–238. [Google Scholar] [CrossRef]
- Humbert, R.; Adler, D.A.; Disteche, C.M.; Hassett, C.; Omiecinski, C.J.; Furlong, C.E. The molecular basis of the human serum paraoxonase activity polymorphism. Nat. Genet. 1993, 3, 73–76. [Google Scholar] [CrossRef]
- Jakubowski, H.; Ambrosius, W.T.; Pratt, J. Genetic determinants of homocysteine thiolactonase activity in humans: Implications for atherosclerosis. FEBS Lett. 2001, 491, 35–39. [Google Scholar] [CrossRef] [PubMed]
- Loscalzo, J. Paraoxonase and coronary heart disease risk: Language misleads, linkage misinforms, function clarifies. Circ. Cardiovasc. Genet. 2008, 1, 79–80. [Google Scholar] [CrossRef]
- Zhang, Q.; Jiang, Z.; Xu, Y. HDL and Oxidation. Adv. Exp. Med. Biol. 2022, 1377, 63–77. [Google Scholar]
- Marsillach, J.; Adorni, M.P.; Zimetti, F.; Papotti, B.; Zuliani, G.; Cervellati, C. HDL Proteome and Alzheimer’s Disease: Evidence of a Link. Antioxidants 2020, 9, 1224. [Google Scholar] [CrossRef]
- Jakubowski, H. Calcium-dependent Human Serum Homocysteine Thiolactone Hydrolase. A protective mechanism against protein N-homocysteinylation. J. Biol. Chem. 2000, 275, 3957–3962. [Google Scholar] [CrossRef] [PubMed]
- Borowczyk, K.; Shih, D.M.; Jakubowski, H. Metabolism and Neurotoxicity of Homocysteine Thiolactone in Mice: Evidence for a Protective Role of Paraoxonase 1. J. Alzheimer’s Dis. 2012, 30, 225–231. [Google Scholar] [CrossRef]
- Jakubowski, H.; Goldman, E. Synthesis of homocysteine thiolactone by methionyl-tRNA synthetase in cultured mammalian cells. FEBS Lett. 1993, 317, 237–240. [Google Scholar] [CrossRef]
- Jakubowski, H. Quality control in tRNA charging. Wiley Interdiscip. Rev. RNA 2012, 3, 295–310. [Google Scholar] [CrossRef] [PubMed]
- Jakubowski, H. Homocysteine Modification in Protein Structure/Function and Human Disease. Physiol. Rev. 2019, 99, 555–604. [Google Scholar] [CrossRef]
- Borowczyk, K.; Piechocka, J.; Głowacki, R.; Dhar, I.; Midtun, O.; Tell, G.S.; Ueland, P.M.; Nygård, O.; Jakubowski, H. Urinary excretion of homocysteine thiolactone and the risk of acute myocardial infarction in coronary artery disease patients: The WENBIT trial. J. Intern. Med. 2019, 285, 232–244. [Google Scholar] [CrossRef]
- Sauls, D.L.; Lockhart, E.; Warren, M.E.; Lenkowski, A.; Wilhelm, S.E.; Hoffman, M. Modification of Fibrinogen by Homocysteine Thiolactone Increases Resistance to Fibrinolysis: A Potential Mechanism of the Thrombotic Tendency in Hyperhomocysteinemia. Biochemistry 2006, 45, 2480–2487. [Google Scholar] [CrossRef] [PubMed]
- Sikora, M.; Skrzydlewski, P.; Perła-Kaján, J.; Jakubowski, H. Homocysteine thiolactone contributes to the prognostic value of fibrin clot structure/function in coronary artery disease. PLoS ONE 2022, 17, e0275956. [Google Scholar] [CrossRef]
- Lacinski, M.; Skorupski, W.; Cieslinski, A.; Sokolowska, J.; Trzeciak, W.H.; Jakubowski, H. Determinants of homocysteine-thiolactonase activity of the paraoxonase-1 (PON1) protein in humans. Cell Mol. Biol. 2004, 50, 885–893. [Google Scholar]
- Domagala, T.; Łacinski, M.; Trzeciak, W.H.; Mackness, B.; Mackness, M.I.; Jakubowski, H. The correlation of homocysteine-thiolactonase activity of the paraoxonase (PON1) protein with coronary heart disease status. Cell Mol. Biol. 2006, 52, 4–10. [Google Scholar]
- Perla-Kajan, J.; Jakubowski, H. Paraoxonase 1 protects against protein N-homocysteinylation in humans. FASEB J. 2010, 24, 931–936. [Google Scholar] [CrossRef] [PubMed]
- Zimny, J.; Sikora, M.; Guranowski, A.; Jakubowski, H. Protective mechanisms against homocysteine toxicity: The role of bleomycin hydrolase. J. Biol. Chem. 2006, 281, 22485–22492. [Google Scholar] [CrossRef] [PubMed]
- Borowczyk, K.; Tisonczyk, J.; Jakubowski, H. Metabolism and neurotoxicity of homocysteine thiolactone in mice: Protective role of bleomycin hydrolase. Amino Acids 2012, 43, 1339–1348. [Google Scholar] [CrossRef]
- Perła-Kaján, J.; Borowczyk, K.; Głowacki, R.; Nygård, O.; Jakubowski, H. Paraoxonase 1 Q192R genotype and activity affect homocysteine thiolactone levels in humans. FASEB J. 2018, 32, 6019–6024. [Google Scholar] [CrossRef] [PubMed]
- Costa, L.G.; Cole, T.B.; Jarvik, G.P.; Furlong, C.E. Functional genomic of the paraoxonase (PON1) polymorphisms: Effects on pesticide sensitivity, cardiovascular disease, and drug metabolism. Annu. Rev. Med. 2003, 54, 371–392. [Google Scholar] [CrossRef]
- Bhattacharyya, T.; Nicholls, S.J.; Topol, E.J.; Zhang, R.; Yang, X.; Schmitt, D.; Fu, X.; Shao, M.; Brennan, D.M.; Ellis, S.G.; et al. Relationship of paraoxonase 1 (PON1) gene polymorphisms and functional activity with systemic oxidative stress and cardiovascular risk. JAMA 2008, 299, 1265–1276. [Google Scholar] [CrossRef]
- Tang, W.W.; Hartiala, J.; Fan, Y.; Wu, Y.; Stewart, A.F.; Erdmann, J.; Kathiresan, S.; The CARDIoGRAM Consortium; Roberts, R.; McPherson, R.; et al. Clinical and Genetic Association of Serum Paraoxonase and Arylesterase Activities with Cardiovascular Risk. Arter. Thromb. Vasc. Biol. 2012, 32, 2803–2812. [Google Scholar] [CrossRef]
- Gan, K.N.; Smolen, A.; Eckerson, H.W.; La Du, B.N. Purification of human serum paraoxonase/arylesterase. Evidence for one esterase catalyzing both activities. Drug Metab. Dispos. 1991, 19, 100–106. [Google Scholar]
- Ben-David, M.; Elias, M.; Filippi, J.-J.; Duñach, E.; Silman, I.; Sussman, J.L.; Tawfik, D.S. Catalytic Versatility and Backups in Enzyme Active Sites: The Case of Serum Paraoxonase 1. J. Mol. Biol. 2012, 418, 181–196. [Google Scholar] [CrossRef]
- Khersonsky, O.; Tawfik, D.S. Structure−Reactivity Studies of Serum Paraoxonase PON1 Suggest that Its Native Activity Is Lactonase. Biochemistry 2005, 44, 6371–6382. [Google Scholar] [CrossRef]
- Draganov, D.I.; Teiber, J.F.; Speelman, A.; Osawa, Y.; Sunahara, R.; La Du, B.N. Human paraoxonases (PON1, PON2, and PON3) are lactonases with overlapping and distinct substrate specificities. J. Lipid Res. 2005, 46, 1239–1247. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, C.J.; Lamichhane, S.; Connolly, J.A.; Soehnlen, S.M.; Khalaf, F.K.; Malhotra, D.; Haller, S.T.; Isailovic, D.; Kennedy, D.J. A PON for All Seasons: Comparing Paraoxonase Enzyme Substrates, Activity and Action including the Role of PON3 in Health and Disease. Antioxidants 2022, 11, 590. [Google Scholar] [CrossRef]
- Eryanni-Levin, S.; Khatib, S.; Levy-Rosenzvig, R.; Tamir, S.; Szuchman-Sapir, A. 5,6-δ-DHTL, a stable metabolite of arachidonic acid, is a potential substrate for paraoxonase 1. Biochim. Et Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2015, 1851, 1118–1122. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.A.S.; Khatib, S.; Szuchman-Sapir, A. Fishing for lipid lactones using selective reaction and characteristic fragmentation pattern. J. Chromatogr. B 2022, 1197, 123201. [Google Scholar] [CrossRef]
- Costa, L.G.; Giordano, G.; Cole, T.B.; Marsillach, J.; Furlong, C.E. Paraoxonase 1 (PON1) as a genetic determinant of susceptibility to organophosphate toxicity. Toxicology 2013, 307, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Shih, D.M.; Gu, L.; Xia, Y.-R.; Navab, M.; Li, W.-F.; Hama, S.; Castellani, L.W.; Furlong, C.E.; Costa, L.G.; Fogelman, A.M.; et al. Mice lacking serum paraoxonase are susceptible to organophosphate toxicity and atherosclerosis. Nature 1998, 394, 284–287. [Google Scholar] [CrossRef]
- Shih, D.M.; Xia, Y.-R.; Wang, X.-P.; Miller, E.; Castellani, L.W.; Subbanagounder, G.; Cheroutre, H.; Faull, K.F.; Berliner, J.A.; Witztum, J.L.; et al. Combined Serum Paraoxonase Knockout/Apolipoprotein E Knockout Mice Exhibit Increased Lipoprotein Oxidation and Atherosclerosis. J. Biol. Chem. 2000, 275, 17527–17535. [Google Scholar] [CrossRef]
- Hong, C.G.; Florida, E.; Li, H.; Parel, P.M.; Mehta, N.N.; Sorokin, A.V. Oxidized low-density lipoprotein associates with cardiovascular disease by a vicious cycle of atherosclerosis and inflammation: A systematic review and meta-analysis. Front. Cardiovasc. Med. 2022, 9, 1023651. [Google Scholar] [CrossRef] [PubMed]
- Mackness, B.; Quarck, R.; Verreth, W.; Mackness, M.; Holvoet, P. Human Paraoxonase-1 Overexpression Inhibits Atherosclerosis in a Mouse Model of Metabolic Syndrome. Arter. Thromb. Vasc. Biol. 2006, 26, 1545–1550. [Google Scholar] [CrossRef]
- She, Z.-G.; Zheng, W.; Wei, Y.-S.; Chen, H.-Z.; Wang, A.-B.; Li, H.-L.; Liu, G.; Zhang, R.; Liu, J.-J.; Stallcup, W.B.; et al. Human Paraoxonase Gene Cluster Transgenic Overexpression Represses Atherogenesis and Promotes Atherosclerotic Plaque Stability in ApoE-Null Mice. Circ. Res. 2009, 104, 1160–1168. [Google Scholar] [CrossRef]
- Kresanov, P.; Vasankari, T.; Ahotupa, M.; Kaikkonen, J.; Hutri-Kähönen, N.; Juonala, M.; Kähönen, M.; Lehtimäki, T.; Viikari, J.; Raitakari, O.T. Paraoxonase-1 and oxidized lipoprotein lipids. The Cardiovascular Risk in Young Finns Study. Atherosclerosis 2015, 241, 502–506. [Google Scholar] [CrossRef] [PubMed]
- Ahotupa, M.; Marniemi, J.; Lehtimäki, T.; Talvinen, K.; Raitakari, O.T.; Vasankari, T.; Viikari, J.; Luoma, J.; Ylä-Herttuala, S. Baseline Diene Conjugation in LDL Lipids as a Direct Measure of In Vivo LDL Oxidation. Clin. Biochem. 1998, 31, 257–261. [Google Scholar] [CrossRef] [PubMed]
- Esterbauer, H.; Striegl, G.; Puhl, H.; Rotheneder, M. Continuous monitoring of in vitro oxidation of human low density lipoprotein. Free. Radic. Res. Commun. 1989, 6, 67–75. [Google Scholar] [CrossRef]
- Breton, C.V.; Yin, F.; Wang, X.; Avol, E.; Gilliland, F.D.; Araujo, J.A. HDL anti-oxidant function associates with LDL level in young adults. Atherosclerosis 2014, 232, 165–170. [Google Scholar] [CrossRef]
- Kuchta, A.; Strzelecki, A.; Ćwiklińska, A.; Totoń, M.; Gruchała, M.; Zdrojewski, Z.; Kortas-Stempak, B.; Gliwińska, A.; Dąbkowski, K.; Jankowski, M. PON-1 Activity and Plasma 8-Isoprostane Concentration in Patients with Angiographically Proven Coronary Artery Disease. Oxidative Med. Cell Longev. 2015, 2016, 5136937. [Google Scholar] [CrossRef]
- Kunutsor, S.K.; Bakker, S.J.; James, R.W.; Dullaart, R.P. Serum paraoxonase-1 activity and risk of incident cardiovascular disease: The PREVEND study and meta-analysis of prospective population studies. Atherosclerosis 2016, 245, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Zuin, M.; Trentini, A.; Marsillach, J.; D’amuri, A.; Bosi, C.; Roncon, L.; Passaro, A.; Zuliani, G.; Mackness, M.; Cervellati, C. Paraoxonase-1 (PON-1) Arylesterase Activity Levels in Patients with Coronary Artery Disease: A Meta-Analysis. Dis. Markers 2022, 2022, 4264314. [Google Scholar] [CrossRef]
- Giusti, B.; Saracini, C.; Bolli, P.; Magi, A.; Sestini, I.; Sticchi, E.; Pratesi, G.; Pulli, R.; Pratesi, C.; Abbate, R. Genetic analysis of 56 polymorphisms in 17 genes involved in methionine metabolism in patients with abdominal aortic aneurysm. J. Med. Genet. 2008, 45, 721–730. [Google Scholar] [CrossRef]
- McCormick, M.L.; Gavrila, D.; Weintraub, N.L. Role of Oxidative Stress in the Pathogenesis of Abdominal Aortic Aneurysms. Arter. Thromb. Vasc. Biol. 2007, 27, 461–469. [Google Scholar] [CrossRef]
- Shih, D.M.; Lusis, A.J. The roles of PON1 and PON2 in cardiovascular disease and innate immunity. Curr. Opin. Infect. Dis. 2009, 20, 288–292. [Google Scholar] [CrossRef]
- Grzegorzewska, A.E.; Adamska, P.; Iwanczyk-Skalska, E.; Ostromecka, K.; Niepolski, L.; Marcinkowski, W.; Mostowska, A.; Warchol, W.; Zaba, C.; Jagodzinski, P.P. Paraoxonase 1 concerning dyslipidaemia, cardiovascular diseases, and mortality in haemodialysis patients. Sci. Rep. 2021, 11, 6773. [Google Scholar] [CrossRef]
- Grzegorzewska, A.E.; Ostromecka, K.; Świderska, M.K.; Adamska, P.; Mostowska, A.; Jagodziński, P.P. Paraoxonase 1 gene variants concerning cardiovascular mortality in conventional cigarette smokers and non-smokers treated with hemodialysis. Sci. Rep. 2021, 11, 19467. [Google Scholar] [CrossRef] [PubMed]
- Yuhanna, I.S.; Zhu, Y.; Cox, B.E.; Hahner, L.D.; Osborne-Lawrence, S.; Lu, P.; Marcel, Y.L.; Anderson, R.G.; Mendelsohn, M.E.; Hobbs, H.H.; et al. High-density lipoprotein binding to scavenger receptor-BI activates endothelial nitric oxide synthase. Nat. Med. 2001, 7, 853–857. [Google Scholar] [CrossRef] [PubMed]
- Trigatti, B.L.; Fuller, M. HDL signaling and protection against coronary artery atherosclerosis in mice. J. Biomed. Res. 2016, 30, 94–100. [Google Scholar] [PubMed]
- Seetharam, D.; Mineo, C.; Gormley, A.K.; Gibson, L.L.; Vongpatanasin, W.; Chambliss, K.L.; Hahner, L.D.; Cummings, M.L.; Kitchens, R.L.; Marcel, Y.L.; et al. High-Density Lipoprotein Promotes Endothelial Cell Migration and Reendothelialization via Scavenger Receptor-B Type I. Circ. Res. 2006, 98, 63–72. [Google Scholar] [CrossRef]
- Besler, C.; Heinrich, K.; Rohrer, L.; Doerries, C.; Riwanto, M.; Shih, D.M.; Chroni, A.; Yonekawa, K.; Stein, S.; Schaefer, N.; et al. Mechanisms underlying adverse effects of HDL on eNOS-activating pathways in patients with coronary artery disease. J. Clin. Investig. 2011, 121, 2693–2708. [Google Scholar] [CrossRef]
- Mackness, M.I.; Durrington, P.N.; Mackness, B. The role of paraoxonase 1 activity in cardiovascular disease: Potential for therapeutic intervention. Am. J. Cardiovasc. Drugs 2004, 4, 211–217. [Google Scholar] [CrossRef]
- Mackness, M.I.; Arrol, S.; Durrington, P.N. Paraoxonase prevents accumulation of lipoperoxides in low-density lipoprotein. FEBS Lett. 1991, 286, 152–154. [Google Scholar] [CrossRef]
- Watson, A.D.; Berliner, J.A.; Hama, S.Y.; La Du, B.N.; Faull, K.F.; Fogelman, A.M.; Navab, M. Protective effect of high density lipoprotein associated paraoxonase. Inhibition of the biological activity of minimally oxidized low density lipoprotein. J. Clin. Investig. 1995, 96, 2882–2891. [Google Scholar] [CrossRef]
- Aviram, M.; Rosenblat, M.; Bisgaier, C.L.; Newton, R.S.; Primo-Parmo, S.L.; La Du, B.N. Paraoxonase inhibits high-density lipoprotein oxidation and preserves its functions. A possible peroxidative role for paraoxonase. J. Clin. Investig. 1998, 101, 1581–1590. [Google Scholar] [CrossRef]
- Aviram, M.; Billecke, S.; Sorenson, R.; Bisgaier, C.; Newton, R.; Rosenblat, M.; Erogul, J.; Hsu, C.; Dunlop, C.; La Du, B. Paraoxonase active site required for protection against LDL oxidation involves its free sulfhydryl group and is different from that required for its arylesterase/paraoxonase activities: Selective action of human paraoxonase allozymes Q and R. Arter. Thromb. Vasc. Biol. 1998, 18, 1617–1624. [Google Scholar] [CrossRef]
- Bayrak, A.; Bayrak, T.; Bodur, E.; Kılınç, K.; Demirpençe, E. The effect of HDL-bound and free PON1 on copper-induced LDL oxidation. Chem. Interact. 2016, 257, 141–146. [Google Scholar] [CrossRef]
- Brophy, V.H.; Jampsa, R.L.; Clendenning, J.B.; McKinstry, L.A.; Jarvik, G.P.; Furlong, C.E. Effects of 5′ Regulatory-Region Polymorphisms on Paraoxonase-Gene (PON1) Expression. Am. J. Hum. Genet. 2001, 68, 1428–1436. [Google Scholar] [CrossRef] [PubMed]
- Sikora, M.; Bretes, E.; Perła-Kaján, J.; Lewandowska, I.; Marczak, L.; Jakubowski, H. Genetic Attenuation of Paraoxonase 1 Activity Induces Proatherogenic Changes in Plasma Proteomes of Mice and Humans. Antioxidants 2020, 9, 1198. [Google Scholar] [CrossRef]
- Marathe, G.K.; Zimmerman, G.A.; McIntyre, T.M. Platelet-activating Factor Acetylhydrolase, and Not Paraoxonase-1, Is the Oxidized Phospholipid Hydrolase of High Density Lipoprotein Particles. J. Biol. Chem. 2003, 278, 3937–3947. [Google Scholar] [CrossRef]
- Teiber, J.F.; Draganov, D.I.; La Du, B.N. Purified human serum PON1 does not protect LDL against oxidation in the in vitro assays initiated with copper or AAPH. J. Lipid Res. 2004, 45, 2260–2268. [Google Scholar] [CrossRef]
- Connelly, P.W.; Draganov, A.; Maguire, G.F. Paraoxonase-1 does not reduce or modify oxidation of phospholipids by peroxynitrite. Free. Radic. Biol. Med. 2005, 38, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Carlson, C.S.; Heagerty, P.J.; Hatsukami, T.S.; Richter, R.J.; Ranchalis, J.; Lewis, J.; Bacus, T.J.; McKinstry, L.A.; Schellenberg, G.D.; Rieder, M.; et al. TagSNP analyses of the PON gene cluster: Effects on PON1 activity, LDL oxidative susceptibility, and vascular disease. J. Lipid Res. 2006, 47, 1014–1024. [Google Scholar] [CrossRef]
- Tanzi, R.E. The genetics of Alzheimer disease. Cold Spring Harb. Perspect. Med. 2012, 2, a006296. [Google Scholar] [CrossRef]
- Sikora, M.; Jakubowski, H. Changes in redox plasma proteome of Pon1−/− mice are exacerbated by a hyperhomocysteinemic diet. Free Radic. Biol. Med. 2021, 169, 169–180. [Google Scholar] [CrossRef] [PubMed]
- de la Torre, J.C. Alzheimer disease as a vascular disorder: Nosological evidence. Stroke 2002, 33, 1152–1162. [Google Scholar] [CrossRef]
- Stampfer, M.J. Cardiovascular disease and Alzheimer’s disease: Common links. J. Intern. Med. 2006, 260, 211–223. [Google Scholar] [CrossRef]
- Menini, T.; Gugliucci, A. Paraoxonase 1 in neurological disorders. Redox Rep. 2013, 19, 49–58. [Google Scholar] [CrossRef]
- Erlich, P.M.; Lunetta, K.L.; Cupples, L.A.; Abraham, C.R.; Green, R.C.; Baldwin, C.T.; Farrer, L.A. Serum paraoxonase activity is associated with variants in the PON gene cluster and risk of Alzheimer disease. Neurobiol. Aging 2012, 33, 1015.e7–1015.e23. [Google Scholar] [CrossRef] [PubMed]
- Bednarska-Makaruk, M.E.; Krzywkowski, T.; Graban, A.; Lipczyńska-Łojkowska, W.; Bochyńska, A.; Rodo, M.; Wehr, H.; Ryglewicz, D.K. Original article Paraoxonase 1 (PON1) gene -108C>T and p.Q192R polymorphisms and arylesterase activity of the enzyme in patients with dementia. Folia Neuropathol. 2013, 2, 111–119. [Google Scholar] [CrossRef]
- Dantoine, T.F.; Debord, J.; Merle, L.; Lacroix-Ramiandrisoa, H.; Bourzeix, L.; Charmes, J.P. Paraoxonase 1 activity: A new vascular marker of dementia? Ann. N. Y. Acad. Sci. 2002, 977, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Paragh, G.; Balla, P.; Katona, E.; Seres, I.; Egerhazi, A.; Degrell, I. Serum paraoxonase activity changes in patients with Alzheimer’s disease and vascular dementia. Eur. Arch. Psychiatry Clin. Neurosci. 2002, 252, 63–67. [Google Scholar] [CrossRef]
- Bednarz-Misa, I.; Berdowska, I.; Zboch, M.; Misiak, B.; Zieliński, B.; Płaczkowska, S.; Fleszar, M.; Wiśniewski, J.; Gamian, A.; Krzystek-Korpacka, M. Paraoxonase 1 decline and lipid peroxidation rise reflect a degree of brain atrophy and vascular impairment in dementia. Adv. Clin. Exp. Med. 2020, 29, 71–78. [Google Scholar] [CrossRef]
- Perla-Kajan, J.; Wloczkowska, O.; Ziola-Frankowska, A.; Frankowski, M.; Smith, A.D.; de Jager, C.A.; Refsum, H.; Jakubowski, H. Paraoxonase 1, B Vitamins Supplementation, and Mild Cognitive Impairment. J. Alzheimer’s Dis. 2021, 81, 1211–1229. [Google Scholar] [CrossRef] [PubMed]
- Narasimhulu, C.A.; Mitra, C.; Bhardwaj, D.; Burge, K.Y.; Parthasarathy, S. Alzheimer’s Disease Markers in Aged ApoE-PON1 Deficient Mice. J. Alzheimer’s Dis. 2019, 67, 1353–1365. [Google Scholar] [CrossRef]
- Salazar, J.G.; Marsillach, J.; Reverte, I.; Mackness, B.; Mackness, M.; Joven, J.; Camps, J.; Colomina, M.T. Paraoxonase-1 and -3 Protein Expression in the Brain of the Tg2576 Mouse Model of Alzheimer’s Disease. Antioxidants 2021, 10, 339. [Google Scholar] [CrossRef]
- Li, H.; Wetten, S.; Li, L.; Jean, P.L.S.; Upmanyu, R.; Surh, L.; Hosford, D.; Barnes, M.R.; Briley, J.D.; Borrie, M.; et al. Candidate Single-Nucleotide Polymorphisms From a Genomewide Association Study of Alzheimer Disease. Arch. Neurol. 2008, 65, 45–53. [Google Scholar] [CrossRef]
- Leduc, V.; Théroux, L.; Dea, D.; Robitaille, Y.; Poirier, J. Involvement of paraoxonase 1 genetic variants in Alzheimer’s disease neuropathology. Eur. J. Neurosci. 2009, 30, 1823–1830. [Google Scholar] [CrossRef]
- Erlich, P.M.; Lunetta, K.L.; Cupples, L.A.; Huyck, M.; Green, R.C.; Baldwin, C.T.; Farrer, L.A.; Auerbach, S.; Akomolafe, A.; Griffith, P.; et al. Polymorphisms in the PON gene cluster are associated with Alzheimer disease. Hum. Mol. Genet. 2006, 15, 77–85. [Google Scholar] [CrossRef]
- Chapuis, J.; Boscher, M.; Bensemain, F.; Cottel, D.; Amouyel, P.; Lambert, J.C. Association study of the paraoxonase 1 gene with the risk of developing Alzheimer’s disease. Neurobiol. Aging 2009, 30, 152–156. [Google Scholar] [CrossRef] [PubMed]
- Nie, Y.; Luo, D.; Yang, M.; Wang, Y.; Xiong, L.; Gao, L.; Liu, Y.; Liu, H. A Meta-Analysis on the Relationship of the PON Genes and Alzheimer Disease. J. Geriatr. Psychiatry Neurol. 2017, 30, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Bacchetti, T.; Vignini, A.; Giulietti, A.; Nanetti, L.; Provinciali, L.; Luzzi, S.; Mazzanti, L.; Ferretti, G. Higher Levels of Oxidized Low Density Lipoproteins in Alzheimer’s Disease Patients: Roles for Platelet Activating Factor Acetyl Hydrolase and Paraoxonase-1. J. Alzheimer’s Dis. 2015, 46, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Cagnin, A.; Leon, A.; Vianello, D.; Colavito, D.; Favaretto, S.; Zarantonello, G.; Stecca, A.; Ermani, M.; Zambon, A. LDL density and oxidation are modulated by PON1 promoter genotype in patients with Alzheimer’s disease. J. Alzheimer’s Dis. 2013, 34, 377–385. [Google Scholar] [CrossRef]
- Gauthier, S.; Reisberg, B.; Zaudig, M.; Petersen, R.C.; Ritchie, K.; Broich, K.; Belleville, S.; Brodaty, H.; Bennett, D.; Chertkow, H.; et al. International Psychogeriatric Association Expert Conference on mild cognitive, i., Mild cognitive impairment. Lancet 2006, 367, 1262–1270. [Google Scholar] [CrossRef]
- Graham, J.E.; Rockwood, K.; Beattie, B.L.; Eastwood, R.; Gauthier, S.; Tuokko, H.; McDowell, I. Prevalence and severity of cognitive impairment with and without dementia in an elderly population. Lancet 1997, 349, 1793–1796. [Google Scholar] [CrossRef]
- Petersen, R.C.; Roberts, R.O.; Knopman, D.S.; Boeve, B.F.; Geda, Y.E.; Ivnik, R.J.; Smith, G.E.; Jack, C.R., Jr. Mild cognitive impairment: Ten years later. Arch. Neurol. 2009, 66, 1447–1455. [Google Scholar] [CrossRef]
- Wehr, H.; Bednarska-Makaruk, M.; Graban, A.; Lipczynska-Lojkowska, W.; Rodo, M.; Bochynska, A.; Ryglewicz, D. Paraoxonase activity and dementia. J. Neurol. Sci. 2009, 283, 107–108. [Google Scholar] [CrossRef]
- Cervellati, C.; Trentini, A.; Romani, A.; Bellini, T.; Bosi, C.; Ortolani, B.; Zurlo, A.; Passaro, A.; Seripa, D.; Zuliani, G. Serum paraoxonase and arylesterase activities of paraoxonase-1 (PON-1), mild cognitive impairment, and 2-year conversion to dementia: A pilot study. J. Neurochem. 2015, 135, 395–401. [Google Scholar] [CrossRef]
- Jakubowski, H.; Zioła-Frankowska, A.; Frankowski, M.; Perła-Kaján, J.; Refsum, H.; de Jager, C.A.; Smith, A.D. B Vitamins Prevent Iron-Associated Brain Atrophy and Domain-Specific Effects of Iron, Copper, Aluminum, and Silicon on Cognition in Mild Cognitive Impairment. J. Alzheimer’s Dis. 2021, 84, 1039–1055. [Google Scholar] [CrossRef]
- Wloczkowska, O.; Perla-Kajan, J.; Smith, A.D.; de Jager, C.; Refsum, H.; Jakubowski, H. Anti-N-homocysteine-protein autoantibodies are associated with impaired cognition. Alzheimer’s Dement. Transl. Res. Clin. Interv. 2021, 7, e12159. [Google Scholar] [CrossRef]
- Hayden, K.M.; Norton, M.C.; Darcey, D.; Ostbye, T.; Zandi, P.P.; Breitner, J.C.S.; Welsh-Bohmer, K.A. Occupational exposure to pesticides increases the risk of incident AD: The Cache County Study. Neurology 2010, 74, 1524–1530. [Google Scholar] [CrossRef] [PubMed]
- Jones, N. Alzheimer disease: Risk of dementia and Alzheimer disease increases with occupational pesticide exposure. Nat. Rev. Neurol. 2010, 6, 353. [Google Scholar] [CrossRef] [PubMed]
- Suszyńska-Zajczyk, J.; Łuczak, M.; Marczak, L.; Jakubowski, H. Inactivation of the Paraoxonase 1 Gene Affects the Expression of Mouse Brain Proteins Involved in Neurodegeneration. J. Alzheimer’s Dis. 2014, 42, 247–260. [Google Scholar] [CrossRef]
- Blatter, M.-C.; James, R.W.; Messmer, S.; Barja, F.; Pometta, D. Identification of a distinct human high-density lipoprotein subspecies defined by a lipoprotein-associated protein, K-45. Identity of K-45 with paraoxonase. JBIC J. Biol. Inorg. Chem. 1993, 211, 871–879. [Google Scholar] [CrossRef]
- Oakley, H.; Cole, S.L.; Logan, S.; Maus, E.; Shao, P.; Craft, J.; Guillozet-Bongaarts, A.; Ohno, M.; Disterhoft, J.; Van Eldik, L.; et al. Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: Potential factors in amyloid plaque formation. J. Neurosci. 2006, 26, 10129–10140. [Google Scholar] [CrossRef] [PubMed]
- Khayati, K.; Antikainen, H.; Bonder, E.M.; Weber, G.F.; Kruger, W.D.; Jakubowski, H.; Dobrowolski, R. The amino acid metabolite homocysteine activates mTORC1 to inhibit autophagy and form abnormal proteins in human neurons and mice. FASEB J. 2017, 31, 598–609. [Google Scholar] [CrossRef]
- Yates, S.C.; Zafar, A.; Hubbard, P.; Nagy, S.; Durant, S.; Bicknell, R.; Wilcock, G.; Christie, S.; Esiri, M.M.; Smith, A.D.; et al. Dysfunction of the mTOR pathway is a risk factor for Alzheimer’s disease. Acta Neuropathol. Commun. 2013, 1, 3. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, S.; Zhou, Y.; Han, Y.; Li, S.; Xu, Q.; Xu, L.; Zhu, Z.; Deng, Y.; Yu, L.; et al. Phf8 histone demethylase deficiency causes cognitive impairments through the mTOR pathway. Nat. Commun. 2018, 9, 114. [Google Scholar] [CrossRef] [PubMed]
- Witucki, Ł.; Jakubowski, H. Homocysteine Metabolites Inhibit Autophagy and Elevate Amyloid Beta by Impairing Phf8/H4K20me1-dependent Epigenetic Regulation of mTOR in Cystathionine β-Synthase-Deficient Mice. bioRxiv. [CrossRef]
- Jakubowski, H.; Perła-Kaján, J.; Finnell, R.H.; Cabrera, R.M.; Wang, H.; Gupta, S.; Kruger, W.D.; Kraus, J.P.; Shih, D.M. Genetic or nutritional disorders in homocysteine or folate metabolism increase protein N-homocysteinylation in mice. FASEB J. 2009, 23, 1721–1727. [Google Scholar] [CrossRef]
- Smith, A.D.; Refsum, H. Homocysteine—From disease biomarker to disease prevention. J. Intern. Med. 2021, 290, 826–854. [Google Scholar] [CrossRef] [PubMed]
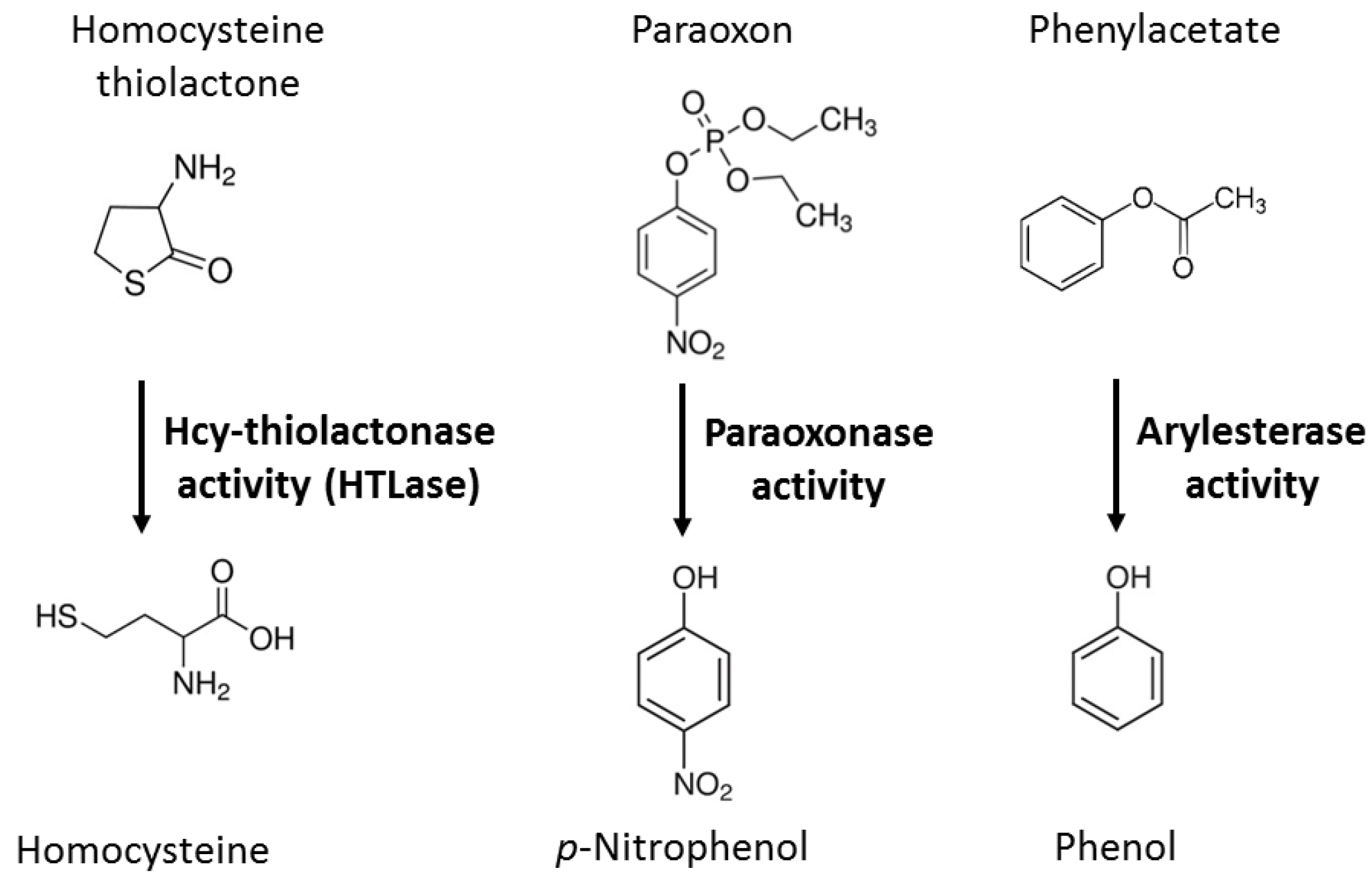
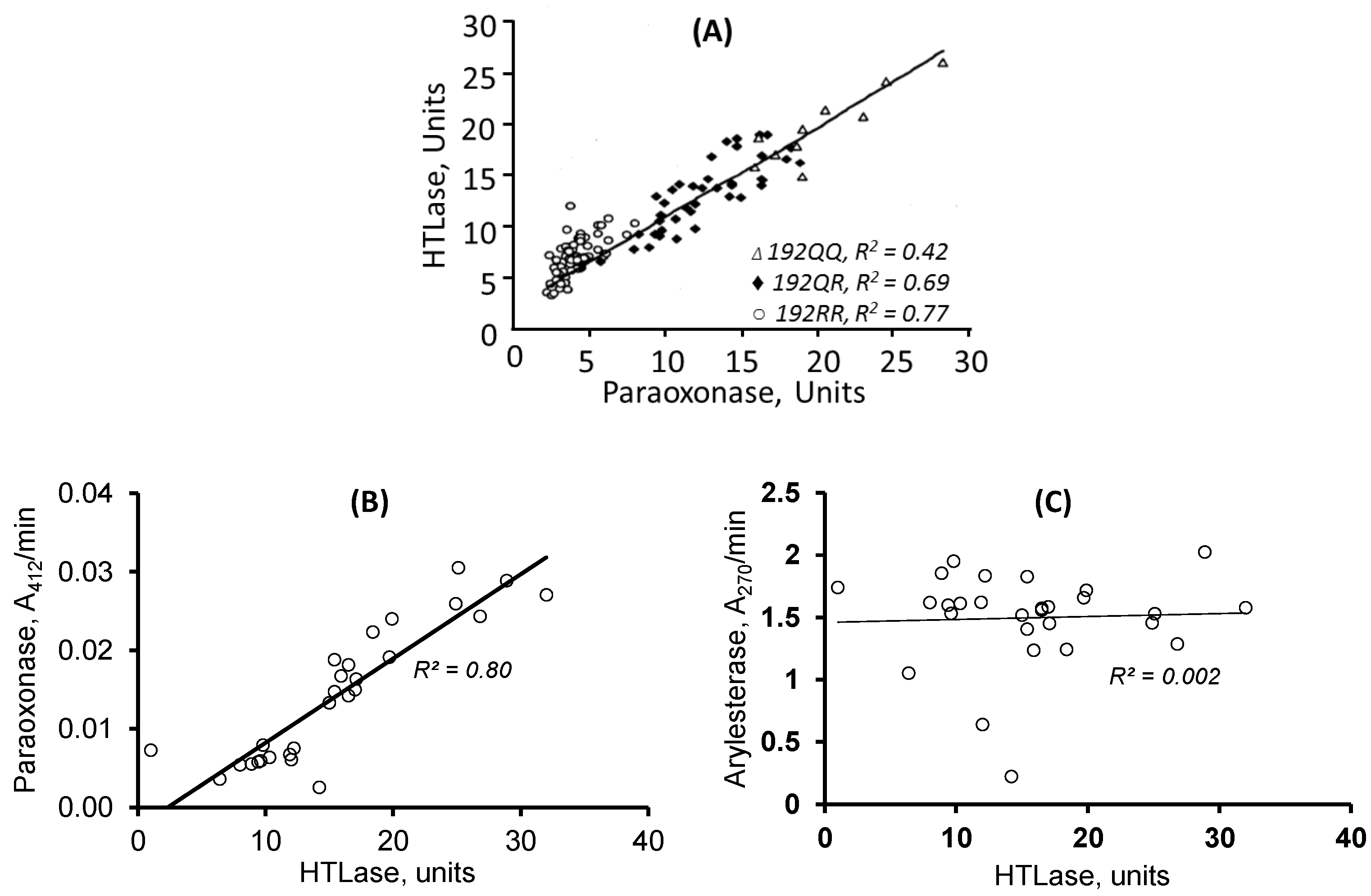
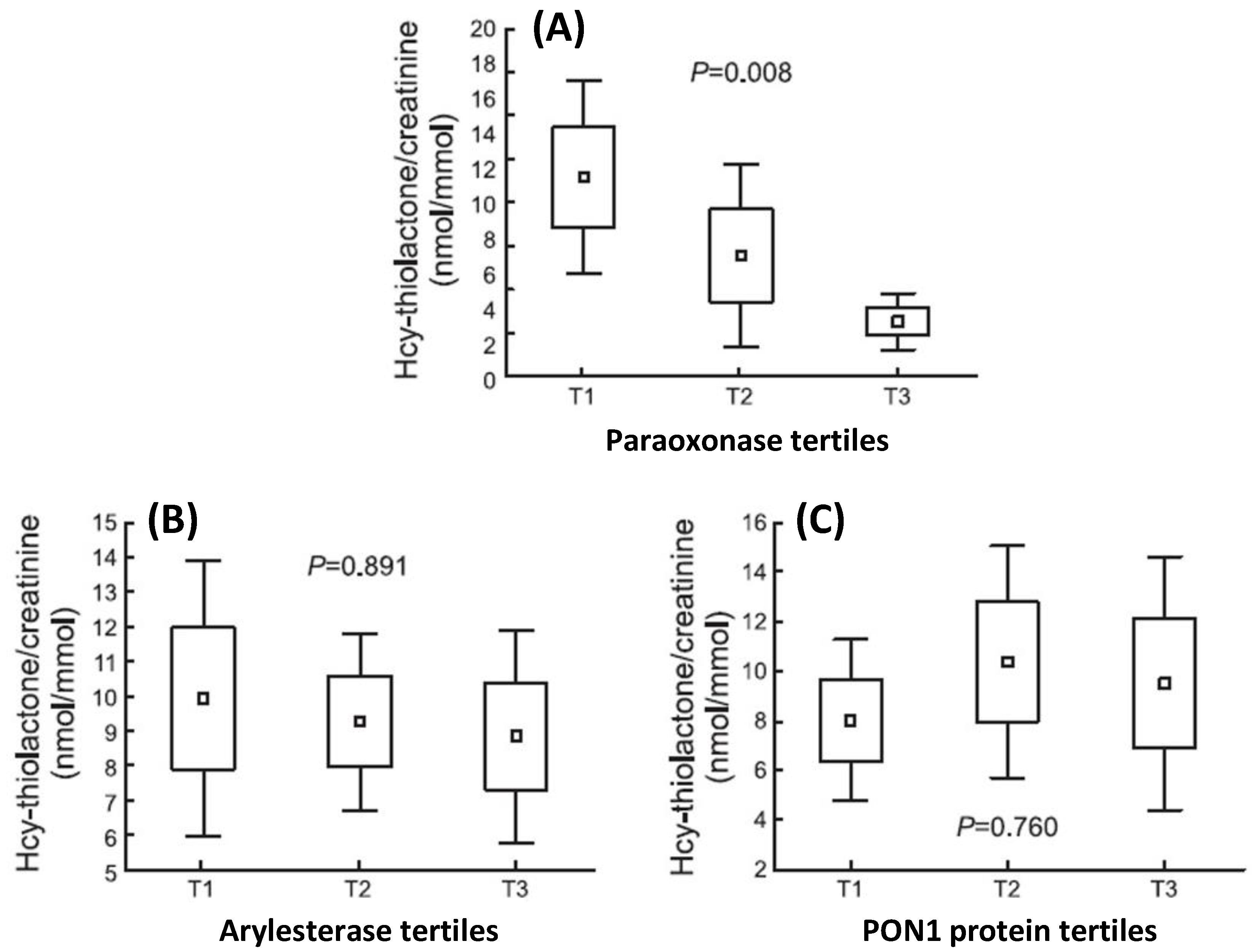
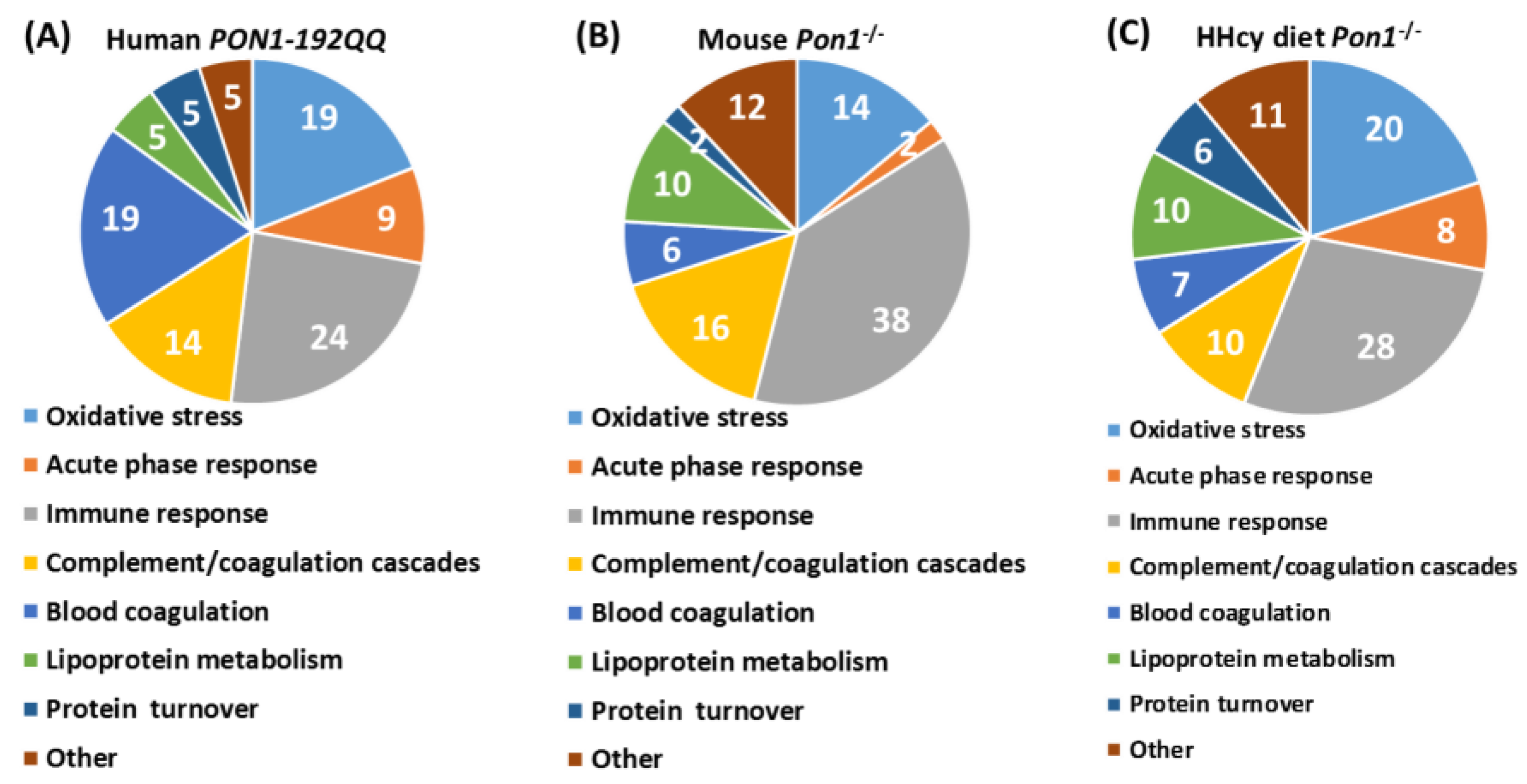
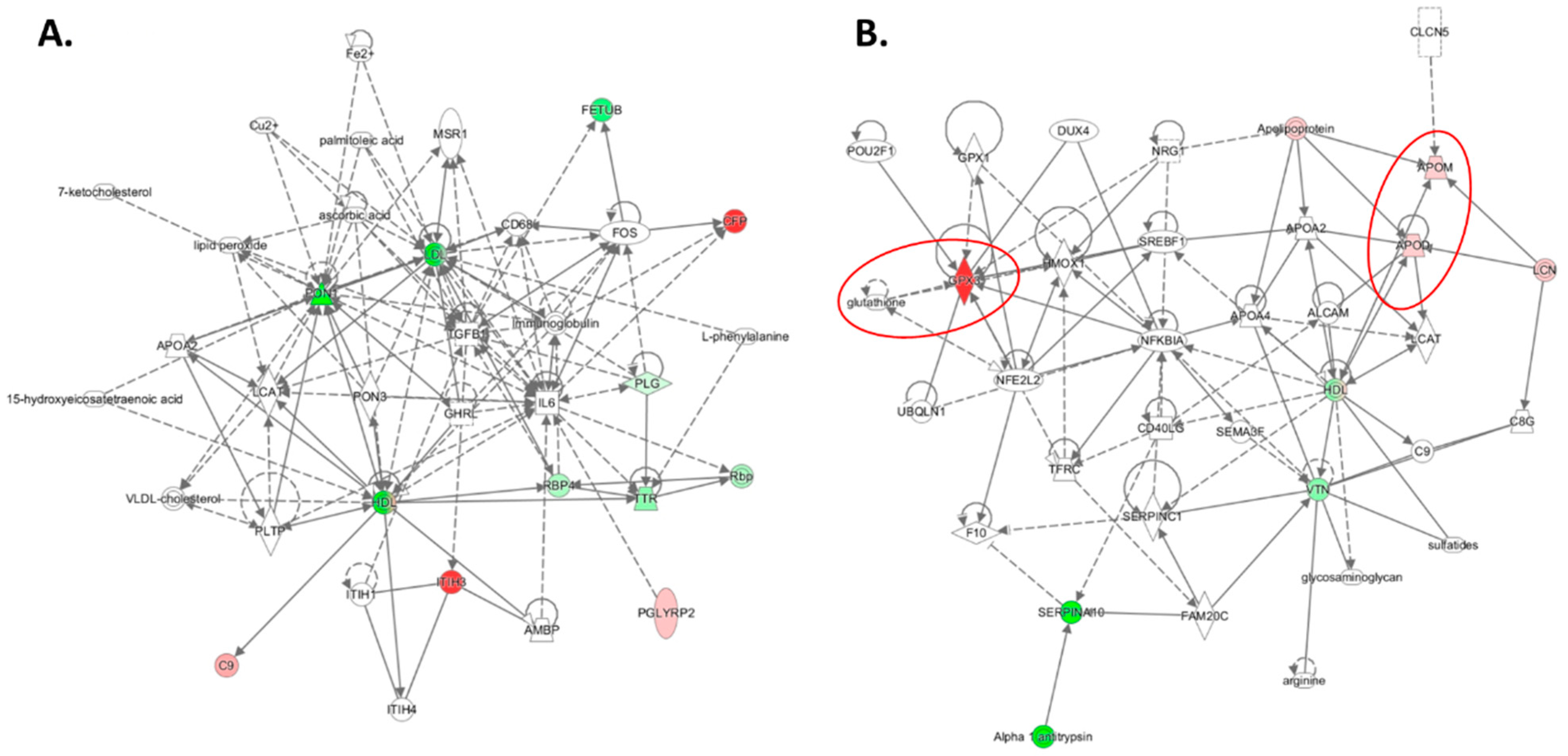

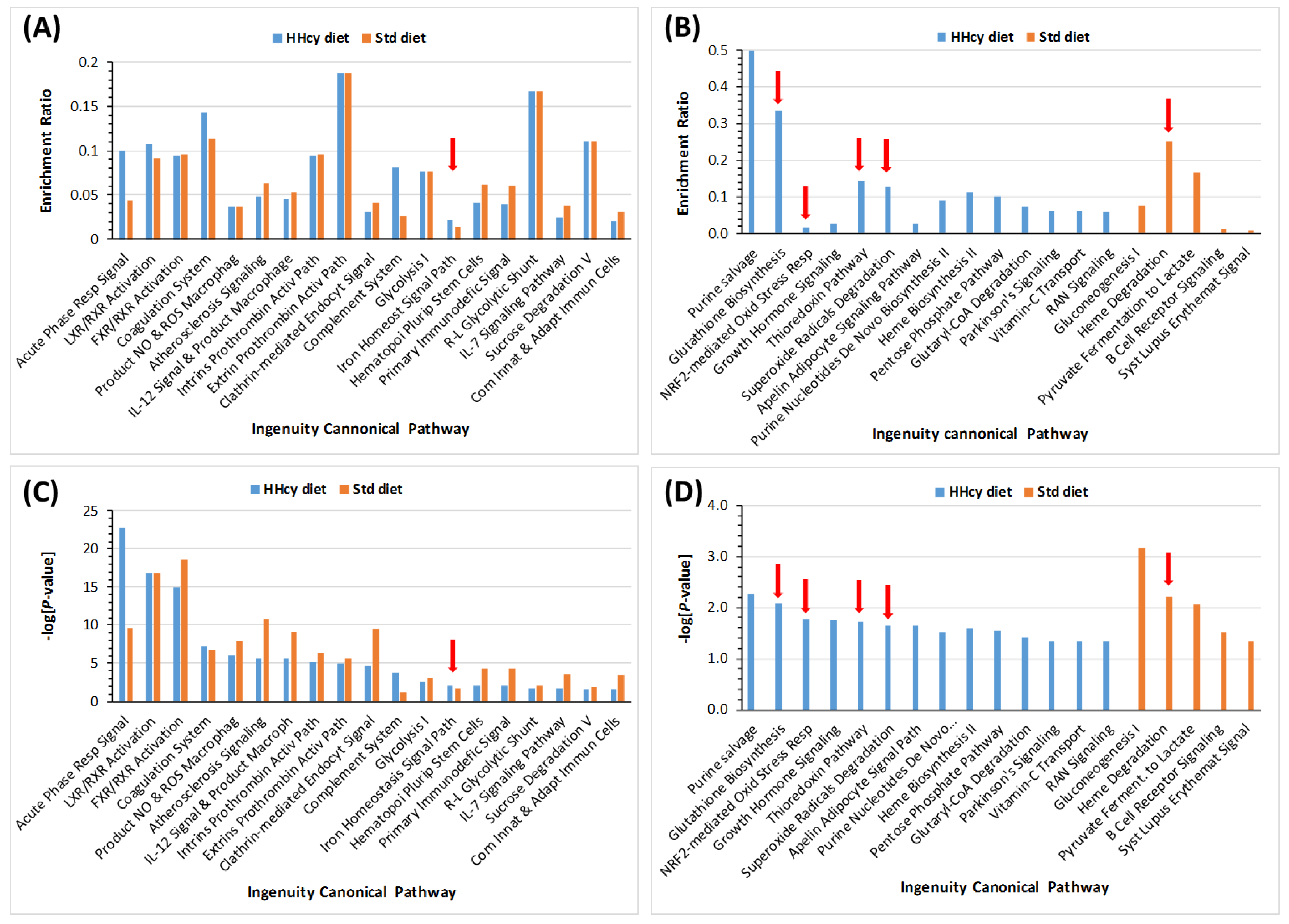
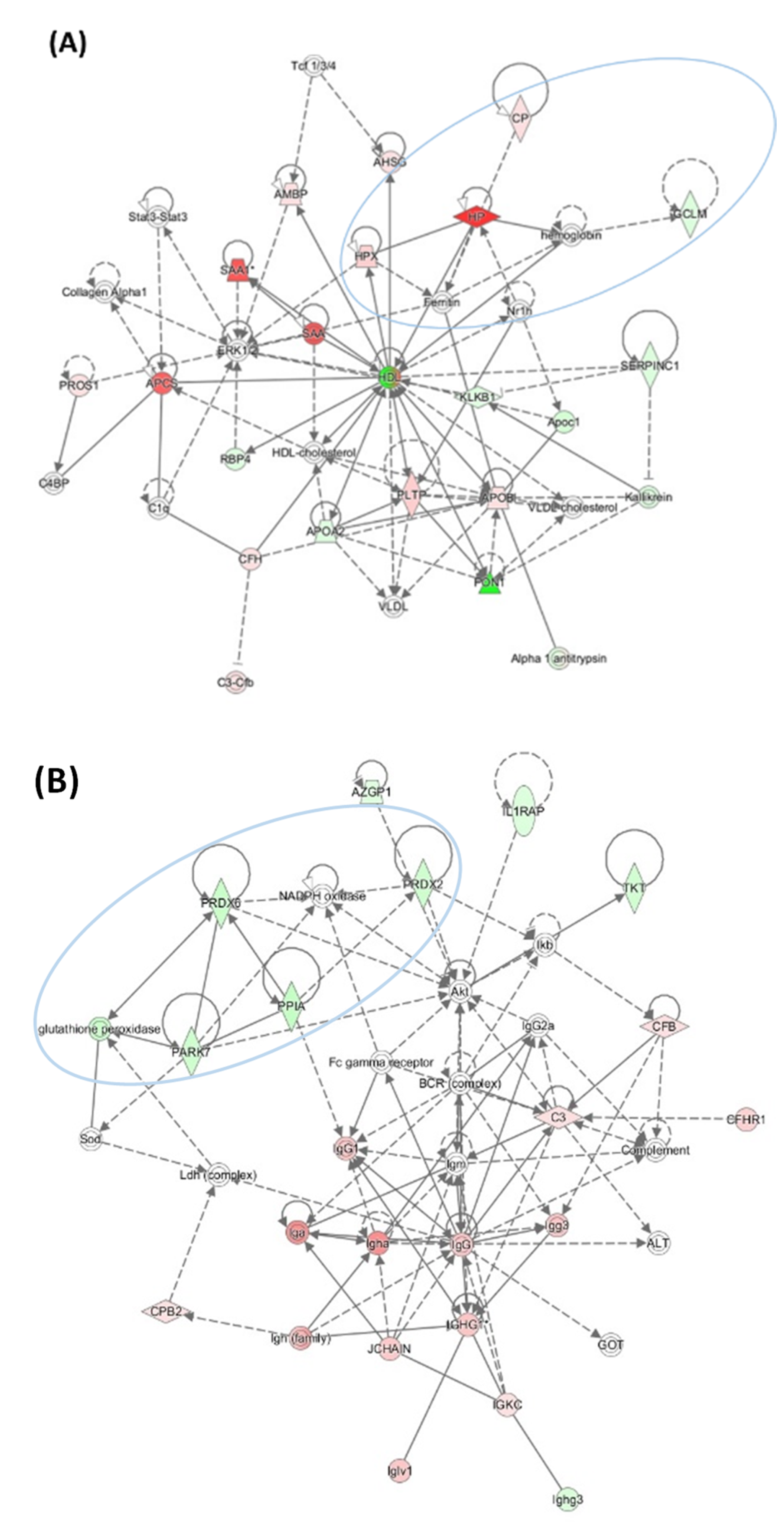

| Unique to Mice (n = 41) | Unique to Humans (n = 12) | Proteins Affected Both in Mice and Humans (n = 9) # |
|---|---|---|
| Oxidative stress † (n = 4): ↓Alb $, ↓Blvrb, ↑α-1-microglubulin (Ambp), ↑Hemopexin (Hpx) | Oxidative stress † (n = 1): ↑Glutathione peroxidase 3 (GPX3) | Oxidative stress † (n = 3): ↑APOD↑, ↑APOM ‡↑, ↑haptoglobin (HP)↓ |
| Immune response (n = 18): ↑Igh (n = 9), ↑Igj, ↑Igk (n = 6), ↑Igl (n = 2) | Immune response (n = 4): ↑CFP, ↓N/A, ↑PGLYRP2, ↑V2-6 (IGL) | Immune response (n = 1): ↓IGHG3↑ |
| Acute phase response (n = 5): ↑Ahsg, ↑Orm1, ↑Orm2, ↑Saa1, ↑Saa2 | Acute phase response (n = 1): ↑Ttr | Acute phase response (n = 1): ↑Ambp |
| Complement/coagulation (n = 7): ↑Al182371, ↑Cfh, ↑Clu $, ↑F2 (prothrombin), ↓Klkb1, ↓Mbl1; ↓Serpinc1 (antithrombin III) | Complement/coagulation (n = 2): ↑C9, ↑V2-17 (IGL) | |
| Blood coagulation (n = 2): ↑Hrg ‡, ↓Itih1 | Blood coagulation (n = 3): ↓PLG, ↓SERPINA10, ↓VTN | Blood coagulation (n = 1): ↓F13B↓ |
| Lipoprotein/lipid metabolism (n = 5): ↓ApoA2, ↓ApoC2, ↓Azgp1, ↓Pgp, ↑Pltp | Lipoprotein metabolism (n = 4): ↓ApoA1, ↑ApoB, ↓ApoC1, ↓Pon1 | |
| Protein turnover (n = 1): ↑Mug1 | Protein turnover (n = 1): ↑FETUB↓ | |
| Other (n = 6): ↓Afm, ↓Aldoa, ↓Bpgm, ↓Ica, ↓Ldha, ↓Lifr | Other (n = 1): ↓RBP4 |
| Unique to HHcy Diet Mice (n = 66) | Unique to Control Diet Mice (n = 27) | Proteins Affected Both in HHcy and Control Diet Mice (n = 23) # |
|---|---|---|
| Oxidative stress (n = 15): ↓Alad, ↑Cp, ↓Gclm, ↓Cat, ↑Ctsb, ↓Gsn, ↑Grn, ↓Prdx2 #, ↓Prdx6, ↓Txn, ↓Igfbp3, ↓Park7 #, ↓Pebp1 #, ↓Ppia, ↓Serpina3k | Oxidative stress (n = 1): ↓Blvrb | Oxidative stress (n = 3): ↓Alb $, ↑Hp, ↑Hpx |
| Immune response (n = 15): ↓Il1rap, Igh (n = 10↑, 1↓), ↑Igk (n = 3), | Immune response (n = 10): ↑Clu $, ↑Igh (n = 3↑, 1↓), ↑Igk (n = 3), ↑Igl, ↑Igm | Immune response (n = 9): ↑Igh (n = 4), ↑Igj, ↑Igk (n = 3), ↑Igl |
| Acute phase response (n = 5): ↑Ahsg, ↑Orm1, ↑Orm2, ↑Saa1, ↑Saa2 | Acute phase response (n = 1): ↑Ttr | Acute phase response (n = 1): ↑Ambp |
| Complement/coagulation (n = 6): ↑A2m $, ↑Apcs, ↓F13a1, ↑C3, ↑Cfb, ↑Cfhr1 | Complement/coagulation (n = 4): ↑AI182371, ↑F2, ↓F13b, ↓Mbl1 | Complement/coagulation (n = 3): ↑Cfh, ↓Klkb1, ↓Serpinc1 |
| Blood coagulation (n = 6): ↑Serpina10, ↓Gp1ba, ↑Gp5, ↑Itih3, ↑Pros1 $, ↓Proz | Blood coagulation (n = 3): ↓Hgfac, ↑Hrg ‡, ↓Itih1 | |
| Lipoprotein/lipid metabolism (n = 5): ↓ApoA2, ↓ApoC2, ↓Azgp1, ↓Pgp, ↑Pltp | Lipoprotein metabolism (n = 4): ↓Afm, ↑ApoD, ↑ApoM, ↑Lcat | Lipoprotein metabolism (n = 4): ↓ApoA1, ↑ApoB, ↓ApoC1, ↓Pon1 |
| Protein turnover (n = 5): ↓Apeh, ↓Mug2, ↓Serpina3m, ↓Uba1, ↓Uba52 | Protein turnover (n = 1): ↑Fetub | Protein turnover (n = 1): ↓Mug1; |
| Other proteins (n = 8): ↓Atic, ↓Nme1, ↓Pnp (purine metabo-lism), ↓Tpi, ↓Tkt (glucose metabolism), ↓Ran # (nucleoplasmic transport), ↓Rbp4 (retinol transport), ↓Spp2 (bone remodeling) | Other proteins (n = 3): ↓Aldoa, ↓Ldha (glucose metabolism), ↓Lifr (tissue regeneration) | Other proteins (n = 2): ↓Bpgm (glucose metabolism), ↓Ica (carbonic anhydrase inhibitor) |
| Variable (n = 82–112) | Global Cognition | Episodic Memory | Attention/Processing Speed | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MMSE_2 1 | TICSm_2 2 | HVLT-TR_2 3 | HVLT-DR_2 4 | Trail_Making_A _2 5 | SDMT_2 6 | SDMT_2 7 | ||||||||
| β | p | β | p | β | p | β | p | β | p | β | p | |||
| Arylesterase activity_1 | −0.24 | 0.034 | −0.24 | 0.027 | −0.19 | 0.046 | −0.32 | 0.012 | 0.24 | 0.015 | −0.18 | 0.008 | ||
| Paraoxonase activity_1 | NS # | NS # | NS # | NS # | −0.33 | 0.028 | ||||||||
| PON1-Q192R | NS | NS | NS | NS | 0.049 | 0.29 | 0.047 | |||||||
| Brain atrophy rate | −0.27 | 0.029 | −0.27 | 0.011 | NS | NS | 0.007 | −0.24 | 0.001 | −0.23 | 0.002 | |||
| MMSE_1 | 0.26 | 0.017 | ||||||||||||
| TICS-m_1 | 0.25 | 0.017 | ||||||||||||
| HVLT-TR_1 | 0.45 | 0.000 | ||||||||||||
| HVLT-DR_1 | 0.46 | 0.000 | ||||||||||||
| Trail Making A_1 | 0.32 | 0.001 | ||||||||||||
| SDMT_1 | 0.78 | 0.000 | 0.66 | 0.000 | ||||||||||
| * Log-transformed data were used in analyses. _1—baseline _2—end of study | p = 0.000, R2 = 0.43 | p = 0.000, R2 = 0.51 | p = 0.000, R2 = 0.54 | p = 0.001, R2 = 0.37 | p = 0.000, R2 = 0.57 | p = 0.000, R2 = 0.78 | p = 0.000, R2 = 0.78 | |||||||
| 1−7 Adjusted for sex, age. Additional adjustment for: 1, 4 Anti-N-Hcy, tHcy_1; 1 BDNF V66M genotype; 3 Creatinine, TCN 776CG genotype; 4 APOE genotype; 5 Fe_1, FA_1, TG_1, COMT V158M and DHFR 19bpins genotypes. # Models with or w/o arylesterase. MMSE—Mini-Mental State Examination; TICS-m—Telephone Inventory for Cognitive Status modified; HVLT-TR_1—Hopkins Verbal Learning Test-revised Total Recall; HVLT-DR—Hopkins Verbal Learning Test-revised, Delayed Recall; SDMT—Symbol Digits Modalities Test. | ||||||||||||||
| Variable (n = 82–112) | Global Cognition | Episodic Memory | Attention/Processing Speed | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MMSE_2 1 | TICSm_2 2 | HVLT-TR_2 3 | HVLT-DR_2 4 | Trail_Making_A _2 5 | SDMT_2 6 | SDMT_2 7 | ||||||||
| β | p | β | p | β | p | β | p | β | p | β | p | |||
| Arylesterase activity_1 | NS | NS | NS | NS | NS | NS | ||||||||
| Paraoxonase activity_1 | NS | |||||||||||||
| PON1-Q192R | NS | NS | ||||||||||||
| Brain atrophy rate | NS | NS | NS | NS | NS | NS | NS | |||||||
| MMSE_1 | 0.52 | 0.000 | ||||||||||||
| TICS-m_1 | 0.38 | 0.002 | ||||||||||||
| HVLT-TR_1 | 0.63 | 0.000 | ||||||||||||
| HVLT-DR_1 | 0.47 | 0.001 | ||||||||||||
| Trail Making_1 | 0.62 | 0.000 | ||||||||||||
| SDMT_1 | 0.65 | 0.000 | 0.66 | 0.000 | ||||||||||
| * Log-transformed data were used in analyses. _1—baseline _2—end of study | p = 0.019, R2 = 0.20 | p = 0.001, R2 = 0.28 | P = 0.000, R2 = 0.38 | p = 0.001, R2 = 0.20 | p = 0.005, R2 = 0.30 | P = 0.000, R2 = 0.61 | p = 0.000, R2 = 0.60 | |||||||
| 1−7 Adjusted for sex, age. Additional adjustment for: 1, 4 Anti-N-Hcy, tHcy_1; 1 BDNF V66M genotype; 3 Creatinine, TCN 776CG genotype; 4 APOE genotype; 5 Fe_1, FA_1, TG_1, COMT V158M and DHFR 19bpins genotypes. Neuropsychological test acronyms defined as in Table 3. | ||||||||||||||
| Protein Name | Change in Pon1−/− vs. Pon1+/+ Brain * | Change in 1% Met Diet vs. Std. Diet Brain * | Change in AD Brain (Other Neuropathy or Animal Model) ** | |
|---|---|---|---|---|
| Std. Diet | 1%-Met Diet | Pon1+/+ | ||
| Brain-specific | ||||
| Ncald | – | ↑ | ↓ | ↓, (↓ in Gls−/− mouse) |
| Nrgn | ↓ | ↑ | ↓ | ↓ |
| Stmn1 | – | ↑ | ↓ | ↓, (↑ in MS, TLE, SMA, schizophrenia), (↑ in HD4 mouse model) |
| Antioxidant defense | ||||
| Sod1 | ↓ | – | – | (↑ in ALS) |
| Prdx2 | – | ↑ | ↓ | ↑ |
| DJ-1 (Park7) | ↓ | ↑ | ↓ | ↑ |
| Energy metabolism | ||||
| Ak1 | – | ↑ | ↓ | ↑ |
| Cell cycle | ||||
| GDI1 | – | ↑ | ↓ | (↑ in rat ischemic brain) |
| Ran | – | ↑ | ↓ | ↑ |
| Cytoskeleton assembly | ||||
| Tbcb | ↓ | ↑ | ↑ | (↑ in GAN) |
| CapZa2 | ↑ | – | ↑ | ↑ CapZb2 # |
| Other proteins | ||||
| Hdhd2 | – | ↑ | – | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jakubowski, H. Proteomic Exploration of Paraoxonase 1 Function in Health and Disease. Int. J. Mol. Sci. 2023, 24, 7764. https://doi.org/10.3390/ijms24097764
Jakubowski H. Proteomic Exploration of Paraoxonase 1 Function in Health and Disease. International Journal of Molecular Sciences. 2023; 24(9):7764. https://doi.org/10.3390/ijms24097764
Chicago/Turabian StyleJakubowski, Hieronim. 2023. "Proteomic Exploration of Paraoxonase 1 Function in Health and Disease" International Journal of Molecular Sciences 24, no. 9: 7764. https://doi.org/10.3390/ijms24097764





