Post-Radiotherapy Exosomal Non-Coding RNA and Hemograms for Early Death Prediction in Patients with Cervical Cancer
Abstract
:1. Introduction
2. Results
2.1. Elevated ANC Is Associated with ED in Patients with Cervical Cancer
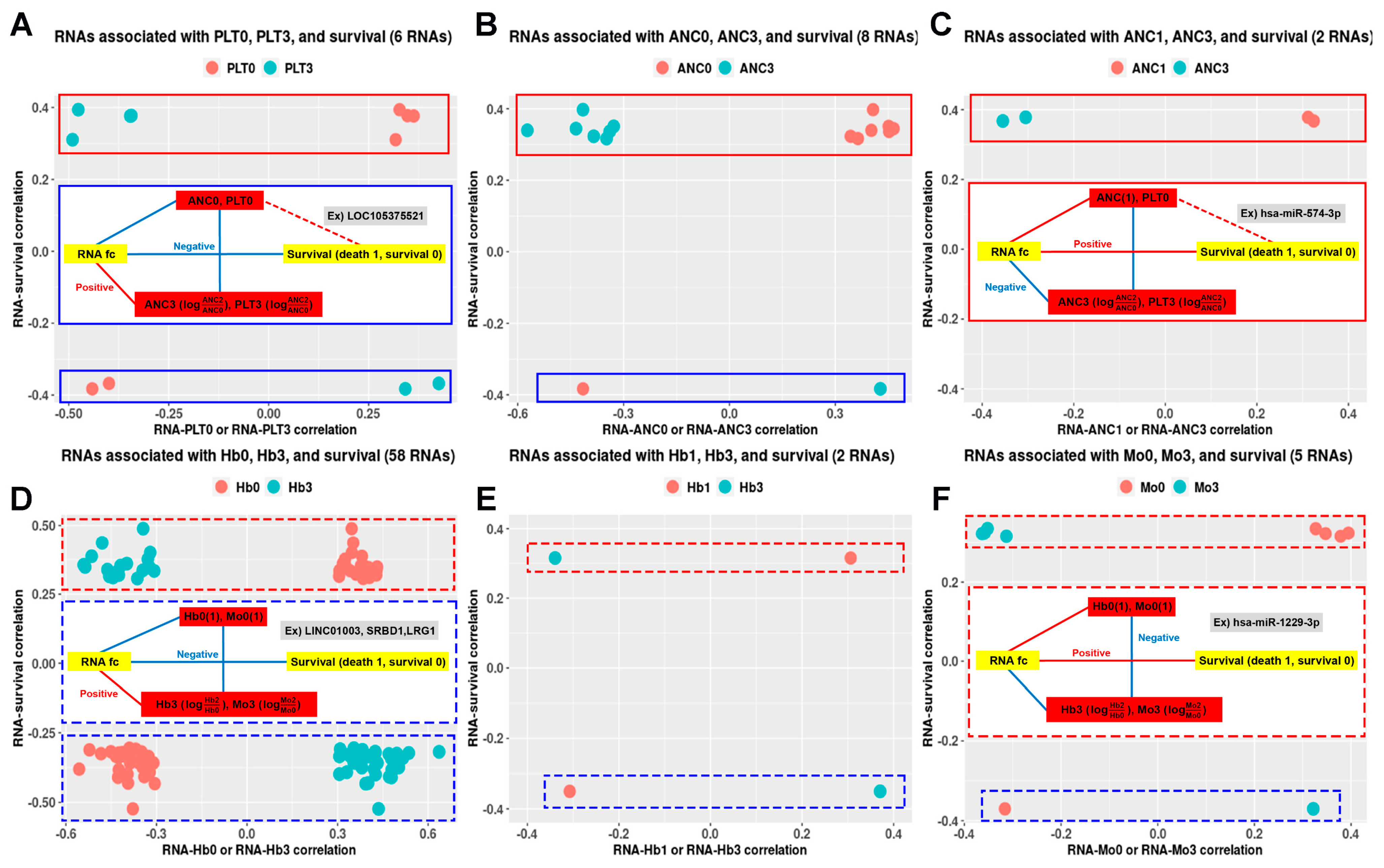
2.2. ncRNAs Associated with ED Were Selected from the CBC-Related Network
2.3. RNAs Which Interplay with CBC Dynamics Are Associated with ED
2.4. RNAs Selected within the Network Can Potentially Predict ED in Patients with Cervical Cancer
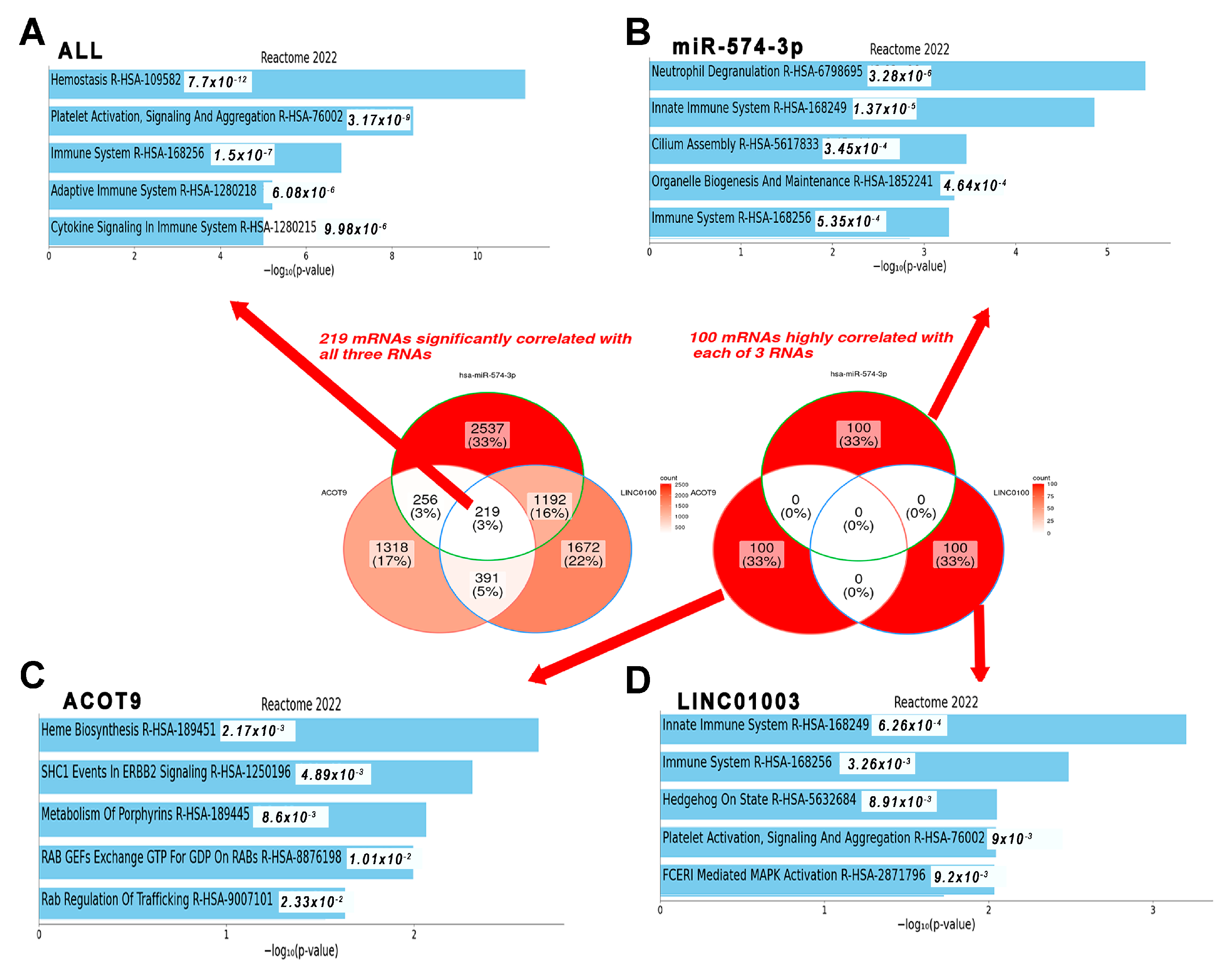
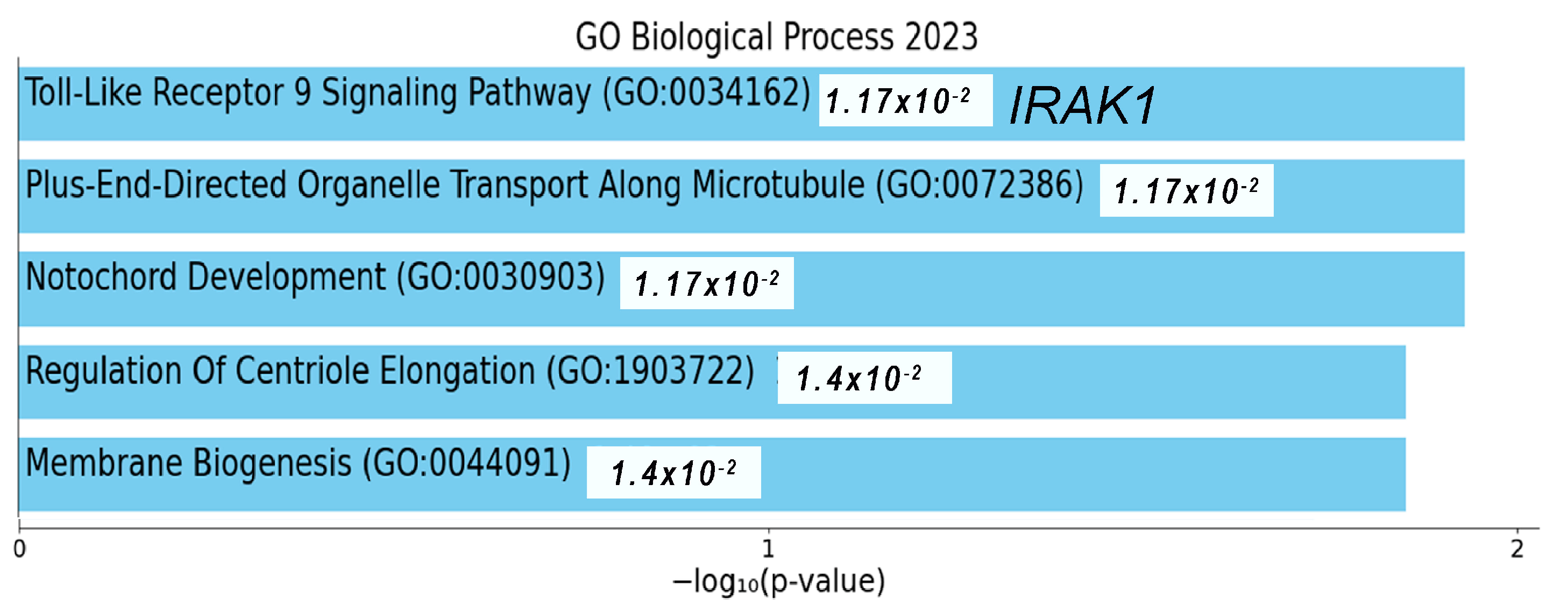
2.5. mRNAs Highly Relevant to Selected ncRNAs Are Linked to the Innate Immune System
3. Discussion
4. Materials and Methods
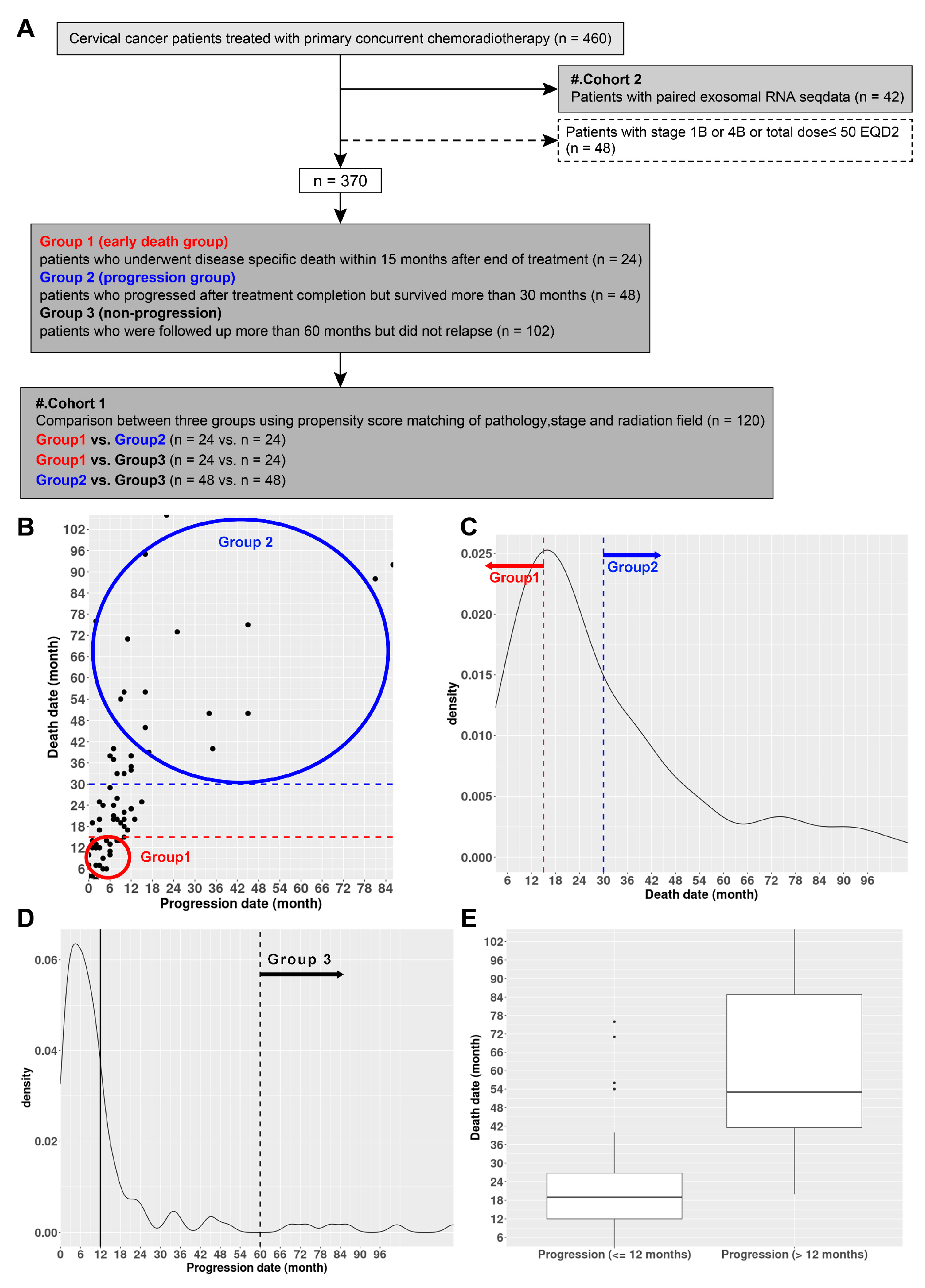
4.1. Study Participants
4.2. Blood Cell Dynamics (CBC)
4.3. Comparison of CBCs between Groups Based on the Timing of DSD
4.4. Log2FC of RNAs
4.5. Network Construction
4.6. Association between CBC Alterations and RNA Expression
4.7. Selection of RNAs as Prognostic Biomarkers of ED
4.8. Survival Analysis
4.9. Biological Function of Selected RNAs
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cho, O.; Chun, M.; Chang, S.J. Exponential Slope from Absolute Lymphocyte Counts during Radio-Chemotherapy Can Predict an Aggressive Course of Cervical Cancer. Cancers 2022, 14, 5109. [Google Scholar] [CrossRef] [PubMed]
- Stapelkamp, C.; Holmberg, L.; Tataru, D.; Møller, H.; Robinson, D. Predictors of early death in female patients with breast cancer in the UK: A cohort study. BMJ Open 2011, 1, e000247. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Xu, G.; Guo, X.; Ma, W.; Xu, Y.; Peltzer, K.; Chekhonin, V.P.; Baklaushev, V.P.; Hu, N.; Wang, X.; et al. Early Death Incidence and Prediction in Stage IV Breast Cancer. Med. Sci. Monit. 2020, 26, e924858. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Fang, Y.; Niu, Y.; Sun, Y. A predictive model for early death in elderly patients with gastric cancer: A population-based study. Front. Oncol. 2022, 12, 972639. [Google Scholar] [CrossRef] [PubMed]
- Christensen, N.L.; Kejs, A.M.T.; Jakobsen, E.; Dalton, S.O.; Rasmussen, T.R. Early death in Danish stage I lung cancer patients: A population-based case study. Acta Oncol. 2018, 57, 1561–1566. [Google Scholar] [CrossRef] [PubMed]
- Talani, C.; Mäkitie, A.; Beran, M.; Holmberg, E.; Laurell, G.; Farnebo, L. Early mortality after diagnosis of cancer of the head and neck – A population-based nationwide study. PLoS ONE 2019, 14, e0223154. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Mao, M.; Xu, G.; Lin, F.; Sun, P.; Baklaushev, V.P.; Chekhonin, V.P.; Peltzer, K.; Zhang, J.; Zhang, C. The incidence, associated factors, and predictive nomogram for early death in stage IV colorectal cancer. Int. J. Color. Dis. 2019, 34, 1189–1201. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Wang, X.; Wang, J.; Wang, S.; Fan, Y.; Fu, T.; Cao, S.; Zhang, X. Preoperative predictors of early death risk in bladder cancer patients treated with robot-assisted radical cystectomy. Cancer Med. 2019, 8, 3447–3452. [Google Scholar] [CrossRef]
- Karagiannis, G.S.; Condeelis, J.S.; Oktay, M.H. Chemotherapy-Induced Metastasis: Molecular Mechanisms, Clinical Manifestations, Therapeutic Interventions. Cancer Res. 2019, 79, 4567–4576. [Google Scholar] [CrossRef]
- Tohme, S.; Simmons, R.L.; Tsung, A. Surgery for Cancer: A Trigger for Metastases. Cancer Res. 2017, 77, 1548–1552. [Google Scholar] [CrossRef]
- Vilalta, M.; Rafat, M.; Graves, E.E. Effects of radiation on metastasis and tumor cell migration. Cell Mol. Life Sci. 2016, 73, 2999–3007. [Google Scholar] [CrossRef] [PubMed]
- Grivennikov, S.I.; Greten, F.R.; Karin, M. Immunity, inflammation, and cancer. Cell 2010, 140, 883–899. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Xie, M.; Jing, X.; Cui, J.; Wu, X.; Shu, Y. Crosstalk between Environmental Inflammatory Stimuli and Non-Coding RNA in Cancer Occurrence and Development. Cancers 2021, 13, 4436. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef] [PubMed]
- Fleshner, M.; Crane, C.R. Exosomes, DAMPs and miRNA: Features of Stress Physiology and Immune Homeostasis. Trends Immunol. 2017, 38, 768–776. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yu, D. Exosomes in cancer development, metastasis, and immunity. Biochim. Et Biophys. Acta (BBA)-Rev. Cancer 2019, 1871, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Della Bella, E.; Koch, J.; Baerenfaller, K. Translation and emerging functions of non-coding RNAs in inflammation and immunity. Allergy 2022, 77, 2025–2037. [Google Scholar] [CrossRef] [PubMed]
- Cho, O.; Kim, D.W.; Cheong, J.Y. Screening Plasma Exosomal RNAs as Diagnostic Markers for Cervical Cancer: An Analysis of Patients Who Underwent Primary Chemoradiotherapy. Biomolecules 2021, 11, 1691. [Google Scholar] [CrossRef]
- Cho, O.; Kim, D.W.; Cheong, J.Y. Plasma Exosomal miRNA Levels after Radiotherapy Are Associated with Early Progression and Metastasis of Cervical Cancer: A Pilot Study. J. Clin. Med. 2021, 10, 2110. [Google Scholar] [CrossRef]
- Pritchard, C.C.; Kroh, E.; Wood, B.; Arroyo, J.D.; Dougherty, K.J.; Miyaji, M.M.; Tait, J.F.; Tewari, M. Blood cell origin of circulating microRNAs: A cautionary note for cancer biomarker studies. Cancer Prev. Res. 2012, 5, 492–497. [Google Scholar] [CrossRef]
- Caro, J.J.; Salas, M.; Ward, A.; Goss, G. Anemia as an independent prognostic factor for survival in patients with cancer. Cancer 2001, 91, 2214–2221. [Google Scholar] [CrossRef] [PubMed]
- Nash, G.; Turner, L.; Scully, M.; Kakkar, A. Platelets and cancer. Lancet Oncol. 2002, 3, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Ocana, A.; Nieto-Jiménez, C.; Pandiella, A.; Templeton, A.J. Neutrophils in cancer: Prognostic role and therapeutic strategies. Mol. Cancer 2017, 16, 137. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-Y.; Choi, C.H.; Sung, C.O.; Do, I.-G.; Huh, S.; Song, T.; Kim, M.K.; Kim, H.-J.; Kim, T.-J.; Lee, J.-W.; et al. Prognostic value of pre-treatment circulating monocyte count in patients with cervical cancer: Comparison with SCC-Ag level. Gynecol. Oncol. 2012, 124, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Cupp, M.A.; Cariolou, M.; Tzoulaki, I.; Aune, D.; Evangelou, E.; Berlanga-Taylor, A.J. Neutrophil to lymphocyte ratio and cancer prognosis: An umbrella review of systematic reviews and meta-analyses of observational studies. BMC Med. 2020, 18, 360. [Google Scholar] [CrossRef] [PubMed]
- Templeton, A.J.; Ace, O.; McNamara, M.G.; Al-Mubarak, M.; Vera-Badillo, F.E.; Hermanns, T.; Seruga, B.; Ocaña, A.; Tannock, I.F.; Amir, E. Prognostic role of platelet to lymphocyte ratio in solid tumors: A systematic review and meta-analysis. Cancer Epidemiol. Biomark. Prev. 2014, 23, 1204–1212. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Li, H.; Chen, L.; Ma, X.; Li, X.; Gao, Y.; Zhang, Y.; Xie, Y.; Zhang, X. Prognostic role of lymphocyte to monocyte ratio for patients with cancer: Evidence from a systematic review and meta-analysis. Oncotarget 2016, 7, 31926–31942. [Google Scholar] [CrossRef]
- Herrero-Cervera, A.; Soehnlein, O.; Kenne, E. Neutrophils in chronic inflammatory diseases. Cell Mol. Immunol. 2022, 19, 177–191. [Google Scholar] [CrossRef]
- Kral, J.B.; Schrottmaier, W.C.; Salzmann, M.; Assinger, A. Platelet Interaction with Innate Immune Cells. Transfus. Med. Hemotherapy 2016, 43, 78–88. [Google Scholar] [CrossRef]
- Rolfes, V.; Ribeiro, L.S.; Hawwari, I.; Bottcher, L.; Rosero, N.; Maasewerd, S.; Santos, M.L.S.; Prochnicki, T.; Silva, C.M.S.; Wanderley, C.W.S.; et al. Platelets Fuel the Inflammasome Activation of Innate Immune Cells. Cell Rep. 2020, 31, 107615. [Google Scholar] [CrossRef]
- Rosales, C. Neutrophils at the crossroads of innate and adaptive immunity. J. Leucoc. Biol. 2020, 108, 377–396. [Google Scholar] [CrossRef] [PubMed]
- Mollinedo, F. Neutrophil Degranulation, Plasticity, and Cancer Metastasis. Trends Immunol. 2019, 40, 228–242. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, K.A.; Kagan, J.C. Toll-like Receptors and the Control of Immunity. Cell 2020, 180, 1044–1066. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Wang, R.R.; Miao, N.J.; Tang, J.J.; Zhang, Y.W.; Lu, X.R.; Yan, P.Y.; Wang, J.; Jia, X.M. PD-L1 negatively regulates antifungal immunity by inhibiting neutrophil release from bone marrow. Nat. Commun. 2022, 13, 6857. [Google Scholar] [CrossRef] [PubMed]
- Furman, D.; Campisi, J.; Verdin, E.; Carrera-Bastos, P.; Targ, S.; Franceschi, C.; Ferrucci, L.; Gilroy, D.W.; Fasano, A.; Miller, G.W.; et al. Chronic inflammation in the etiology of disease across the life span. Nat. Med. 2019, 25, 1822–1832. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Li, N.; Lin, Y.; Gupta, C.; Jiang, F. Circulating Neutrophil MicroRNAs as Biomarkers for the Detection of Lung Cancer. Biomark. Cancer 2016, 8, BIC-S37333. [Google Scholar] [CrossRef]
- Uematsu, S.; Sato, S.; Yamamoto, M.; Hirotani, T.; Kato, H.; Takeshita, F.; Matsuda, M.; Coban, C.; Ishii, K.J.; Kawai, T.; et al. Interleukin-1 receptor-associated kinase-1 plays an essential role for Toll-like receptor (TLR)7- and TLR9-mediated interferon-alpha induction. J. Exp. Med. 2005, 201, 915–923. [Google Scholar] [CrossRef]
- Lam, L.K.M.; Murphy, S.; Kokkinaki, D.; Venosa, A.; Sherrill-Mix, S.; Casu, C.; Rivella, S.; Weiner, A.; Park, J.; Shin, S.; et al. DNA binding to TLR9 expressed by red blood cells promotes innate immune activation and anemia. Sci. Transl Med. 2021, 13, eabj1008. [Google Scholar] [CrossRef]
- Anderson, H.L.; Brodsky, I.E.; Mangalmurti, N.S. The Evolving Erythrocyte: Red Blood Cells as Modulators of Innate Immunity. J. Immunol. 2018, 201, 1343–1351. [Google Scholar] [CrossRef]
- Kopinski, P.K.; Singh, L.N.; Zhang, S.; Lott, M.T.; Wallace, D.C. Mitochondrial DNA variation and cancer. Nat. Rev. Cancer 2021, 21, 431–445. [Google Scholar] [CrossRef]
- Kang, T.H.; Mao, C.P.; Kim, Y.S.; Kim, T.W.; Yang, A.; Lam, B.; Tseng, S.H.; Farmer, E.; Park, Y.M.; Hung, C.F. TLR9 acts as a sensor for tumor-released DNA to modulate anti-tumor immunity after chemotherapy. J. Immunother. Cancer 2019, 7, 260. [Google Scholar] [CrossRef] [PubMed]
- Guan, X.; Sun, Y.; Zhang, C. LncRNAs in blood cells: Roles in cell development and potential pathogenesis in hematological malignancies. Crit. Rev. Oncol. Hematol. 2022, 180, 103849. [Google Scholar] [CrossRef] [PubMed]
- Danesh, A.; Inglis, H.C.; Jackman, R.P.; Wu, S.; Deng, X.; Muench, M.O.; Heitman, J.W.; Norris, P.J. Exosomes from red blood cell units bind to monocytes and induce proinflammatory cytokines, boosting T-cell responses in vitro. Blood 2014, 123, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Buzas, E.I. The roles of extracellular vesicles in the immune system. Nat. Rev. Immunol. 2023, 23, 236–250. [Google Scholar] [CrossRef]
- Steensels, S.; Qiao, J.; Zhang, Y.; Maner-Smith, K.M.; Kika, N.; Holman, C.D.; Corey, K.E.; Bracken, W.C.; Ortlund, E.A.; Ersoy, B.A. Acyl-Coenzyme A Thioesterase 9 Traffics Mitochondrial Short-Chain Fatty Acids Toward De Novo Lipogenesis and Glucose Production in the Liver. Hepatology 2020, 72, 857–872. [Google Scholar] [CrossRef]
- Pernes, G.; Flynn, M.C.; Lancaster, G.I.; Murphy, A.J. Fat for fuel: Lipid metabolism in haematopoiesis. Clin. Transl. Immunol. 2019, 8, e1098. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef]



| PSM | Time | ANC | PLT | Hb | ALC | Mo | NLR | PLR | LMR |
|---|---|---|---|---|---|---|---|---|---|
| Group 1 | CBC0 | ●(↑) (↓) | ●(↑) (↓) | ●(↑) (↓) | |||||
| versus | Min CBC | ●(↑) (↓) | |||||||
| Group 2 | CBC1 | ●(↑) (↓) | ●(↑) (↓) | ||||||
| Group 1 | CBC0 | ▲(↓) (↑) | ●(↓) (↑) | ●(↑) (↓) | ●(↑) (↓) | ||||
| versus | Min CBC | ●(↑) (↓) | ●(↑) (↓) | ||||||
| Group 3 | CBC1 | ●(↑) (↓) | ●(↑) (↓) | ●(↑) (↓) | ●(↑) (↓) | ||||
| Group 2 | CBC0 | ●(↓) (↑) | ▲(↓) (↑) | ●(↓) (↑) | |||||
| versus | Min CBC | ▲(↓) (↑) | ▲(↓) (↑) | ||||||
| Group 3 | CBC1 | ●(↓) (↑) | ●(↓) (↑) | ●(↓) (↑) | ●(↑) (↓) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cho, O. Post-Radiotherapy Exosomal Non-Coding RNA and Hemograms for Early Death Prediction in Patients with Cervical Cancer. Int. J. Mol. Sci. 2024, 25, 126. https://doi.org/10.3390/ijms25010126
Cho O. Post-Radiotherapy Exosomal Non-Coding RNA and Hemograms for Early Death Prediction in Patients with Cervical Cancer. International Journal of Molecular Sciences. 2024; 25(1):126. https://doi.org/10.3390/ijms25010126
Chicago/Turabian StyleCho, Oyeon. 2024. "Post-Radiotherapy Exosomal Non-Coding RNA and Hemograms for Early Death Prediction in Patients with Cervical Cancer" International Journal of Molecular Sciences 25, no. 1: 126. https://doi.org/10.3390/ijms25010126





