ROS Consumers or Producers? Interpreting Transcriptomic Data by AlphaFold Modeling Provides Insights into Class III Peroxidase Functions in Response to Biotic and Abiotic Stresses
Abstract
:1. Introduction
2. Results
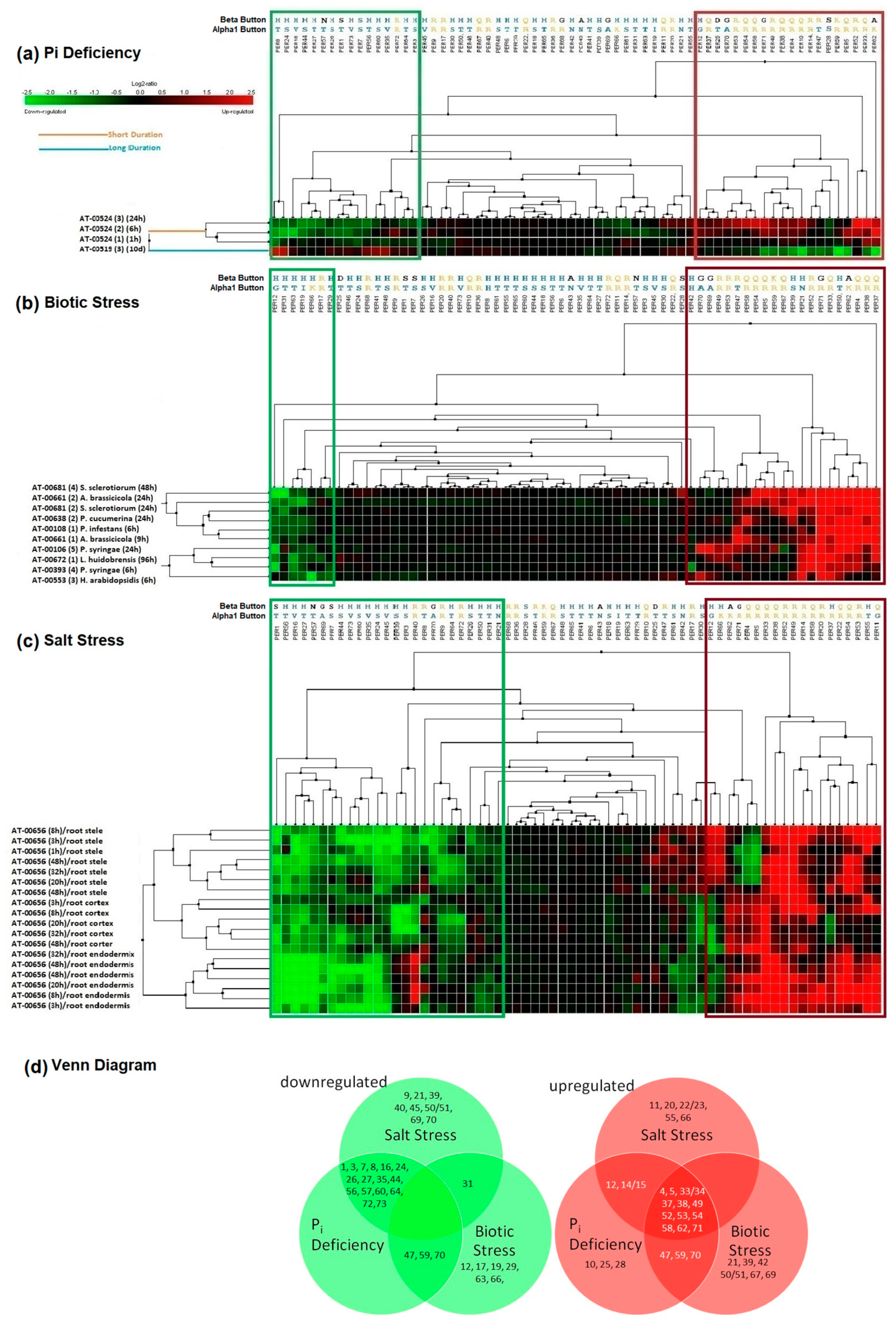
2.1. Transcriptomic Meta-Analysis Reveals Similarity of Gene Expression Patterns between Biotic and Short-Term Abiotic Stress Responses
2.2. Bioinformatic Analysis Reveals Three Key Amino Acid Positions That Determine Membership in Either of Two Groups of PERs: Those That Consume and Those That Produce H2O2/ROS
2.3. Phylogeny Reveals Correlation between Gene Expression and Evolutionary Relationship
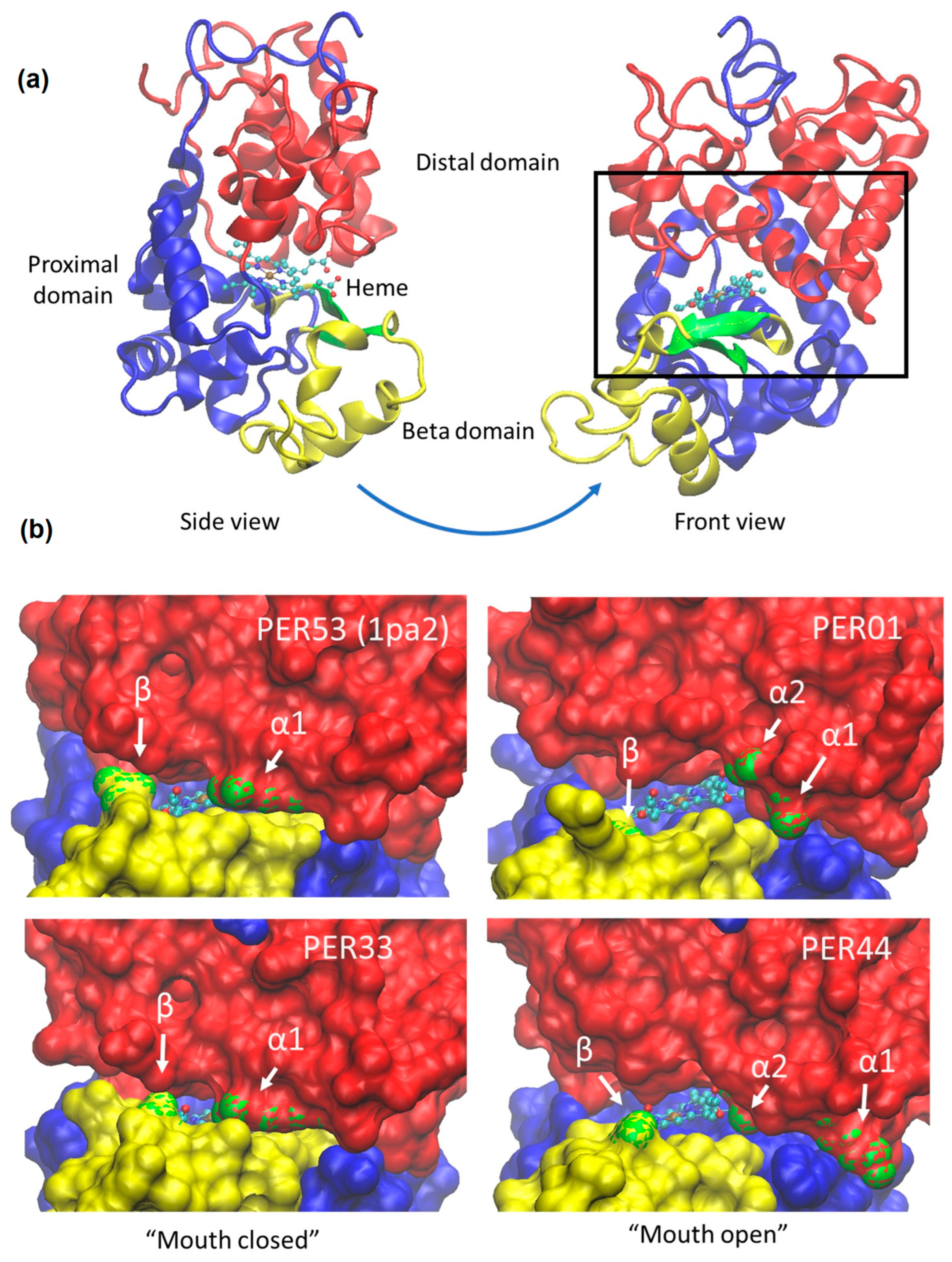
2.4. Crystal Structures and AlphaFold Modeling Reveals the Alpha and Beta Buttons as Possible Gate Keepers That Modulate Access to the Heme
3. Discussion
4. Materials and Methods
4.1. Meta-Analysis of Gene Expression
4.2. Multiple Sequence Alignment (MSA) and Site-Specific Analysis to Identify Key Amino Acids
4.3. Phylogeny
4.4. Structural Analysis of Crystal Structures and AlphaFold 3D Protein Models
- (1)
- We removed the N-terminal signal peptide regions from the downloaded models (typically 20 to 40 residues). N-terminal signal peptides are removed in the formation of the mature (secreted) protein but were nevertheless present in all the models. In all the models inspected, these peptides protruded unnaturally and were visually removed, and these identifications were additionally supported by the signal peptide program SignalP (v6.0) [46].
- (2)
- The models were pairwise structurally aligned to the crystal structure of PER53 (PDB code: 1pa2) [17], using LGA [32,33]. The alignments were obtained through “local-global” structural comparisons without a preassigned residue correspondence (option 4) and with the default distance cutoff of 5 Å. The coordinates of the heme molecule were simply copied over to the model from the 1pa2 coordinate file after the structural alignment of the model.
- (3)
- The aligned models were analyzed using the VMD program [47], which enabled us to check for clashes, identify hydrogen bonds (protein–protein and between protein and heme), as well as to visualize the mouth and heme accessibility (see Figure 4 and Supplementary Figure S4).
- (4)
- In order to make the correct correspondence of the buttons in each PER, we used the following simple motifs based on our MSA (see Supplementary Materials). These motifs are valid for the entire Arabidopsis PER family:
- Alpha1 and Alpha2: 1xxx2xxRxxfhDC, where 1 and 2 correspond to Alpha1 and Alpha2, respectively, and RxxfhDC is the highly conserved active site. Uppercase letters are 100% conserved, lowercase letters are mostly conserved, and “x” is variable.
- Beta: HtxGxxBCxxxxxR, where B corresponds to the Beta Button.
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pascual, L.S.; Segarra-Medina, C.; Gomez-Cadenas, A.; Lopez-Climent, M.F.; Vives-Peris, V.; Zandalinas, S.I. Climate change-associated multifactorial stress combination: A present challenge for our ecosystems. J. Plant Physiol. 2022, 276, 153764. [Google Scholar] [CrossRef] [PubMed]
- Kidwai, M.; Ahmad, I.Z.; Chakrabarty, D. Class III peroxidase: An indispensable enzyme for biotic/abiotic stress tolerance and a potent candidate for crop improvement. Plant Cell Rep. 2020, 39, 1381–1393. [Google Scholar] [CrossRef] [PubMed]
- Mbadinga Mbadinga, D.L.; Li, Q.; Ranocha, P.; Martinez, Y.; Dunand, C. Global analysis of non-animal peroxidases provides insights into the evolution of this gene family in the green lineage. J. Exp. Bot. 2020, 71, 3350–3360. [Google Scholar] [CrossRef]
- Passardi, F.; Cosio, C.; Penel, C.; Dunand, C. Peroxidases have more functions than a Swiss army knife. Plant Cell Rep. 2005, 24, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Cosio, C.; Dunand, C. Specific functions of individual class III peroxidase genes. J. Exp. Bot. 2009, 60, 391–408. [Google Scholar] [CrossRef]
- Shigeto, J.; Tsutsumi, Y. Diverse functions and reactions of class III peroxidases. New Phytol. 2016, 209, 1395–1402. [Google Scholar] [CrossRef]
- Noctor, G.; Reichheld, J.-P.; Foyer, C.H. ROS-related redox regulation and signaling in plants. Semin. Cell Dev. Biol. 2018, 80, 3–12. [Google Scholar] [CrossRef]
- Meng, G.; Fan, W.Y.; Rasmussen, S.K. Characterisation of the class III peroxidase gene family in carrot taproots and its role in anthocyanin and lignin accumulation. Plant Physiol. Biochem. 2021, 167, 245–256. [Google Scholar] [CrossRef]
- Passardi, F.; Penel, C.; Dunand, C. Performing the paradoxical: How plant peroxidases modify the cell wall. Trends Plant Sci. 2004, 9, 534–540. [Google Scholar] [CrossRef]
- Eljebbawi, A.; Savelli, B.; Libourel, C.; Estevez, J.M.; Dunand, C. Class III Peroxidases in Response to Multiple Abiotic Stresses in Arabidopsis thaliana Pyrenean Populations. Int. J. Mol. Sci. 2022, 23, 3960. [Google Scholar] [CrossRef]
- Almagro, L.; Gomez Ros, L.V.; Belchi-Navarro, S.; Bru, R.; Ros Barcelo, A.; Pedreno, M.A. Class III peroxidases in plant defence reactions. J. Exp. Bot. 2009, 60, 377–390. [Google Scholar] [CrossRef] [PubMed]
- Minibayeva, F.; Beckett, R.P.; Kranner, I. Roles of apoplastic peroxidases in plant response to wounding. Phytochemistry 2015, 112, 122–129. [Google Scholar] [CrossRef]
- Shin, R.; Berg, R.; Schachtman, D. Reactive oxygen species and root hairs in Arabidopsis root response to nitrogen, phosphorus and potassium deficiency. Plant Cell Physiol. 2005, 46, 1350–1357. [Google Scholar] [CrossRef]
- Duroux, L.; Welinder, K.G. The peroxidase gene family in plants: A phylogenetic overview. J. Mol. Evol. 2003, 57, 397–407. [Google Scholar] [CrossRef]
- Schuller, D.J.; Ban, N.; Huystee, R.B.; McPherson, A.; Poulos, T.L. The crystal structure of peanut peroxidase. Structure 1996, 4, 311–321. [Google Scholar] [CrossRef]
- Gajhede, M.; Schuller, D.J.; Henriksen, A.; Smith, A.T.; Poulos, T.L. Crystal structure of horseradish peroxidase C at 2.15 A resolution. Nat. Struct. Biol. 1997, 4, 1032–1038. [Google Scholar] [CrossRef]
- Ostergaard, L.; Teilum, K.; Mirza, O.; Mattsson, O.; Petersen, M.; Welinder, K.G.; Mundy, J.; Gajhede, M.; Henriksen, A. Arabidopsis ATP A2 peroxidase. Expression and high-resolution structure of a plant peroxidase with implications for lignification. Plant Mol. Biol. 2000, 44, 231–243. [Google Scholar] [CrossRef]
- Mirza, O.; Henriksen, A.; Ostergaard, L.; Welinder, K.G.; Gajhede, M. Arabidopsis thaliana peroxidase N: Structure of a novel neutral peroxidase. Acta Cryst. D Biol. Cryst. 2000, 56, 372–375. [Google Scholar] [CrossRef]
- Welinder, K.G.; Gajhede, M. Structure & Evolution of Peroxidases. In Plant Peroxidases: Biochemistry and Physiology; Welinder, K.G., Rasmussen, S.K., Penel, C., Greppin, H., Eds.; University of Geneva: Geneva, Switzerland, 1993. [Google Scholar]
- Singh, S.; Pandey, V.P.; Naaz, H.; Dwivedi, U.N. Phylogenetic analysis, molecular modeling, substrate-inhibitor specificity, and active site comparison of bacterial, fungal, and plant heme peroxidases. Biotechnol. Appl. Biochem. 2012, 59, 283–294. [Google Scholar] [CrossRef]
- Welinder, K.G.; Justesen, A.F.; Kjaersgard, I.V.; Jensen, R.B.; Rasmussen, S.K.; Jespersen, H.M.; Duroux, L. Structural diversity and transcription of class III peroxidases from Arabidopsis thaliana. Eur. J. Biochem. 2002, 269, 6063–6081. [Google Scholar] [CrossRef]
- Nielsen, K.L.; Indiani, C.; Henriksen, A.; Feis, A.; Becucci, M.; Gajhede, M.; Smulevich, G.; Welinder, K.G. Differential activity and structure of highly similar peroxidases. Spectroscopic, crystallographic, and enzymatic analyses of lignifying Arabidopsis thaliana peroxidase A2 and horseradish peroxidase A2. Biochemistry 2001, 40, 11013–11021. [Google Scholar] [CrossRef] [PubMed]
- Gajhede, M. Plant peroxidases: Substrate complexes with mechanistic implications. Biochem. Soc. Trans. 2001, 29, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Moural, T.W.; Lewis, K.M.; Barnaba, C.; Zhu, F.; Palmer, N.A.; Sarath, G.; Scully, E.D.; Jones, J.P.; Sattler, S.E.; Kang, C. Characterization of Class III Peroxidases from Switchgrass. Plant Physiol. 2017, 173, 417–433. [Google Scholar] [CrossRef]
- Douroupi, T.G.; Papassideri, I.S.; Stravopodis, D.J.; Margaritis, L.H. Molecular cloning and tissue-specific transcriptional regulation of the first peroxidase family member, Udp1, in stinging nettle (Urtica dioica). Gene 2005, 362, 57–69. [Google Scholar] [CrossRef]
- Jumper, J.; Hassabis, D. Protein structure predictions to atomic accuracy with AlphaFold. Nat. Methods 2022, 19, 11–12. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Hruz, T.; Laule, O.; Szabo, G.; Wessendorp, F.; Bleuler, S.; Oertle, L.; Widmayer, P.; Gruissem, W.; Zimmermann, P. Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinform. 2008, 2008, 420747. [Google Scholar] [CrossRef]
- Zimmermann, P.; Hirsch-Hoffmann, M.; Hennig, L.; Gruissem, W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004, 136, 2621–2632. [Google Scholar] [CrossRef]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef]
- Zemla, A. LGA: A method for finding 3D similarities in protein structures. Nucleic Acids Res. 2003, 31, 3370–3374. [Google Scholar] [CrossRef] [PubMed]
- Zemla, A.; Zhou, C.E.; Slezak, T.; Kuczmarski, T.; Rama, D.; Torres, C.; Sawicka, D.; Barsky, D. AS2TS system for protein structure modeling and analysis. Nucleic Acids Res. 2005, 33, W111–W115. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.; Moody, P.C.E.; Raven, E. Understanding the Reactivity and Interactions od Peroxidases with Substrates. In Heme Peroxidases; Raven, E., Dunford, B.H., Eds.; Royal Society of Chemistry: London, UK, 2015. [Google Scholar]
- Passardi, F.; Tognolli, M.; De Meyer, M.; Penel, C.; Dunand, C. Two cell wall associated peroxidases from Arabidopsis influence root elongation. Planta 2006, 223, 965–974. [Google Scholar] [CrossRef] [PubMed]
- Bindschedler, L.V.; Dewdney, J.; Blee, K.A.; Stone, J.M.; Asai, T.; Plotnikov, J.; Denoux, C.; Hayes, T.; Gerrish, C.; Davies, D.R.; et al. Peroxidase-dependent apoplastic oxidative burst in Arabidopsis required for pathogen resistance. Plant J. 2006, 47, 851–863. [Google Scholar] [CrossRef] [PubMed]
- Bolwell, G.P.; Bindschedler, L.V.; Blee, K.A.; Butt, V.S.; Davies, D.R.; Gardner, S.L.; Gerrish, C.; Minibayeva, F. The apoplastic oxidative burst in response to biotic stress in plants: A three-component system. J. Exp. Bot. 2002, 53, 1367–1376. [Google Scholar]
- Survila, M.; Davidsson, P.R.; Pennanen, V.; Kariola, T.; Broberg, M.; Sipari, N.; Heino, P.; Palva, E.T. Peroxidase-Generated Apoplastic ROS Impair Cuticle Integrity and Contribute to DAMP-Elicited Defenses. Front. Plant Sci. 2016, 7, 1945. [Google Scholar] [CrossRef]
- Raggi, S.; Ferrarini, A.; Delledonne, M.; Dunand, C.; Ranocha, P.; De Lorenzo, G.; Cervone, F.; Ferrari, S. The Arabidopsis Class III Peroxidase AtPRX71 Negatively Regulates Growth under Physiological Conditions and in Response to Cell Wall Damage. Plant Physiol. 2015, 169, 2513–2525. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Zdobnov, E.M.; Apweiler, R. InterProScan—An integration platform for the signature-recognition methods in InterPro. Bioinformatics 2001, 17, 847–848. [Google Scholar] [CrossRef]
- Guindon, S.; Gascuel, O. A Simple, Fast, and Accurate Algorithm to Estimate Large Phylogenies by Maximum Likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef]
- Anisimova, M.; Gascuel, O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst. Biol. 2006, 55, 539–552. [Google Scholar] [CrossRef]
- Chevenet, F.; Brun, C.; Bañuls, A.L.; Jacq, B.; Christen, R. TreeDyn: Towards dynamic graphics and annotations for analyses of trees. BMC Bioinform. 2006, 7, 439. [Google Scholar] [CrossRef]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gislason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]


| Model | RMSD (Å) | Residues Aligned |
|---|---|---|
| PER01 | 1.42 | 295 |
| PER33 | 0.85 | 304 |
| PER44 | 1.25 | 289 |
| PER53 (AlphaFold) | 0.25 | 303 |
| PER59 (PDB code: 1qgj) | 1.05 | 297 |
| Stress-Regulation | Mouth Prediction | Alpha1 Button | Alpha2 Button | Beta Button |
|---|---|---|---|---|
| Upregulated | Closed | R(12), k | S(10), g, k | Q(8), R(3), a, g |
| Downregulated | Open | S(6), T(6), V(3), r | A(10), K(3) *, p, s | H(12), s, n, r |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
New, J.; Barsky, D.; Uhde-Stone, C. ROS Consumers or Producers? Interpreting Transcriptomic Data by AlphaFold Modeling Provides Insights into Class III Peroxidase Functions in Response to Biotic and Abiotic Stresses. Int. J. Mol. Sci. 2023, 24, 8297. https://doi.org/10.3390/ijms24098297
New J, Barsky D, Uhde-Stone C. ROS Consumers or Producers? Interpreting Transcriptomic Data by AlphaFold Modeling Provides Insights into Class III Peroxidase Functions in Response to Biotic and Abiotic Stresses. International Journal of Molecular Sciences. 2023; 24(9):8297. https://doi.org/10.3390/ijms24098297
Chicago/Turabian StyleNew, James, Daniel Barsky, and Claudia Uhde-Stone. 2023. "ROS Consumers or Producers? Interpreting Transcriptomic Data by AlphaFold Modeling Provides Insights into Class III Peroxidase Functions in Response to Biotic and Abiotic Stresses" International Journal of Molecular Sciences 24, no. 9: 8297. https://doi.org/10.3390/ijms24098297





