Phafins Are More Than Phosphoinositide-Binding Proteins
Abstract
:1. Introduction
2. The Phafin Protein Family
2.1. Overview of Phafin Protein Family
2.2. The Availability of Phafins among Animal Species
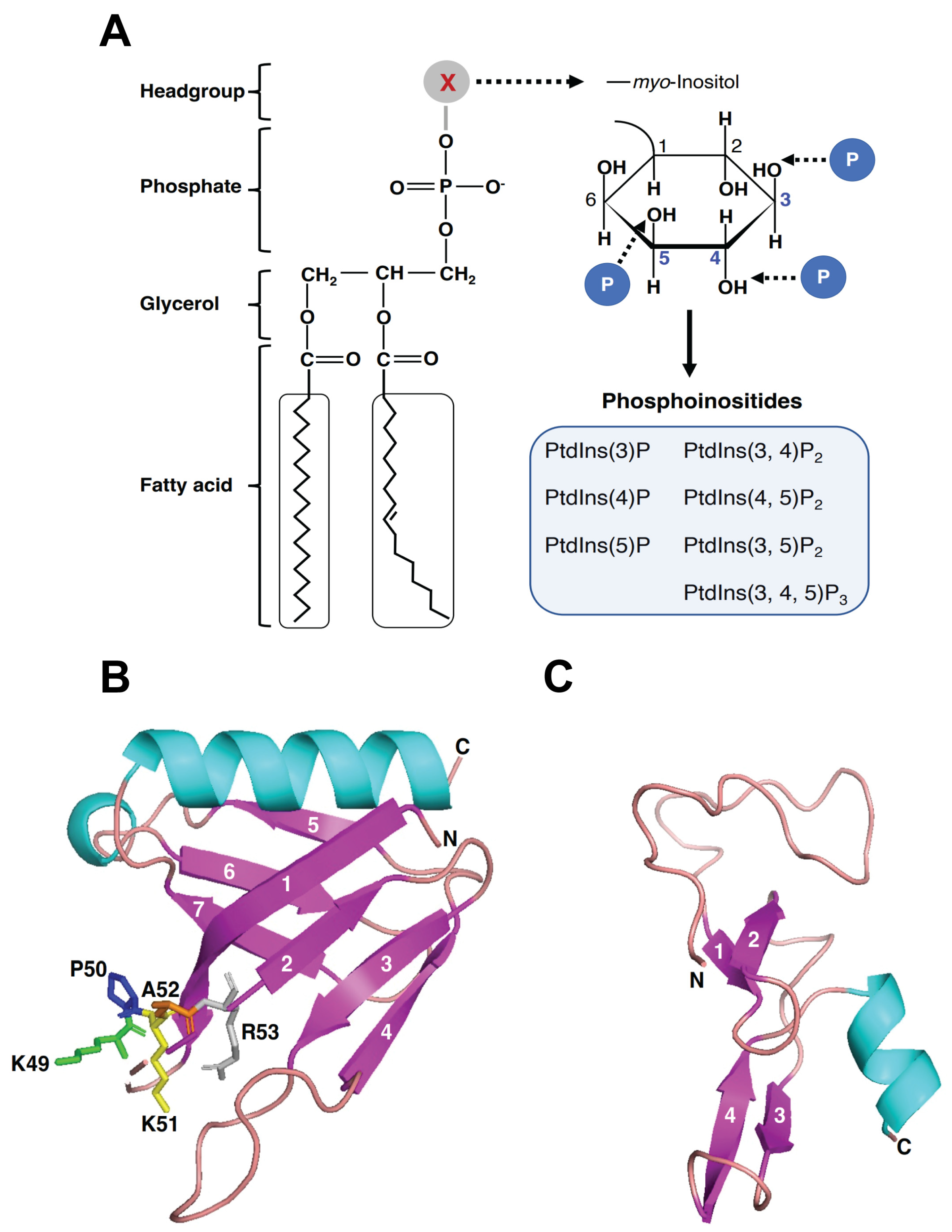
3. PIPs and PIP-Binding Domains
4. The PIP-Binding Domains of Phafin Proteins
4.1. The PH Domain
4.2. The FYVE Domain
5. The Functions of Phafin Proteins in the Apoptotic, Autophagic, and Endocytic Pathways
5.1. Apoptosis
5.2. Endosomal Cargo Transport
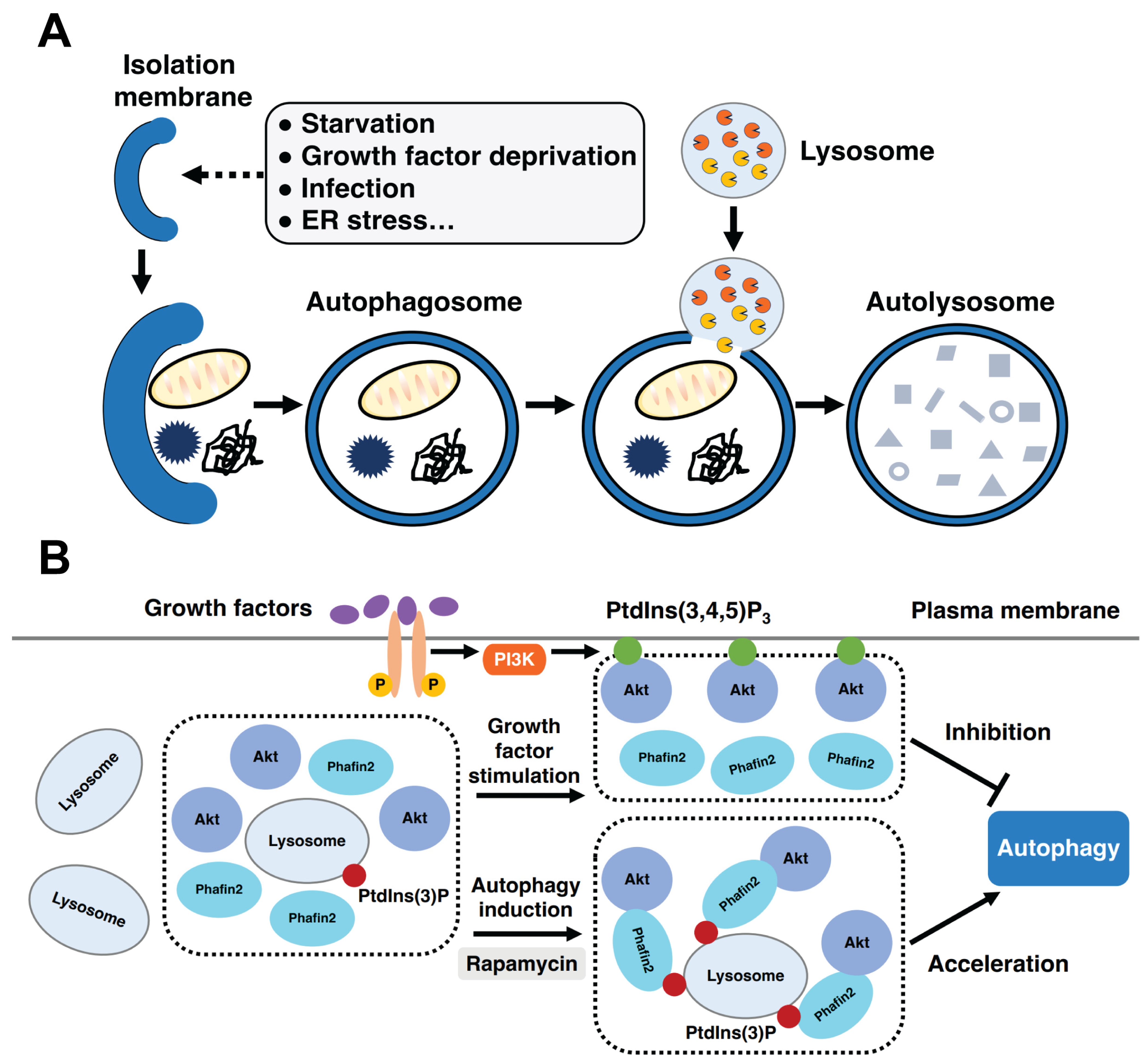
5.3. Autophagy
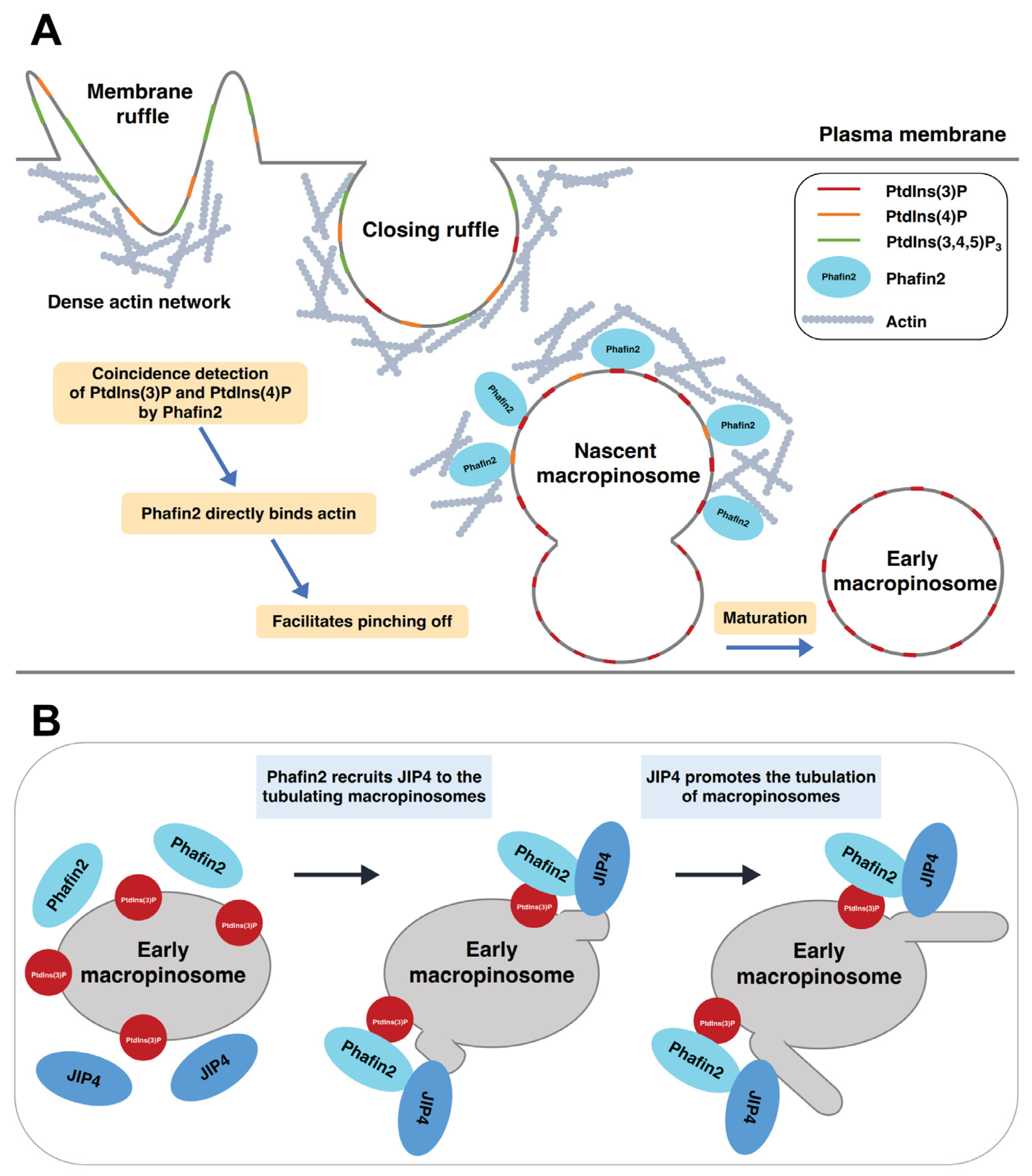
5.4. Macropinocytosis
6. Physiological and Pathological Functions of Phafins
6.1. Immunity
6.2. Cancer
7. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Akt/PKB | protein kinase B |
| BC | breast cancer |
| Btk | Bruton’s tyrosine kinase |
| EEA1 | early endosomal antigen 1 |
| EGF | epidermal growth factor |
| EGFR | epidermal growth factor receptor |
| FYVE | Fab1, YOTB, Vac1, and EEA1 |
| GDI | guanosine diphosphate (GDP) dissociation inhibitor |
| Grp1 | General receptor for phosphoinositides isoform 1 |
| HCC | human hepatocellular carcinoma |
| InsR | insulin receptor |
| JIP4 | JNK-interacting protein 4 |
| PH | Pleckstrin Homology |
| PKC | Protein Kinase C |
| PLC-δ1 | phospholipase C-δ1 |
| polyD | aspartic acid-rich |
| PtdIns(3)P | phosphatidylinositol 3-phosphate |
| PtdIns(4)P | phosphatidylinositol 4-phosphate |
| PtdIns(5)P | phosphatidylinositol 5-phosphate |
| PtdIns(3,4)P2 | phosphatidylinositol (3,4)-bisphosphate |
| PtdIns(3,5)P2 | phosphatidylinositol (3,5)-bisphosphate |
| PtdIns(4,5)P2 | phosphatidylinositol (4,5)-bisphosphate |
| PtdIns(3,4,5)P3 | phosphatidylinositol (3,4,5)-trisphosphate |
| TORC1 | target of rapamycin complex 1 |
References
- Conner, S.D.; Schmid, S.L. Regulated portals of entry into the cell. Nature 2003, 422, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Doherty, G.J.; McMahon, H.T. Mechanisms of Endocytosis. Annu. Rev. Biochem. 2009, 78, 857–902. [Google Scholar] [CrossRef] [PubMed]
- Platta, H.W.; Stenmark, H. Endocytosis and signaling. Curr. Opin. Cell Biol. 2011, 23, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Yim, W.W.-Y.; Mizushima, N. Lysosome biology in autophagy. Cell Discov. 2020, 6, 6. [Google Scholar] [CrossRef]
- Birgisdottir, Å.B.; Johansen, T. Autophagy and endocytosis-interconnections and interdependencies. J. Cell Sci. 2020, 133, jcs228114. [Google Scholar] [CrossRef]
- Peña-Martinez, C.; Rickman, A.D.; Heckmann, B.L. Beyond autophagy: LC3-associated phagocytosis and endocytosis. Sci. Adv. 2022, 8, eabn1702. [Google Scholar] [CrossRef]
- Wang, H.; Lo, W.-T.; Haucke, V. Phosphoinositide switches in endocytosis and in the endolysosomal system. Curr. Opin. Cell Biol. 2019, 59, 50–57. [Google Scholar] [CrossRef]
- Knævelsrud, H.; Søreng, K.; Raiborg, C.; Håberg, K.; Rasmuson, F.; Brech, A.; Liestøl, K.; Rusten, T.E.; Stenmark, H.; Neufeld, T.P.; et al. Membrane remodeling by the PX-BAR protein SNX18 promotes autophagosome formation. J. Cell Biol. 2013, 202, 331–349. [Google Scholar] [CrossRef]
- Jarsch, I.K.; Daste, F.; Gallop, J.L. Membrane curvature in cell biology: An integration of molecular mechanisms. J. Cell Biol. 2016, 214, 375–387. [Google Scholar] [CrossRef]
- Schink, K.O.; Tan, K.-W.; Stenmark, H. Phosphoinositides in Control of Membrane Dynamics. Annu. Rev. Cell Dev. Biol. 2016, 32, 143–171. [Google Scholar] [CrossRef]
- Di Paolo, G.; De Camilli, P. Phosphoinositides in cell regulation and membrane dynamics. Nature 2006, 443, 651–657. [Google Scholar] [CrossRef]
- Posor, Y.; Jang, W.; Haucke, V. Phosphoinositides as membrane organizers. Nat. Rev. Mol. Cell Biol. 2022, 23, 797–816. [Google Scholar] [CrossRef]
- Lemmon, M.A. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 2008, 9, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.-J.; Yang, C.-Y.; Lin, Y.-C.; Tsai, M.-C.; Yang, C.-W.; Tung, C.-Y.; Ho, P.-Y.; Kao, F.-J.; Lin, C.-H. Phafin2 modulates the structure and function of endosomes by a Rab5-dependent mechanism. Biochem. Biophys. Res. Commun. 2010, 391, 1043–1048. [Google Scholar] [CrossRef]
- Lin, W.-J.; Yang, C.-Y.; Li, L.-L.; Yi, Y.-H.; Chen, K.-W.; Lin, Y.-C.; Liu, C.-C.; Lin, C.-H. Lysosomal targeting of phafin1 mediated by Rab7 induces autophagosome formation. Biochem. Biophys. Res. Commun. 2012, 417, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.-X.; Finkielstein, C.V.; Capelluto, D.G.S. The C-terminal acidic motif of Phafin2 inhibits PH domain binding to phosphatidylinositol 3-phosphate. Biochim. Biophys. Acta (BBA) Biomembr. 2020, 1862, 183230. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.X.; Jo, A.; Deng, J.; Ellena, J.F.; Lazar, I.M.; Davis, R.M.; Capelluto, D.G. Structural, thermodynamic, and phosphatidylinositol 3-phosphate binding properties of Phafin2. Protein Sci. 2017, 26, 814–823. [Google Scholar] [CrossRef]
- Hasan, M.; Capelluto, D.G.S. The PH Domain and C-Terminal polyD Motif of Phafin2 Exhibit a Unique Concurrence in Animals. Membranes 2022, 12, 696. [Google Scholar] [CrossRef]
- Fagerberg, L.; Hallström, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the Human Tissue-specific Expression by Genome-wide Integration of Transcriptomics and Antibody-based Proteomics. Mol. Cell. Proteom. 2014, 13, 397–406. [Google Scholar] [CrossRef]
- Li, C.; Liu, Q.; Li, N.; Chen, W.; Wang, L.; Wang, Y.; Yu, Y.; Cao, X. EAPF/Phafin-2, a novel endoplasmic reticulum-associated protein, facilitates TNF-α-triggered cellular apoptosis through endoplasmic reticulum-mitochondrial apoptotic pathway. J. Mol. Med. 2008, 86, 471–484. [Google Scholar] [CrossRef]
- Schink, K.O.; Tan, K.W.; Spangenberg, H.; Martorana, D.; Sneeggen, M.; Stévenin, V.; Enninga, J.; Campsteijn, C.; Raiborg, C.; Stenmark, H. The phosphoinositide coincidence detector Phafin2 promotes macropinocytosis by coordinating actin organisation at forming macropinosomes. Nat. Commun. 2021, 12, 6577. [Google Scholar] [CrossRef]
- Tan, K.W.; Nähse, V.; Campsteijn, C.; Brech, A.; Schink, K.O.; Stenmark, H. JIP4 is recruited by the phosphoinositide-binding protein Phafin2 to promote recycling tubules on macropinosomes. J. Cell Sci. 2021, 134, jcs258495. [Google Scholar] [CrossRef]
- Howe, K.L.; Achuthan, P.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; Bhai, J. Ensembl 2021. Nucleic Acids Res. 2021, 49, D884–D891. [Google Scholar] [CrossRef] [PubMed]
- Simonsen, A.; Wurmser, A.E.; Emr, S.D.; Stenmark, H. The role of phosphoinositides in membrane transport. Curr. Opin. Cell Biol. 2001, 13, 485–492. [Google Scholar] [CrossRef] [PubMed]
- Wallroth, A.; Haucke, V. Phosphoinositide conversion in endocytosis and the endolysosomal system. J. Biol. Chem. 2018, 293, 1526–1535. [Google Scholar] [CrossRef] [PubMed]
- Olivença, D.V.; Uliyakina, I.; Fonseca, L.L.; Amaral, M.D.; Voit, E.O.; Pinto, F.R. A Mathematical Model of the Phosphoinositide Pathway. Sci. Rep. 2018, 8, 3904. [Google Scholar] [CrossRef]
- Balla, T. Phosphoinositides: Tiny Lipids With Giant Impact on Cell Regulation. Physiol. Rev. 2013, 93, 1019–1137. [Google Scholar] [CrossRef]
- Kutateladze, T.G. Translation of the phosphoinositide code by PI effectors. Nat. Chem. Biol. 2010, 6, 507–513. [Google Scholar] [CrossRef]
- Poccia, D.; Larijani, B. Phosphatidylinositol metabolism and membrane fusion. Biochem. J. 2009, 418, 233–246. [Google Scholar] [CrossRef]
- Picas, L.; Gaits-Iacovoni, F.; Goud, B. The emerging role of phosphoinositide clustering in intracellular trafficking and signal transduction. F1000Res 2016, 5, 422. [Google Scholar] [CrossRef]
- Tsuji, T.; Takatori, S.; Fujimoto, T. Definition of phosphoinositide distribution in the nanoscale. Curr. Opin. Cell Biol. 2019, 57, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Balla, T.; Szentpetery, Z.; Kim, Y.J. Phosphoinositide Signaling: New Tools and Insights. Physiology 2009, 24, 231–244. [Google Scholar] [CrossRef] [PubMed]
- Antonietta De Matteis, M.; Di Campli, A.; Godi, A. The role of the phosphoinositides at the Golgi complex. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2005, 1744, 396–405. [Google Scholar] [CrossRef] [PubMed]
- Roth, M.G. Phosphoinositides in Constitutive Membrane Traffic. Physiol. Rev. 2004, 84, 699–730. [Google Scholar] [CrossRef]
- Mandal, K. Review of PIP2 in Cellular Signaling, Functions and Diseases. Int. J. Mol. Sci. 2020, 21, 8342. [Google Scholar] [CrossRef]
- Lystad, A.H.; Simonsen, A. Phosphoinositide-binding proteins in autophagy. FEBS Lett. 2016, 590, 2454–2468. [Google Scholar] [CrossRef]
- Downes, C.P.; Gray, A.; Lucocq, J.M. Probing phosphoinositide functions in signaling and membrane trafficking. Trends Cell Biol. 2005, 15, 259–268. [Google Scholar] [CrossRef]
- Hurley, J.H.; Meyer, T. Subcellular targeting by membrane lipids. Curr. Opin. Cell Biol. 2001, 13, 146–152. [Google Scholar] [CrossRef]
- Legendre-Guillemin, V.; Wasiak, S.; Hussain, N.K.; Angers, A.; McPherson, P.S. ENTH/ANTH proteins and clathrin-mediated membrane budding. J. Cell Sci. 2004, 117, 9–18. [Google Scholar] [CrossRef]
- Gallop, J.L.; Jao, C.C.; Kent, H.M.; Butler, P.J.G.; Evans, P.R.; Langen, R.; McMahon, H.T. Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J. 2006, 25, 2898–2910. [Google Scholar] [CrossRef]
- Raftopoulou, M.; Etienne-Manneville, S.; Self, A.; Nicholls, S.; Hall, A. Regulation of Cell Migration by the C2 Domain of the Tumor Suppressor PTEN. Science 2004, 303, 1179–1181. [Google Scholar] [CrossRef] [PubMed]
- Itoh, T.; Koshiba, S.; Kigawa, T.; Kikuchi, A.; Yokoyama, S.; Takenawa, T. Role of the ENTH Domain in Phosphatidylinositol-4,5-Bisphosphate Binding and Endocytosis. Science 2001, 291, 1047–1051. [Google Scholar] [CrossRef]
- Gaullier, J.-M.; Simonsen, A.; D’Arrigo, A.; Bremnes, B.; Stenmark, H.; Aasland, R. FYVE fingers bind PtdIns(3)P. Nature 1998, 394, 432–433. [Google Scholar] [CrossRef] [PubMed]
- Musacchio, A.; Gibson, T.; Rice, P.; Thompson, J.; Saraste, M. The PH domain: A common piece in the structural pathcwork of signalling proteins. Trends Biochem. Sci. 1993, 18, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Gopaldass, N.; Fauvet, B.; Lashuel, H.; Roux, A.; Mayer, A. Membrane scission driven by the PROPPIN Atg18. EMBO J. 2017, 36, 3274–3291. [Google Scholar] [CrossRef]
- Uhlik, M.T.; Temple, B.; Bencharit, S.; Kimple, A.J.; Siderovski, D.P.; Johnson, G.L. Structural and Evolutionary Division of Phosphotyrosine Binding (PTB) Domains. J. Mol. Biol. 2005, 345, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Chandra, M.; Chin, Y.K.Y.; Mas, C.; Feathers, J.R.; Paul, B.; Datta, S.; Chen, K.-E.; Jia, X.; Yang, Z.; Norwood, S.J.; et al. Classification of the human phox homology (PX) domains based on their phosphoinositide binding specificities. Nat. Commun. 2019, 10, 1528. [Google Scholar] [CrossRef]
- Lemmon, M.A. Phosphoinositide Recognition Domains. Traffic 2003, 4, 201–213. [Google Scholar] [CrossRef]
- Hammond, G.R.V.; Balla, T. Polyphosphoinositide binding domains: Key to inositol lipid biology. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2015, 1851, 746–758. [Google Scholar] [CrossRef]
- Kutateladze, T.G. Molecular Analysis of Protein–Phosphoinositide Interactions. In Phosphoinositides and Disease; Falasca, M., Ed.; Springer Netherlands: Dordrecht, The Netherlands, 2012; pp. 111–126. [Google Scholar]
- Pemberton, J.G.; Balla, T. Polyphosphoinositide-Binding Domains: Insights from Peripheral Membrane and Lipid-Transfer Proteins. In Protein Reviews–Purinergic Receptors: Volume 20; Atassi, M.Z., Ed.; Springer International Publishing: Cham, Switzerland, 2019; Volume 20, pp. 77–137. [Google Scholar]
- Kutateladze, T.G.; Capelluto, D.G.S.; Ferguson, C.G.; Cheever, M.L.; Kutateladze, A.G.; Prestwich, G.D.; Overduin, M. Multivalent Mechanism of Membrane Insertion by the FYVE Domain. J. Biol. Chem. 2004, 279, 3050–3057. [Google Scholar] [CrossRef]
- Prakash, S.; Krishna, A.; Sengupta, D. Cofilin-Membrane Interactions: Electrostatic Effects in Phosphoinositide Lipid Binding. ChemPhysChem 2023, 24, e202200509. [Google Scholar] [CrossRef] [PubMed]
- Cho, W.; Stahelin, R.V. Membrane-Protein Interactions in Cell Signaling and Membrane Trafficking. Annu. Rev. Biophys. Biomol. Struct. 2005, 34, 119–151. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, E.; Domański, J.; Naughton, F.B.; Best, R.B.; Kalli, A.C.; Stansfeld, P.J.; Sansom, M.S.P. Multiple lipid binding sites determine the affinity of PH domains for phosphoinositide-containing membranes. Sci. Adv. 2020, 6, eaay5736. [Google Scholar] [CrossRef]
- Lemmon, M.A. Pleckstrin homology (PH) domains and phosphoinositides. Biochem. Soc. Symp. 2007, 74, 81–93. [Google Scholar] [CrossRef] [PubMed]
- LEMMON, M.A.; FERGUSON, K.M. Signal-dependent membrane targeting by pleckstrin homology (PH) domains. Biochem. J. 2000, 350, 1–18. [Google Scholar] [CrossRef]
- Thomas, C.C.; Deak, M.; Alessi, D.R.; van Aalten, D.M.F. High-Resolution Structure of the Pleckstrin Homology Domain of Protein Kinase B/Akt Bound to Phosphatidylinositol (3,4,5)-Trisphosphate. Curr. Biol. 2002, 12, 1256–1262. [Google Scholar] [CrossRef]
- Yoon, H.S.; Hajduk, P.J.; Petros, A.M.; Olejniczak, E.T.; Meadows, R.P.; Fesik, S.W. Solution structure of a pleckstrin-homology domain. Nature 1994, 369, 672–675. [Google Scholar] [CrossRef]
- Auguin, D.; Barthe, P.; Augé-Sénégas, M.-T.; Stern, M.-H.; Noguchi, M.; Roumestand, C. Solution Structure and Backbone Dynamics of the Pleckstrin Homology Domain of the Human Protein Kinase B (PKB/Akt). Interaction with Inositol Phosphates. J. Biomol. NMR 2004, 28, 137–155. [Google Scholar] [CrossRef]
- Lenoir, M.; Coskun, Ü.; Grzybek, M.; Cao, X.; Buschhorn, S.B.; James, J.; Simons, K.; Overduin, M. Structural basis of wedging the Golgi membrane by FAPP pleckstrin homology domains. EMBO Rep. 2010, 11, 279–284. [Google Scholar] [CrossRef]
- Harlan, J.E.; Hajduk, P.J.; Yoon, H.S.; Fesik, S.W. Pleckstrin homology domains bind to phosphatidylinositol-4, 5-bisphosphate. Nature 1994, 371, 168–170. [Google Scholar] [CrossRef]
- Lemmon, M.A.; Ferguson, K.M.; O’Brien, R.; Sigler, P.B.; Schlessinger, J. Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc. Natl. Acad. Sci. USA 1995, 92, 10472–10476. [Google Scholar] [CrossRef] [PubMed]
- Manna, D.; Albanese, A.; Park, W.S.; Cho, W. Mechanistic basis of differential cellular responses of phosphatidylinositol 3, 4-bisphosphate-and phosphatidylinositol 3, 4, 5-trisphosphate-binding pleckstrin homology domains. J. Biol. Chem. 2007, 282, 32093–32105. [Google Scholar] [CrossRef]
- He, J.; Haney, R.M.; Vora, M.; Verkhusha, V.V.; Stahelin, R.V.; Kutateladze, T.G. Molecular mechanism of membrane targeting by the GRP1 PH domain. J. Lipid Res. 2008, 49, 1807–1815. [Google Scholar] [CrossRef]
- Stephens, L.; Anderson, K.; Stokoe, D.; Erdjument-Bromage, H.; Painter, G.F.; Holmes, A.B.; Gaffney, P.R.; Reese, C.B.; McCormick, F.; Tempst, P. Protein kinase B kinases that mediate phosphatidylinositol 3, 4, 5-trisphosphate-dependent activation of protein kinase B. Science 1998, 279, 710–714. [Google Scholar] [CrossRef] [PubMed]
- Frech, M.; Andjelkovic, M.; Ingley, E.; Reddy, K.K.; Falck, J.R.; Hemmings, B.A. High affinity binding of inositol phosphates and phosphoinositides to the pleckstrin homology domain of RAC/protein kinase B and their influence on kinase activity. J. Biol. Chem. 1997, 272, 8474–8481. [Google Scholar] [CrossRef] [PubMed]
- Agorio, A.; Giraudat, J.; Bianchi, M.W.; Marion, J.; Espagne, C.; Castaings, L.; Lelièvre, F.; Curie, C.; Thomine, S.; Merlot, S. Phosphatidylinositol 3-phosphate–binding protein AtPH1 controls the localization of the metal transporter NRAMP1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, E3354–E3363. [Google Scholar] [CrossRef]
- Sugiki, T.; Takeuchi, K.; Yamaji, T.; Takano, T.; Tokunaga, Y.; Kumagai, K.; Hanada, K.; Takahashi, H.; Shimada, I. Structural Basis for the Golgi Association by the Pleckstrin Homology Domain of the Ceramide Trafficking Protein (CERT) *. J. Biol. Chem. 2012, 287, 33706–33718. [Google Scholar] [CrossRef]
- Zwolak, A.; Yang, C.; Feeser, E.A.; Michael Ostap, E.; Svitkina, T.; Dominguez, R. CARMIL leading edge localization depends on a non-canonical PH domain and dimerization. Nat. Commun. 2013, 4, 2523. [Google Scholar] [CrossRef]
- Ghai, R.; Du, X.; Wang, H.; Dong, J.; Ferguson, C.; Brown, A.J.; Parton, R.G.; Wu, J.-W.; Yang, H. ORP5 and ORP8 bind phosphatidylinositol-4, 5-biphosphate (PtdIns (4, 5) P 2) and regulate its level at the plasma membrane. Nat. Commun. 2017, 8, 757. [Google Scholar] [CrossRef]
- Matsuda-Lennikov, M.; Suizu, F.; Hirata, N.; Hashimoto, M.; Kimura, K.; Nagamine, T.; Fujioka, Y.; Ohba, Y.; Iwanaga, T.; Noguchi, M. Lysosomal Interaction of Akt with Phafin2: A Critical Step in the Induction of Autophagy. PLoS ONE 2014, 9, e79795. [Google Scholar] [CrossRef]
- Singh, N.; Reyes-Ordoñez, A.; Compagnone, M.A.; Moreno, J.F.; Leslie, B.J.; Ha, T.; Chen, J. Redefining the specificity of phosphoinositide-binding by human PH domain-containing proteins. Nat. Commun. 2021, 12, 4339. [Google Scholar] [CrossRef] [PubMed]
- Gray, A.; Van Der Kaay, J.; Downes, C.P. The pleckstrin homology domains of protein kinase B and GRP1 (general receptor for phosphoinositides-1) are sensitive and selective probes for the cellular detection of phosphatidylinositol 3,4-bisphosphate and/or phosphatidylinositol 3,4,5-trisphosphate in vivo. Biochem. J. 1999, 344, 929–936. [Google Scholar] [CrossRef] [PubMed]
- Idevall-Hagren, O.; De Camilli, P. Detection and manipulation of phosphoinositides. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2015, 1851, 736–745. [Google Scholar] [CrossRef] [PubMed]
- Pant, S.; Tajkhorshid, E. Microscopic Characterization of GRP1 PH Domain Interaction with Anionic Membranes. J. Comput. Chem. 2020, 41, 489–499. [Google Scholar] [CrossRef]
- Scheffzek, K.; Welti, S. Pleckstrin homology (PH) like domains–versatile modules in protein–protein interaction platforms. FEBS Lett. 2012, 586, 2662–2673. [Google Scholar] [CrossRef] [PubMed]
- Agamasu, C.; Ghanam, R.H.; Saad, J.S. Structural and Biophysical Characterization of the Interactions between Calmodulin and the Pleckstrin Homology Domain of Akt *. J. Biol. Chem. 2015, 290, 27403–27413. [Google Scholar] [CrossRef]
- Yao, L.; Suzuki, H.; Ozawa, K.; Deng, J.; Lehel, C.; Fukamachi, H.; Anderson, W.B.; Kawakami, Y.; Kawakami, T. Interactions between Protein Kinase C and Pleckstrin Homology Domains: Inhibition by phosphatidylinositol 4,5-bisphosphate and phorbol 12-myristate 13-acetate. J. Biol. Chem. 1997, 272, 13033–13039. [Google Scholar] [CrossRef]
- Gailite, I.; Egger-Adam, D.; Wodarz, A. The phosphoinositide-associated protein Rush hour regulates endosomal trafficking in Drosophila. Mol. Biol. Cell 2012, 23, 433–447. [Google Scholar] [CrossRef]
- Pedersen, N.M.; Raiborg, C.; Brech, A.; Skarpen, E.; Roxrud, I.; Platta, H.W.; Liestøl, K.; Stenmark, H. The PtdIns3P-Binding Protein Phafin 2 Mediates Epidermal Growth Factor Receptor Degradation by Promoting Endosome Fusion. Traffic 2012, 13, 1547–1563. [Google Scholar] [CrossRef]
- Stenmark, H.; Aasland, R. FYVE-finger proteins-effectors of an inositol lipid. J. Cell Sci. 1999, 112, 4175–4183. [Google Scholar] [CrossRef]
- Kutateladze, T.G. Phosphatidylinositol 3-phosphate recognition and membrane docking by the FYVE domain. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2006, 1761, 868–877. [Google Scholar] [CrossRef] [PubMed]
- Stenmark, H.; Aasland, R.; Toh, B.-H.; D’Arrigo, A. Endosomal Localization of the Autoantigen EEA1 Is Mediated by a Zinc-binding FYVE Finger. J. Biol. Chem. 1996, 271, 24048–24054. [Google Scholar] [CrossRef] [PubMed]
- Gillooly, D.J.; Morrow, I.C.; Lindsay, M.; Gould, R.; Bryant, N.J.; Gaullier, J.M.; Parton, R.G.; Stenmark, H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. Embo. J. 2000, 19, 4577–4588. [Google Scholar] [CrossRef] [PubMed]
- Nascimbeni, A.C.; Codogno, P.; Morel, E. Local detection of PtdIns3P at autophagosome biogenesis membrane platforms. Autophagy 2017, 13, 1602–1612. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Jian, Y.; Sun, X.; Yang, C.; Gao, Z.; Zhang, Z.; Liu, X.; Li, Y.; Xu, J.; Jing, Y.; et al. Negative regulation of phosphatidylinositol 3-phosphate levels in early-to-late endosome conversion. J. Cell Biol. 2016, 212, 181–198. [Google Scholar] [CrossRef]
- Misra, S.; Hurley, J.H. Crystal Structure of a Phosphatidylinositol 3-Phosphate-Specific Membrane-Targeting Motif, the FYVE Domain of Vps27p. Cell 1999, 97, 657–666. [Google Scholar] [CrossRef]
- Mao, Y.; Nickitenko, A.; Duan, X.; Lloyd, T.E.; Wu, M.N.; Bellen, H.; Quiocho, F.A. Crystal Structure of the VHS and FYVE Tandem Domains of Hrs, a Protein Involved in Membrane Trafficking and Signal Transduction. Cell 2000, 100, 447–456. [Google Scholar] [CrossRef]
- Blatner, N.R.; Stahelin, R.V.; Diraviyam, K.; Hawkins, P.T.; Hong, W.; Murray, D.; Cho, W. The Molecular Basis of the Differential Subcellular Localization of FYVE Domains. J. Biol. Chem. 2004, 279, 53818–53827. [Google Scholar] [CrossRef]
- Kutateladze, T.; Overduin, M. Structural Mechanism of Endosome Docking by the FYVE Domain. Science 2001, 291, 1793–1796. [Google Scholar] [CrossRef]
- Lee, S.A.; Eyeson, R.; Cheever, M.L.; Geng, J.; Verkhusha, V.V.; Burd, C.; Overduin, M.; Kutateladze, T.G. Targeting of the FYVE domain to endosomal membranes is regulated by a histidine switch. Proc. Natl. Acad. Sci. USA 2005, 102, 13052–13057. [Google Scholar] [CrossRef]
- He, J.; Vora, M.; Haney, R.M.; Filonov, G.S.; Musselman, C.A.; Burd, C.G.; Kutateladze, A.G.; Verkhusha, V.V.; Stahelin, R.V.; Kutateladze, T.G. Membrane insertion of the FYVE domain is modulated by pH. Proteins Struct. Funct. Bioinform. 2009, 76, 852–860. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.-X.; Xiong, W.; Finkielstein, C.V.; Capelluto, D.G.S. Identification of Lipid Binding Modulators Using the Protein-Lipid Overlay Assay. In Proteomics for Drug Discovery: Methods and Protocols; Lazar, I.M., Kontoyianni, M., Lazar, A.C., Eds.; Springer: New York, NY, USA, 2017; pp. 197–206. [Google Scholar]
- GILLOOLY, D.J.; SIMONSEN, A.; STENMARK, H. Cellular functions of phosphatidylinositol 3-phosphate and FYVE domain proteins. Biochem. J. 2001, 355, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Dumas, J.J.; Merithew, E.; Sudharshan, E.; Rajamani, D.; Hayes, S.; Lawe, D.; Corvera, S.; Lambright, D.G. Multivalent endosome targeting by homodimeric EEA1. Mol. Cell 2001, 8, 947–958. [Google Scholar] [CrossRef]
- Hengartner, M.O. The biochemistry of apoptosis. Nature 2000, 407, 770–776. [Google Scholar] [CrossRef] [PubMed]
- Strasser, A.; O’Connor, L.; Dixit, V.M. Apoptosis signaling. Annu. Rev. Biochem. 2000, 69, 217–245. [Google Scholar] [CrossRef] [PubMed]
- Green, D.R. The Coming Decade of Cell Death Research: Five Riddles. Cell 2019, 177, 1094–1107. [Google Scholar] [CrossRef]
- Carneiro, B.A.; El-Deiry, W.S. Targeting apoptosis in cancer therapy. Nat. Rev. Clin. Oncol. 2020, 17, 395–417. [Google Scholar] [CrossRef]
- Chaabane, W.; Cieślar-Pobuda, A.; El-Gazzah, M.; Jain, M.V.; Rzeszowska-Wolny, J.; Rafat, M.; Stetefeld, J.; Ghavami, S.; Łos, M.J. Human-Gyrovirus-Apoptin Triggers Mitochondrial Death Pathway—Nur77 is Required for Apoptosis Triggering. Neoplasia 2014, 16, 679–693. [Google Scholar] [CrossRef]
- Bertheloot, D.; Latz, E.; Franklin, B.S. Necroptosis, pyroptosis and apoptosis: An intricate game of cell death. Cell. Mol. Immunol. 2021, 18, 1106–1121. [Google Scholar] [CrossRef]
- Leonard, B.C.; Johnson, D.E. Signaling by cell surface death receptors: Alterations in head and neck cancer. Adv. Biol. Regul. 2018, 67, 170–178. [Google Scholar] [CrossRef]
- Kim, B.-C.; Kim, H.-T.; Mamura, M.; Ambudkar, I.S.; Choi, K.-S.; Kim, S.-J. Tumor Necrosis Factor Induces Apoptosis in Hepatoma Cells by Increasing Ca2+ Release from the Endoplasmic Reticulum and Suppressing Bcl-2 Expression. J. Biol. Chem. 2002, 277, 31381–31389. [Google Scholar] [CrossRef] [PubMed]
- Gould, G.W.; Lippincott-Schwartz, J. New roles for endosomes: From vesicular carriers to multi-purpose platforms. Nat. Rev. Mol. Cell Biol. 2009, 10, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Simonsen, A.; Lippe, R.; Christoforidis, S.; Gaullier, J.-M.; Brech, A.; Callaghan, J.; Toh, B.-H.; Murphy, C.; Zerial, M.; Stenmark, H. EEA1 links PI (3) K function to Rab5 regulation of endosome fusion. Nature 1998, 394, 494–498. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Chuang, J.-Z.; Xu, K.; McGraw, T.G.; Sung, C.-H. SARA, a FYVE domain protein, affects Rab5-mediated endocytosis. J. Cell Sci. 2002, 115, 4755–4763. [Google Scholar] [CrossRef]
- Kundu, M.; Thompson, C.B. Autophagy: Basic Principles and Relevance to Disease. Annu. Rev. Pathol. Mech. Dis. 2008, 3, 427–455. [Google Scholar] [CrossRef]
- Levine, B.; Kroemer, G. Autophagy in the Pathogenesis of Disease. Cell 2008, 132, 27–42. [Google Scholar] [CrossRef]
- Levine, B.; Deretic, V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat. Rev. Immunol. 2007, 7, 767–777. [Google Scholar] [CrossRef]
- Levine, B.; Mizushima, N.; Virgin, H.W. Autophagy in immunity and inflammation. Nature 2011, 469, 323–335. [Google Scholar] [CrossRef]
- Kawabata, T.; Yoshimori, T. Beyond starvation: An update on the autophagic machinery and its functions. J. Mol. Cell. Cardiol. 2016, 95, 2–10. [Google Scholar] [CrossRef]
- Jang, D.-J.; Lee, J.-A. The roles of phosphoinositides in mammalian autophagy. Arch. Pharmacal Res. 2016, 39, 1129–1136. [Google Scholar] [CrossRef]
- Boya, P.; Reggiori, F.; Codogno, P. Emerging regulation and functions of autophagy. Nat. Cell Biol. 2013, 15, 713–720. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, M.; Hirata, N.; Tanaka, T.; Suizu, F.; Nakajima, H.; Chiorini, J.A. Autophagy as a modulator of cell death machinery. Cell Death Dis. 2020, 11, 517. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Levine, B.; Cuervo, A.M.; Klionsky, D.J. Autophagy fights disease through cellular self-digestion. Nature 2008, 451, 1069–1075. [Google Scholar] [CrossRef] [PubMed]
- Shintani, T.; Klionsky, D.J. Autophagy in Health and Disease: A Double-Edged Sword. Science 2004, 306, 990–995. [Google Scholar] [CrossRef]
- Choi, A.M.K.; Ryter, S.W.; Levine, B. Autophagy in Human Health and Disease. N. Engl. J. Med. 2013, 368, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Mowers, E.E.; Sharifi, M.N.; Macleod, K.F. Functions of autophagy in the tumor microenvironment and cancer metastasis. FEBS J. 2018, 285, 1751–1766. [Google Scholar] [CrossRef]
- Hara, T.; Nakamura, K.; Matsui, M.; Yamamoto, A.; Nakahara, Y.; Suzuki-Migishima, R.; Yokoyama, M.; Mishima, K.; Saito, I.; Okano, H.; et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006, 441, 885–889. [Google Scholar] [CrossRef]
- Keller, M.D.; Torres, V.J.; Cadwell, K. Autophagy and microbial pathogenesis. Cell Death Differ. 2020, 27, 872–886. [Google Scholar] [CrossRef]
- Nishino, I.; Fu, J.; Tanji, K.; Yamada, T.; Shimojo, S.; Koori, T.; Mora, M.; Riggs, J.E.; Oh, S.J.; Koga, Y.; et al. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease). Nature 2000, 406, 906–910. [Google Scholar] [CrossRef]
- Lipinski, M.M.; Zheng, B.; Lu, T.; Yan, Z.; Py, B.F.; Ng, A.; Xavier, R.J.; Li, C.; Yankner, B.A.; Scherzer, C.R.; et al. Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2010, 107, 14164–14169. [Google Scholar] [CrossRef]
- Mizushima, N.; Yoshimori, T.; Levine, B. Methods in Mammalian Autophagy Research. Cell 2010, 140, 313–326. [Google Scholar] [CrossRef]
- Baskaran, S.; Ragusa, M.J.; Boura, E.; Hurley, J.H. Two-Site Recognition of Phosphatidylinositol 3-Phosphate by PROPPINs in Autophagy. Mol. Cell 2012, 47, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Nascimbeni, A.C.; Codogno, P.; Morel, E. Phosphatidylinositol-3-phosphate in the regulation of autophagy membrane dynamics. FEBS J. 2017, 284, 1267–1278. [Google Scholar] [CrossRef]
- Hirata, N.; Suizu, F.; Matsuda-Lennikov, M.; Tanaka, T.; Edamura, T.; Ishigaki, S.; Donia, T.; Lithanatudom, P.; Obuse, C.; Iwanaga, T.; et al. Functional characterization of lysosomal interaction of Akt with VRK2. Oncogene 2018, 37, 5367–5386. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, M.; Hirata, N.; Suizu, F. The links between AKT and two intracellular proteolytic cascades: Ubiquitination and autophagy. Biochim. Biophys. Acta (BBA) Rev. Cancer 2014, 1846, 342–352. [Google Scholar] [CrossRef] [PubMed]
- Manning, B.D.; Toker, A. AKT/PKB Signaling: Navigating the Network. Cell 2017, 169, 381–405. [Google Scholar] [CrossRef]
- Basu, A.; Lambring, C.B. Akt Isoforms: A Family Affair in Breast Cancer. Cancers 2021, 13, 3445. [Google Scholar] [CrossRef]
- Ebner, M.; Lučić, I.; Leonard, T.A.; Yudushkin, I. PI (3, 4, 5) P3 engagement restricts Akt activity to cellular membranes. Mol. Cell 2017, 65, 416–431.e416. [Google Scholar] [CrossRef]
- Swanson, J.A. Shaping cups into phagosomes and macropinosomes. Nat. Rev. Mol. Cell Biol. 2008, 9, 639–649. [Google Scholar] [CrossRef]
- Lim, J.P.; Gleeson, P.A. Macropinocytosis: An endocytic pathway for internalising large gulps. Immunol. Cell Biol. 2011, 89, 836–843. [Google Scholar] [CrossRef]
- West, M.A.; Bretscher, M.S.; Watts, C. Distinct endocytotic pathways in epidermal growth factor-stimulated human carcinoma A431 cells. J. Cell Biol. 1989, 109, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Mercer, J.; Helenius, A. Virus entry by macropinocytosis. Nat. Cell Biol. 2009, 11, 510–520. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, M.; Terasaka, S.; Mochizuki, Y.; Kawai, K.; Ikeda, Y.; Araki, N.; Skolnik, E.Y.; Taguchi, T.; Arai, H. Sequential breakdown of 3-phosphorylated phosphoinositides is essential for the completion of macropinocytosis. Proc. Natl. Acad. Sci. USA 2014, 111, E978–E987. [Google Scholar] [CrossRef] [PubMed]
- Ellena, J.F.; Tang, T.-X.; Shanaiah, N.; Capelluto, D.G.S. Backbone 1H, 15N, and 13C resonance assignments of the Phafin2 pleckstrin homology domain. Biomol. NMR Assign. 2022, 16, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Kelkar, N.; Standen, C.L.; Davis, R.J. Role of the JIP4 scaffold protein in the regulation of mitogen-activated protein kinase signaling pathways. Mol. Cell. Biol. 2005, 25, 2733–2743. [Google Scholar] [CrossRef]
- Ren, Y.; Wang, C.; Wang, H.; Chang, Q.; Guo, D.; Wang, X. Identification of zebrafish PLEKHF2 presents in egg/embryos as an antibacterial protein. Fish Shellfish Immunol. 2022, 127, 925–932. [Google Scholar] [CrossRef]
- Wu, C.; Jin, X.; Tsueng, G.; Afrasiabi, C.; Su, A.I. BioGPS: Building your own mash-up of gene annotations and expression profiles. Nucleic Acids Res. 2016, 44, D313–D316. [Google Scholar] [CrossRef]
- Weisz, A.; Basile, W.; Scafoglio, C.; Altucci, L.; Bresciani, F.; Facchiano, A.; Sismondi, P.; Cicatiello, L.; De Bortoli, M. Molecular identification of ERα-positive breast cancer cells by the expression profile of an intrinsic set of estrogen regulated genes. J. Cell. Physiol. 2004, 200, 440–450. [Google Scholar] [CrossRef]
- Pufall, M.A.; Graves, B.J. Autoinhibitory domains: Modular effectors of cellular regulation. Annu. Rev. Cell Dev. Biol. 2002, 18, 421–462. [Google Scholar] [CrossRef]
- Peterson, J.R.; Golemis, E.A. Autoinhibited proteins as promising drug targets. J. Cell. Biochem. 2004, 93, 68–73. [Google Scholar] [CrossRef]
- Trudeau, T.; Nassar, R.; Cumberworth, A.; Wong, E.T.C.; Woollard, G.; Gsponer, J. Structure and Intrinsic Disorder in Protein Autoinhibition. Structure 2013, 21, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.; Cunningham, K.W. A LAPF/phafin1-like protein regulates TORC1 and lysosomal membrane permeabilization in response to endoplasmic reticulum membrane stress. Mol. Biol. Cell 2015, 26, 4631–4645. [Google Scholar] [CrossRef] [PubMed]

| Proteins | Binding Partners | Functional Domains | References |
|---|---|---|---|
| Phafin1 | PtdIns(3)P | PH and FYVE domains | [15] |
| Phafin2 | PtdIns(3)P, PtdIns(4)P | PH and FYVE domains | [21] |
| Rush * | GDI | - | [80] |
| Phafin2 | EEA1 | - | [81] |
| Phafin2 | Akt | PH and FYVE domains | [72] |
| Phafin2 | Actin | PH domain | [21] |
| Phafin2 | JIP4 | PH domain | [22] |
| Phafins | Cellular Functions | References |
|---|---|---|
| Rush * | Endosomal cargo transport | [80] |
| Phafin1 | Autophagy | [15] |
| Phafin2 | Macropinocytosis | [21] |
| Apoptotic Pathways | [20] | |
| Endosomal cargo transport | [14,81] | |
| Autophagy | [72] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, T.; Hasan, M.; Capelluto, D.G.S. Phafins Are More Than Phosphoinositide-Binding Proteins. Int. J. Mol. Sci. 2023, 24, 8096. https://doi.org/10.3390/ijms24098096
Tang T, Hasan M, Capelluto DGS. Phafins Are More Than Phosphoinositide-Binding Proteins. International Journal of Molecular Sciences. 2023; 24(9):8096. https://doi.org/10.3390/ijms24098096
Chicago/Turabian StyleTang, Tuoxian, Mahmudul Hasan, and Daniel G. S. Capelluto. 2023. "Phafins Are More Than Phosphoinositide-Binding Proteins" International Journal of Molecular Sciences 24, no. 9: 8096. https://doi.org/10.3390/ijms24098096





