Circular RNAs: Promising Treatment Targets and Biomarkers of Ischemic Stroke
Abstract
:1. Introduction
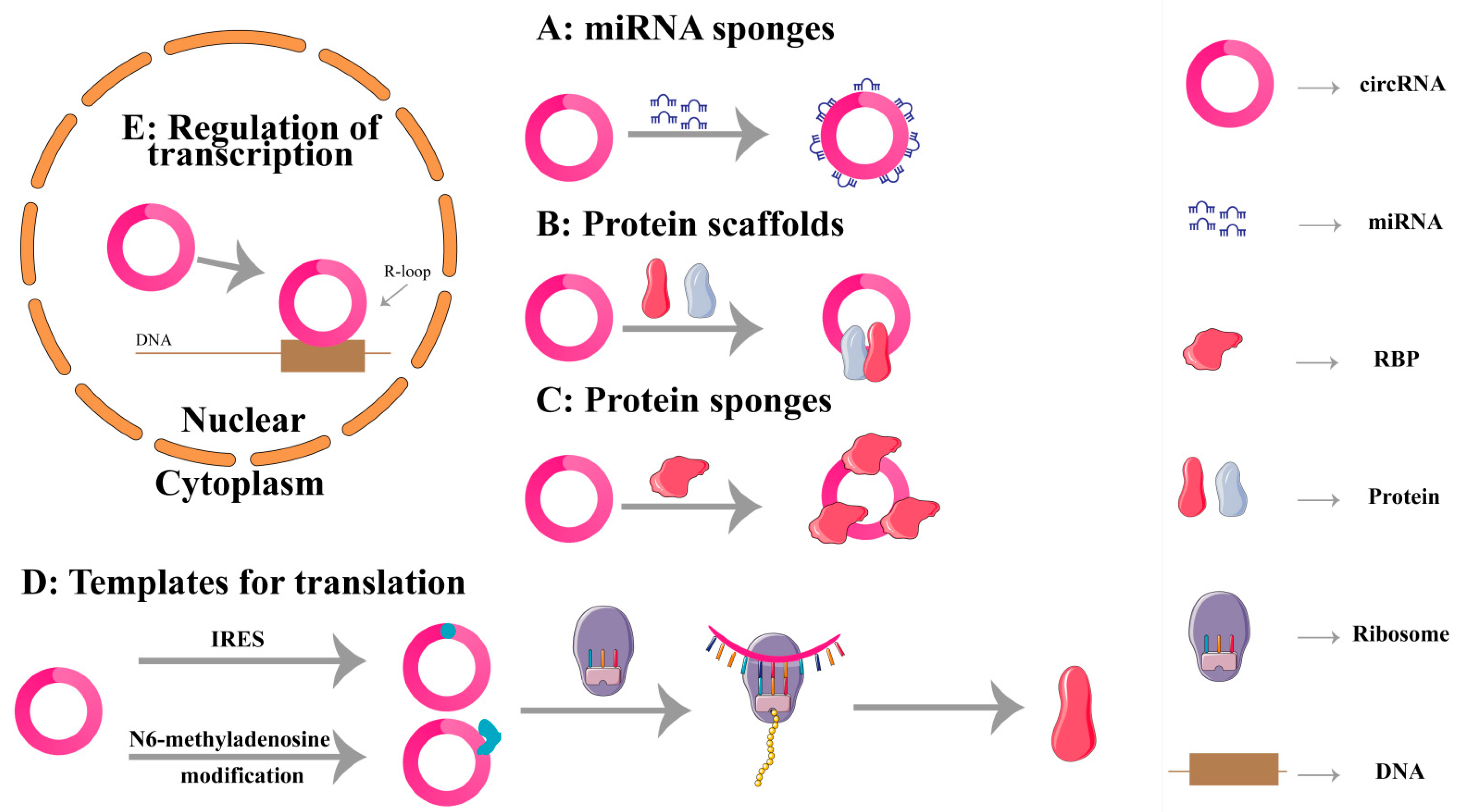
2. Biological Functions of circRNAs
2.1. miRNA Sponges
2.2. Protein Scaffolds
2.3. Protein Sponges
2.4. Translation
2.5. Regulating Transcription
3. CircRNAs and Stroke
3.1. Apoptosis
3.1.1. CircFOXP1
3.1.2. CircCDC14A
3.1.3. CircOGDH
3.1.4. CircPRDX3
3.1.5. Circ_0000566
3.1.6. Circ_0000647
3.1.7. CircFUNDC1
3.1.8. CircTLK1
3.1.9. CircUSP36
3.1.10. CircHIPK3
3.1.11. Circ_0010729
3.1.12. CircPUM1
3.1.13. CircHECTD1
3.1.14. CircPHC3
3.1.15. CircTTC3
3.1.16. CircCCDC9
3.1.17. CircUCK2
3.1.18. Circ_016719
3.1.19. CircLIFR
3.1.20. Circ_0025984
3.1.21. CDR1as
3.2. Autophagy
3.2.1. Circ-FoxO3
3.2.2. CircSHOC2
3.3. Inflammation
3.3.1. Circ_0000831
3.3.2. CircDLGAP4
3.3.3. CircRps5
3.4. Oxidative Stress
3.4.1. Circ-Memo1
3.4.2. CircDLGAP4
3.4.3. CircCTNNB1
3.5. Angiogenesis
3.5.1. CircPDS5B
3.5.2. CircHECTD1
3.5.3. CircFUNDC1
3.5.4. Circ_0006768
3.5.5. CircPHKA2
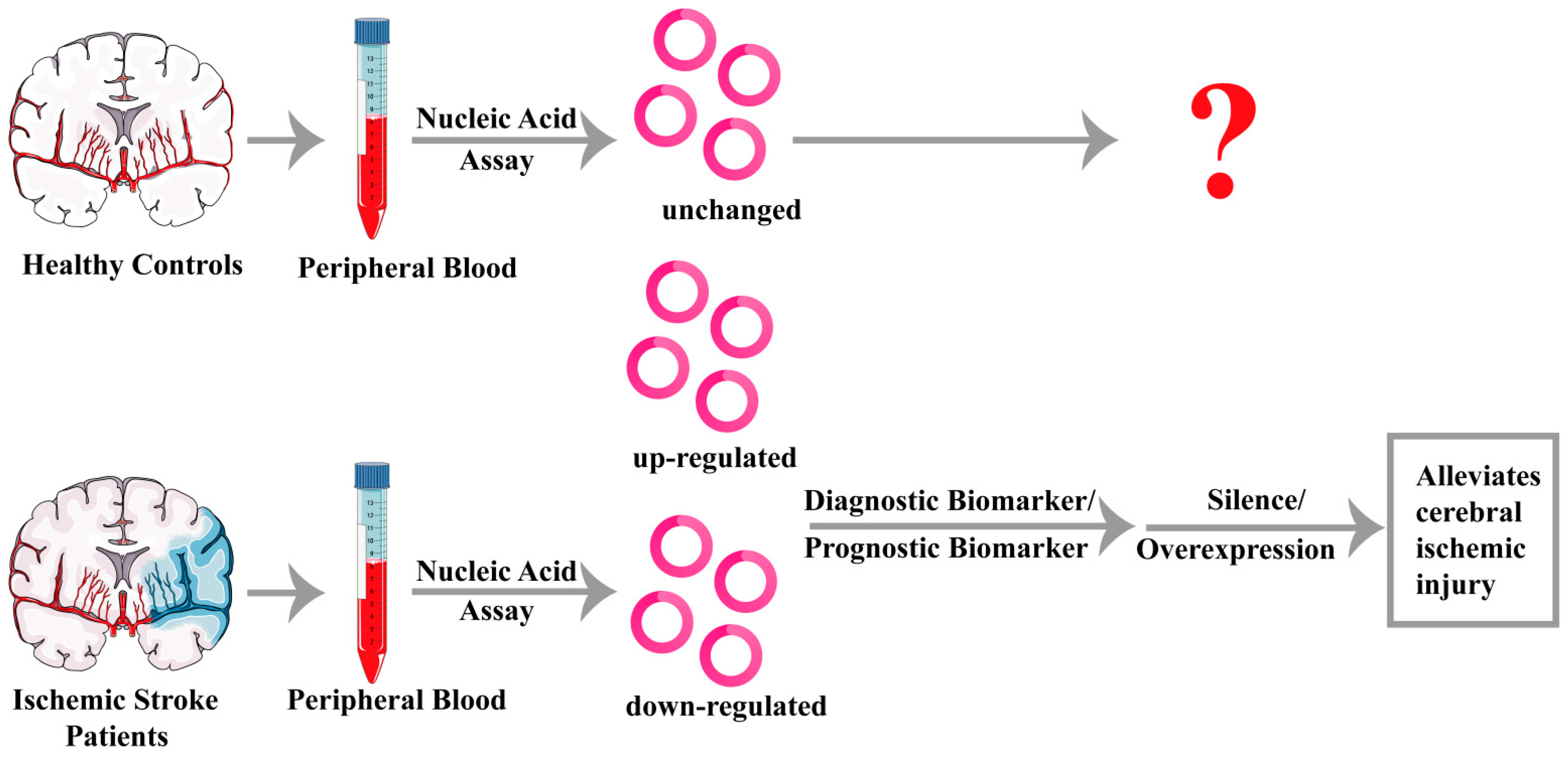
4. Potential Clinical Applications of circRNAs in Ischemic Stroke
4.1. CircRNAs Act as Biomarkers for Ischemic Stroke
4.2. CircRNAs Act as Treatment Targets for Ischemic Stroke
5. Conclusions and Perspective
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids Are Single-Stranded Covalently Closed Circular RNA Molecules Existing as Highly Base-Paired Rod-Like Structures. Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856. [Google Scholar] [CrossRef]
- Hsu, M.T.; Coca-Prados, M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 1979, 280, 339–340. [Google Scholar] [CrossRef]
- Ju, J.; Song, Y.N.; Chen, X.Z.; Wang, T.; Liu, C.Y.; Wang, K. circRNA is a potential target for cardiovascular diseases treatment. Mol. Cell Biochem. 2022, 477, 417–430. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, J.; Ma, J.; Sun, T.; Zhou, Q.; Wang, W.; Wang, G.; Wu, P.; Wang, H.; Jiang, L.; et al. Exosomal circRNAs: Biogenesis, effect and application in human diseases. Mol. Cancer 2019, 18, 116. [Google Scholar] [CrossRef]
- Fan, C.; Li, Y.; Lan, T.; Wang, W.; Long, Y.; Yu, S.Y. Microglia secrete miR-146a-5p-containing exosomes to regulate neurogenesis in depression. Mol. Ther. 2022, 30, 1300–1314. [Google Scholar] [CrossRef]
- Tuo, B.; Chen, Z.; Dang, Q.; Chen, C.; Zhang, H.; Hu, S.; Sun, Z. Roles of exosomal circRNAs in tumour immunity and cancer progression. Cell Death Dis. 2022, 13, 539. [Google Scholar] [CrossRef]
- Sun, P.; Hamblin, M.H.; Yin, K.J. Non-coding RNAs in the regulation of blood-brain barrier functions in central nervous system disorders. Fluids Barriers CNS 2022, 19, 27. [Google Scholar] [CrossRef]
- Tu, W.J.; Zhao, Z.; Yin, P.; Cao, L.; Zeng, J.; Chen, H.; Fan, D.; Fang, Q.; Gao, P.; Gu, Y.; et al. Estimated Burden of Stroke in China in 2020. JAMA Netw. Open 2023, 6, e231455. [Google Scholar] [CrossRef]
- Hankey, G.J. Stroke. Lancet 2017, 389, 641–654. [Google Scholar] [CrossRef]
- Panda, A.C. Circular RNAs Act as miRNA Sponges. Adv. Exp. Med. Biol. 2018, 1087, 67–79. [Google Scholar] [CrossRef]
- Okholm, T.L.H.; Sathe, S.; Park, S.S.; Kamstrup, A.B.; Rasmussen, A.M.; Shankar, A.; Chua, Z.M.; Fristrup, N.; Nielsen, M.M.; Vang, S.; et al. Transcriptome-wide profiles of circular RNA and RNA-binding protein interactions reveal effects on circular RNA biogenesis and cancer pathway expression. Genome Med. 2020, 12, 112. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Yang, Y.; Shen, P.; Ma, J.; Fang, B.; Wang, Q.; Wang, K.; Shi, P.; Fan, S.; Fang, X. circPDE4B prevents articular cartilage degeneration and promotes repair by acting as a scaffold for RIC8A and MID1. Ann. Rheum. Dis. 2021, 80, 1209–1219. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, C.; Du, Y.; Li, Z.; Li, M.; Hou, P.; Shen, Z.; Chu, S.; Zheng, J.; Bai, J. Expanding uncapped translation and emerging function of circular RNA in carcinomas and noncarcinomas. Mol. Cancer 2022, 21, 13. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Qi, M.; Fei, X.; Wang, X.; Wang, K. Hsa_circRNA_0088036 acts as a ceRNA to promote bladder cancer progression by sponging miR-140-3p. Cell Death Dis. 2022, 13, 322. [Google Scholar] [CrossRef] [PubMed]
- Fu, B.; Liu, W.; Zhu, C.; Li, P.; Wang, L.; Pan, L.; Li, K.; Cai, P.; Meng, M.; Wang, Y.; et al. Circular RNA circBCBM1 promotes breast cancer brain metastasis by modulating miR-125a/BRD4 axis. Int. J. Biol. Sci. 2021, 17, 3104–3117. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, T.; Li, C.; Li, W.; Zhou, X.; Li, Y.; Luo, D.; Zhang, N.; Chen, B.; Wang, L.; et al. CircRNA-CREIT inhibits stress granule assembly and overcomes doxorubicin resistance in TNBC by destabilizing PKR. J. Hematol. Oncol. 2022, 15, 122. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Du, W.W.; Awan, F.M.; Dong, J.; Yang, B.B. The circular RNA circ-Ccnb1 dissociates Ccnb1/Cdk1 complex suppressing cell invasion and tumorigenesis. Cancer Lett. 2019, 459, 216–226. [Google Scholar] [CrossRef]
- Ji, H.; Kim, T.W.; Lee, W.J.; Jeong, S.D.; Cho, Y.B.; Kim, H.H. Two circPPFIA1s negatively regulate liver metastasis of colon cancer via miR-155-5p/CDX1 and HuR/RAB36. Mol. Cancer 2022, 21, 197. [Google Scholar] [CrossRef]
- Qiu, S.; Li, B.; Xia, Y.; Xuan, Z.; Li, Z.; Xie, L.; Gu, C.; Lv, J.; Lu, C.; Jiang, T.; et al. CircTHBS1 drives gastric cancer progression by increasing INHBA mRNA expression and stability in a ceRNA- and RBP-dependent manner. Cell Death Dis. 2022, 13, 266. [Google Scholar] [CrossRef]
- Wen, S.Y.; Qadir, J.; Yang, B.B. Circular RNA translation: Novel protein isoforms and clinical significance. Trends Mol. Med. 2022, 28, 405–420. [Google Scholar] [CrossRef] [PubMed]
- Pamudurti, N.R.; Bartok, O.; Jens, M.; Ashwal-Fluss, R.; Stottmeister, C.; Ruhe, L.; Hanan, M.; Wyler, E.; Perez-Hernandez, D.; Ramberger, E.; et al. Translation of CircRNAs. Mol. Cell 2017, 66, 9–21.e27. [Google Scholar] [CrossRef] [PubMed]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell 2017, 66, 22–37.e29. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.L.; Wang, Y.; et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017, 27, 626–641. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.L.; Chen, W.; Xie, J.J.; Zhang, M.L.; Nie, R.C.; Liang, H.; Mei, J.; Han, K.; Xiang, Z.C.; Wang, F.W.; et al. A novel peptide encoded by N6-methyladenosine modified circMAP3K4 prevents apoptosis in hepatocellular carcinoma. Mol. Cancer 2022, 21, 93. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 2020, 21, 475–490. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhang, J.; Tian, Y.; Gao, Y.; Dong, X.; Chen, W.; Yuan, X.; Yin, W.; Xu, J.; Chen, K.; et al. CircRNA inhibits DNA damage repair by interacting with host gene. Mol. Cancer 2020, 19, 128. [Google Scholar] [CrossRef]
- Hollensen, A.K.; Thomsen, H.S.; Lloret-Llinares, M.; Kamstrup, A.B.; Jensen, J.M.; Luckmann, M.; Birkmose, N.; Palmfeldt, J.; Jensen, T.H.; Hansen, T.B.; et al. circZNF827 nucleates a transcription inhibitory complex to balance neuronal differentiation. Elife 2020, 9, e58478. [Google Scholar] [CrossRef]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef]
- Yang, J.; He, W.; Gu, L.; Zhu, L.; Liang, T.; Liang, X.; Zhong, Q.; Zhang, R.; Nan, A.; Su, L. CircFOXP1 alleviates brain injury after acute ischemic stroke by regulating STAT3/apoptotic signaling. Transl. Res. 2023, 257, 15–29. [Google Scholar] [CrossRef]
- Huo, H.; Hu, C.; Lu, Y.; Zhou, J.; Mai, Z. Silencing of circCDC14A prevents cerebral ischemia-reperfusion injury via miR-23a-3p/CXCL12 axis. J. Biochem. Mol. Toxicol. 2022, 36, e22982. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, Y.; Zang, J.; Zhang, T.; Li, Y.; Tan, Z.; Ma, D.; Zhang, T.; Wang, S.; Zhang, Y.; et al. CircOGDH Is a Penumbra Biomarker and Therapeutic Target in Acute Ischemic Stroke. Circ. Res. 2022, 130, 907–924. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zhang, Y.; Yin, M.; Cheng, Z.; Li, D.; Luo, X.; Liu, X.; Tu, J. Circular RNA circPRDX3 mediates neuronal survival apoptosis in ischemic stroke by targeting miR-641 and NPR3. Brain. Res. 2022, 1797, 148114. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Xiao, H.; Liu, J.; Zhang, Y.; Li, J.; Zhang, T.; Chen, H. Circ_0000566 contributes oxygen-glucose deprivation and reoxygenation (OGD/R)-induced human brain microvascular endothelial cell injury via regulating miR-18a-5p/ACVR2B axis. Metab. Brain. Dis. 2023, 38, 1273–1284. [Google Scholar] [CrossRef]
- Dai, Y.; Sheng, Y.; Deng, Y.; Wang, H.; Zhao, Z.; Yu, X.; Xu, T. Circ_0000647 promotes cell injury by modulating miR-126-5p/TRAF3 axis in oxygen-glucose deprivation and reperfusion-induced SK-N-SH cell model. Int. Immunopharmacol. 2022, 104, 108464. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Liu, J.; Wang, D.; Yao, Y.; Jiao, X. Knockdown of circ_0007290 alleviates oxygen-glucose deprivation-induced neuronal injury by regulating miR-496/PDCD4 axis. Metab. Brain. Dis. 2022, 37, 807–818. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Yun, Q.; Zhang, J.; Wang, Z.; Zhang, X.; Bao, J. Knockdown of circular RNA tousled-like kinase 1 relieves ischemic stroke in middle cerebral artery occlusion mice and oxygen-glucose deprivation and reoxygenation-induced N2a cell damage. Bioengineered 2022, 13, 3434–3449. [Google Scholar] [CrossRef]
- Yang, J.; He, W.; Gu, L.; Long, J.; Zhu, L.; Zhang, R.; Zhao, Z.; Xu, B.; Nan, A.; Su, L. CircUSP36 attenuates ischemic stroke injury through the miR-139-3p/SMAD3/Bcl2 signal axis. Clin. Sci. 2022, 136, 953–971. [Google Scholar] [CrossRef]
- Chen, G.; Shan, X.; Li, L.; Dong, L.; Huang, G.; Tao, H. circHIPK3 regulates apoptosis and mitochondrial dysfunction induced by ischemic stroke in mice by sponging miR-148b-3p via CDK5R1/SIRT1. Exp. Neurol. 2022, 355, 114115. [Google Scholar] [CrossRef]
- Ouyang, X.; Shi, G.; Wang, S.; Chen, L.; Xu, J.; Xie, D. Hsa_circ_0010729 is Involved in Oxygen-Glucose Deprivation/Reoxygenation-Induced Human Microvascular Endothelial Cell Deprivation by Targeting miR-665/ING5. Biochem. Genet. 2022, 60, 2455–2470. [Google Scholar] [CrossRef]
- Hu, T.; Li, D.; Fan, T.; Zhao, X.; Chen, Z. Circular RNA PUM1 performs as a competing endogenous RNA of microRNA-340-5p to mediate DEAD-box helicase 5 to mitigate cerebral ischemia-reperfusion injury. Bioengineered 2022, 13, 11564–11578. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; He, J.; Wang, B. Circular RNA circ_HECTD1 regulates cell injury after cerebral infarction by miR-27a-3p/FSTL1 axis. Cell Cycle 2021, 20, 914–926. [Google Scholar] [CrossRef] [PubMed]
- Dai, Q.; Ma, Y.; Xu, Z.; Zhang, L.; Yang, H.; Liu, Q.; Wang, J. Downregulation of circular RNA HECTD1 induces neuroprotection against ischemic stroke through the microRNA-133b/TRAF3 pathway. Life Sci. 2021, 264, 118626. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Wu, Z.; Qiu, H.; Wu, J. Circular RNA circPHC3 Promotes Cell Death and Apoptosis in Human BMECs After Oxygen Glucose Deprivation via miR-455-5p/TRAF3 Axis in vitro. Neuropsychiatr. Dis. Treat. 2021, 17, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Zang, L.; Cui, J.; Wei, L. Circular RNA TTC3 regulates cerebral ischemia-reperfusion injury and neural stem cells by miR-372-3p/TLR4 axis in cerebral infarction. Stem. Cell Res. Ther. 2021, 12, 125. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Xu, H.; Zhang, W.; Chen, Z.; Li, W.; Ke, W. Circular RNA circCCDC9 alleviates ischaemic stroke ischaemia/reperfusion injury via the Notch pathway. J. Cell Mol. Med. 2020, 24, 14152–14159. [Google Scholar] [CrossRef]
- Chen, W.; Wang, H.; Feng, J.; Chen, L. Overexpression of circRNA circUCK2 Attenuates Cell Apoptosis in Cerebral Ischemia-Reperfusion Injury via miR-125b-5p/GDF11 Signaling. Mol. Ther. Nucleic. Acids 2020, 22, 673–683. [Google Scholar] [CrossRef]
- Tang, C.; Ou, J.; Kou, L.; Deng, J.; Luo, S. Circ_016719 plays a critical role in neuron cell apoptosis induced by I/R via targeting miR-29c/Map2k6. Mol. Cell Probes 2020, 49, 101478. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, J.; Li, J.; Xu, L.; Lian, W. The decreased circular RNA hsa_circ_0072309 promotes cell apoptosis of ischemic stroke by sponging miR-100. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 4420–4429. [Google Scholar] [CrossRef]
- Zhou, D.; Huang, Z.; Zhu, X.; Hong, T.; Zhao, Y. Circular RNA 0025984 Ameliorates Ischemic Stroke Injury and Protects Astrocytes Through miR-143-3p/TET1/ORP150 Pathway. Mol. Neurobiol. 2021, 58, 5937–5953. [Google Scholar] [CrossRef]
- Mehta, S.L.; Chokkalla, A.K.; Bathula, S.; Arruri, V.; Chelluboina, B.; Vemuganti, R. CDR1as regulates alpha-synuclein-mediated ischemic brain damage by controlling miR-7 availability. Mol. Ther. Nucleic Acids 2023, 31, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Huang, C.; Wen, X.; Liu, W.; Huang, X.; Li, Y.; Zang, J.; Weng, Z.; Lu, D.; Tsang, C.K.; et al. Circular RNA circ-FoxO3 attenuates blood-brain barrier damage by inducing autophagy during ischemia/reperfusion. Mol. Ther. 2022, 30, 1275–1287. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Wang, H.; Zhu, Z.; Feng, J.; Chen, L. Exosome-Shuttled circSHOC2 from IPASs Regulates Neuronal Autophagy and Ameliorates Ischemic Brain Injury via the miR-7670-3p/SIRT1 Axis. Mol. Ther. Nucleic Acids 2020, 22, 657–672. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Zhang, W.; Li, W.; Gao, Y.; Zheng, D.; Bi, G. Overexpressing circ_0000831 is sufficient to inhibit neuroinflammation and vertigo in cerebral ischemia through a miR-16-5p-dependent mechanism. Exp. Neurol. 2022, 353, 114047. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.; He, J.; Chen, H.; Xu, X.; Tao, Y. CircDLGAP4 overexpression relieves oxygen-glucose deprivation-induced neuronal injury by elevating NEGR1 through sponging miR-503-3p. J. Mol. Histol. 2022, 53, 321–332. [Google Scholar] [CrossRef]
- Yang, H.; Tu, Z.; Yang, D.; Hu, M.; Zhou, L.; Li, Q.; Yu, B.; Hou, S. Exosomes from hypoxic pre-treated ADSCs attenuate acute ischemic stroke-induced brain injury via delivery of circ-Rps5 and promote M2 microglia/macrophage polarization. Neurosci. Lett. 2022, 769, 136389. [Google Scholar] [CrossRef]
- Ren, X.; Jing, Y.X.; Zhou, Z.W.; Yang, J.W. Knockdown of circRNA-Memo1 Reduces Hypoxia/Reoxygenation Injury in Human Brain Endothelial Cells Through miRNA-17-5p/SOS1 Axis. Mol. Neurobiol. 2022, 59, 2085–2097. [Google Scholar] [CrossRef]
- Liu, H.; Zheng, W.; Song, Z. circDlgap4 Alleviates Cerebral Ischaemic Injury by Binding to AUF1 to Suppress Oxidative Stress and Neuroinflammation. Mol. Neurobiol. 2022, 59, 3218–3232. [Google Scholar] [CrossRef]
- Chen, C.; Chang, X.; Zhang, S.; Zhao, Q.; Lei, C. CircRNA CTNNB1 (circCTNNB1) ameliorates cerebral ischemia/reperfusion injury by sponging miR-96-5p to up-regulate scavenger receptor class B type 1 (SRB1) expression. Bioengineered 2022, 13, 10258–10273. [Google Scholar] [CrossRef]
- Jiang, Z.; Jiang, Y. Circular RNA CircPDS5B impairs angiogenesis following ischemic stroke through its interaction with hnRNPL to inactivate VEGF-A. Neurobiol. Dis. 2023, 181, 106080. [Google Scholar] [CrossRef]
- He, G.H.; Wang, Z.; Xu, W.; Song, K.P.; Xiao, H. Knockdown of circHECTD1 inhibits oxygen-glucose deprivation and reperfusion induced endothelial-mesenchymal transition. Metab. Brain. Dis. 2022, 37, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Liu, X.; Wu, H.; Feng, J.; Chen, H.; Zhou, D. CircFUNDC1 knockdown alleviates oxygen-glucose deprivation-induced human brain microvascular endothelial cell injuries by inhibiting PTEN via miR-375. Neurosci. Lett. 2022, 770, 136381. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, J.; Wang, Z. Circ_0006768 upregulation attenuates oxygen-glucose deprivation/reoxygenation-induced human brain microvascular endothelial cell injuries by upregulating VEZF1 via miR-222-3p inhibition. Metab. Brain. Dis. 2021, 36, 2521–2534. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Li, X.; Zhong, C.; Peng, J.; Pang, J.; Peng, T.; Wan, W.; Li, X. Circular RNA circPHKA2 Relieves OGD-Induced Human Brain Microvascular Endothelial Cell Injuries through Competitively Binding miR-574-5p to Modulate SOD2. Oxid. Med. Cell Longev. 2021, 2021, 3823122. [Google Scholar] [CrossRef] [PubMed]
- Tuo, Q.Z.; Zhang, S.T.; Lei, P. Mechanisms of neuronal cell death in ischemic stroke and their therapeutic implications. Med. Res. Rev. 2022, 42, 259–305. [Google Scholar] [CrossRef] [PubMed]
- Bertheloot, D.; Latz, E.; Franklin, B.S. Necroptosis, pyroptosis and apoptosis: An intricate game of cell death. Cell Mol. Immunol. 2021, 18, 1106–1121. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.P.; Yuan, H.; Xu, Y.; Liu, R.M.; Luo, Y.; Xiao, J.H. Protective effects of Ligularia fischeri root extracts against ulcerative colitis in mice through activation of Bcl-2/Bax signalings. Phytomedicine 2022, 99, 154006. [Google Scholar] [CrossRef]
- Eskandari, E.; Eaves, C.J. Paradoxical roles of caspase-3 in regulating cell survival, proliferation, and tumorigenesis. J. Cell Biol. 2022, 221, e202201159. [Google Scholar] [CrossRef]
- Radak, D.; Katsiki, N.; Resanovic, I.; Jovanovic, A.; Sudar-Milovanovic, E.; Zafirovic, S.; Mousad, S.A.; Isenovic, E.R. Apoptosis and Acute Brain Ischemia in Ischemic Stroke. Curr. Vasc. Pharmacol. 2017, 15, 115–122. [Google Scholar] [CrossRef]
- Shafiey, S.I.; Ahmed, K.A.; Abo-Saif, A.A.; Abo-Youssef, A.M.; Mohamed, W.R. Galantamine mitigates testicular injury and disturbed spermatogenesis in adjuvant arthritic rats via modulating apoptosis, inflammatory signals, and IL-6/JAK/STAT3/SOCS3 signaling. Inflammopharmacology, 2023; Online ahead of print. [Google Scholar] [CrossRef]
- Zuo, L.; Xie, J.; Liu, Y.; Leng, S.; Zhang, Z.; Yan, F. Down-regulation of circular RNA CDC14A peripherally ameliorates brain injury in acute phase of ischemic stroke. J. Neuroinflamm. 2021, 18, 283. [Google Scholar] [CrossRef]
- Feng, B.; Meng, L.; Luan, L.; Fang, Z.; Zhao, P.; Zhao, G. Upregulation of Extracellular Vesicles-Encapsulated miR-132 Released from Mesenchymal Stem Cells Attenuates Ischemic Neuronal Injury by Inhibiting Smad2/c-jun Pathway via Acvr2b Suppression. Front. Cell Dev. Biol. 2020, 8, 568304. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Zhao, P.; Lian, Y.; Li, S.; Chen, Y.; Li, L. MiR-340-5p alleviates oxygen-glucose deprivation/reoxygenation-induced neuronal injury via PI3K/Akt activation by targeting PDCD4. Neurochem. Int. 2020, 134, 104650. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Gui, Q.; Hui, X.; Wang, X.; Jiang, J.; Ding, L.; Sun, X.; Wang, Y.; Chen, H. TGF-beta1/Smad3 Signaling Pathway Suppresses Cell Apoptosis in Cerebral Ischemic Stroke Rats. Med. Sci. Monit. 2017, 23, 366–376. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, S.C.; Dou, B.K.; Zou, Y.X.; Han, H.Z.; Liu, D.X.; Ke, Z.J.; Wang, Z.F. Cycloastragenol upregulates SIRT1 expression, attenuates apoptosis and suppresses neuroinflammation after brain ischemia. Acta Pharmacol. Sin. 2020, 41, 1025–1032. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhou, J.; Zhang, M.; Yi, Y.; He, B. Upregulation of miR-376c-3p alleviates oxygen-glucose deprivation-induced cell injury by targeting ING5. Cell Mol. Biol. Lett. 2019, 24, 67. [Google Scholar] [CrossRef]
- Tanaka, K.; Tanaka, T.; Nakano, T.; Hozumi, Y.; Yanagida, M.; Araki, Y.; Iwazaki, K.; Takagi, M.; Goto, K. Knockdown of DEAD-box RNA helicase DDX5 selectively attenuates serine 311 phosphorylation of NF-kappaB p65 subunit and expression level of anti-apoptotic factor Bcl-2. Cell Signal. 2020, 65, 109428. [Google Scholar] [CrossRef] [PubMed]
- Hacker, H.; Tseng, P.H.; Karin, M. Expanding TRAF function: TRAF3 as a tri-faced immune regulator. Nat. Rev. Immunol. 2011, 11, 457–468. [Google Scholar] [CrossRef] [PubMed]
- Seki, M.; Powers, J.C.; Maruyama, S.; Zuriaga, M.A.; Wu, C.L.; Kurishima, C.; Kim, L.; Johnson, J.; Poidomani, A.; Wang, T.; et al. Acute and Chronic Increases of Circulating FSTL1 Normalize Energy Substrate Metabolism in Pacing-Induced Heart Failure. Circ. Heart Fail 2018, 11, e004486. [Google Scholar] [CrossRef]
- Han, B.; Zhang, Y.; Zhang, Y.; Bai, Y.; Chen, X.; Huang, R.; Wu, F.; Leng, S.; Chao, J.; Zhang, J.H.; et al. Novel insight into circular RNA HECTD1 in astrocyte activation via autophagy by targeting MIR142-TIPARP: Implications for cerebral ischemic stroke. Autophagy 2018, 14, 1164–1184. [Google Scholar] [CrossRef]
- Arumugam, T.V.; Baik, S.H.; Balaganapathy, P.; Sobey, C.G.; Mattson, M.P.; Jo, D.G. Notch signaling and neuronal death in stroke. Prog. Neurobiol. 2018, 165–167, 103–116. [Google Scholar] [CrossRef]
- Ohshiro, K.; Chen, J.; Srivastav, J.; Mishra, L.; Mishra, B. Alterations in TGF-beta signaling leads to high HMGA2 levels potentially through modulation of PJA1/SMAD3 in HCC cells. Genes Cancer 2020, 11, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, K.; Yamana, H.; Kitanaka, C.; Mochizuki, T.; Kokubu, A.; Kuchino, Y. Differential role of the JNK and p38 MAPK pathway in c-Myc- and s-Myc-mediated apoptosis. Biochem. Biophys. Res. Commun. 2000, 267, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Toyoshima, F.; Moriguchi, T.; Nishida, E. Fas induces cytoplasmic apoptotic responses and activation of the MKK7-JNK/SAPK and MKK6-p38 pathways independent of CPP32-like proteases. J. Cell Biol. 1997, 139, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Sanson, M.; Ingueneau, C.; Vindis, C.; Thiers, J.C.; Glock, Y.; Rousseau, H.; Sawa, Y.; Bando, Y.; Mallat, Z.; Salvayre, R.; et al. Oxygen-regulated protein-150 prevents calcium homeostasis deregulation and apoptosis induced by oxidized LDL in vascular cells. Cell Death Differ. 2008, 15, 1255–1265. [Google Scholar] [CrossRef] [PubMed]
- Piwecka, M.; Glazar, P.; Hernandez-Miranda, L.R.; Memczak, S.; Wolf, S.A.; Rybak-Wolf, A.; Filipchyk, A.; Klironomos, F.; Cerda Jara, C.A.; Fenske, P.; et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 2017, 357, eaam8526. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Ebbesen, K.K.; Sokol, M.; Jakobsen, T.; Korsgaard, U.; Eriksen, A.C.; Hansen, T.B.; Kjems, J.; Hager, H. Spatial expression analyses of the putative oncogene ciRS-7 in cancer reshape the microRNA sponge theory. Nat. Commun. 2020, 11, 4551. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Mehta, S.L.; Kaimal, B.; Lyons, K.; Dempsey, R.J.; Vemuganti, R. Poststroke Induction of alpha-Synuclein Mediates Ischemic Brain Damage. J. Neurosci. 2016, 36, 7055–7065. [Google Scholar] [CrossRef]
- Real, S.; Meo-Evoli, N.; Espada, L.; Tauler, A. E2F1 regulates cellular growth by mTORC1 signaling. PLoS ONE 2011, 6, e16163. [Google Scholar] [CrossRef]
- Wang, P.; Guan, Y.F.; Du, H.; Zhai, Q.W.; Su, D.F.; Miao, C.Y. Induction of autophagy contributes to the neuroprotection of nicotinamide phosphoribosyltransferase in cerebral ischemia. Autophagy 2012, 8, 77–87. [Google Scholar] [CrossRef]
- Duris, K.; Splichal, Z.; Jurajda, M. The Role of Inflammatory Response in Stroke Associated Programmed Cell Death. Curr. Neuropharmacol. 2018, 16, 1365–1374. [Google Scholar] [CrossRef]
- Li, Q.; Tian, Z.; Wang, M.; Kou, J.; Wang, C.; Rong, X.; Li, J.; Xie, X.; Pang, X. Luteoloside attenuates neuroinflammation in focal cerebral ischemia in rats via regulation of the PPARgamma/Nrf2/NF-kappaB signaling pathway. Int. Immunopharmacol. 2019, 66, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Zhang, Y.; Han, B.; Yang, L.; Chen, X.; Huang, R.; Wu, F.; Chao, J.; Liu, P.; Hu, G.; et al. Circular RNA DLGAP4 Ameliorates Ischemic Stroke Outcomes by Targeting miR-143 to Regulate Endothelial-Mesenchymal Transition Associated with Blood-Brain Barrier Integrity. J. Neurosci. 2018, 38, 32–50. [Google Scholar] [CrossRef] [PubMed]
- Orihuela, R.; McPherson, C.A.; Harry, G.J. Microglial M1/M2 polarization and metabolic states. Br. J. Pharmacol. 2016, 173, 649–665. [Google Scholar] [CrossRef] [PubMed]
- White, E.J.; Matsangos, A.E.; Wilson, G.M. AUF1 regulation of coding and noncoding RNA. Wiley Interdiscip. Rev. RNA 2017, 8, wrna.1393. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Fang, E.; Mei, H.; Chen, Y.; Li, H.; Li, D.; Song, H.; Wang, J.; Hong, M.; Xiao, W.; et al. Cis-Acting circ-CTNNB1 Promotes beta-Catenin Signaling and Cancer Progression via DDX3-Mediated Transactivation of YY1. Cancer Res. 2019, 79, 557–571. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Wang, Z.; Miao, C.Y. Angiogenesis after ischemic stroke. Acta Pharmacol. Sin. 2023, 44, 1305–1321. [Google Scholar] [CrossRef]
- Giraud, M.; Jmari, N.; Du, L.; Carallis, F.; Nieland, T.J.; Perez-Campo, F.M.; Bensaude, O.; Root, D.E.; Hacohen, N.; Mathis, D.; et al. An RNAi screen for Aire cofactors reveals a role for Hnrnpl in polymerase release and Aire-activated ectopic transcription. Proc. Natl. Acad. Sci. USA 2014, 111, 1491–1496. [Google Scholar] [CrossRef]
- Huang, M.; Huang, X.; Jiang, B.; Zhang, P.; Guo, L.; Cui, X.; Zhou, S.; Ren, L.; Zhang, M.; Zeng, J.; et al. linc00174-EZH2-ZNF24/Runx1-VEGFA Regulatory Mechanism Modulates Post-burn Wound Healing. Mol. Ther. Nucleic Acids 2020, 21, 824–836. [Google Scholar] [CrossRef]
- Han, X.; Zhang, G.; Chen, G.; Wu, Y.; Xu, T.; Xu, H.; Liu, B.; Zhou, Y. Buyang Huanwu Decoction promotes angiogenesis in myocardial infarction through suppression of PTEN and activation of the PI3K/Akt signalling pathway. J. Ethnopharmacol. 2022, 287, 114929. [Google Scholar] [CrossRef]
- AlAbdi, L.; He, M.; Yang, Q.; Norvil, A.B.; Gowher, H. The transcription factor Vezf1 represses the expression of the antiangiogenic factor Cited2 in endothelial cells. J. Biol. Chem. 2018, 293, 11109–11118. [Google Scholar] [CrossRef]
- Dosunmu-Ogunbi, A.M.; Wood, K.C.; Novelli, E.M.; Straub, A.C. Decoding the role of SOD2 in sickle cell disease. Blood Adv. 2019, 3, 2679–2687. [Google Scholar] [CrossRef] [PubMed]
- He, A.T.; Liu, J.; Li, F.; Yang, B.B. Targeting circular RNAs as a therapeutic approach: Current strategies and challenges. Signal Transduct Target Ther. 2021, 6, 185. [Google Scholar] [CrossRef]
- Lukiw, W.J. Circular RNA (circRNA) in Alzheimer's disease (AD). Front. Genet. 2013, 4, 307. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Wang, J.; Li, M.; Fang, J.; Ding, H.; Meng, J.; Zhang, J.; Fang, X.; Liu, H.; Ma, C.; et al. CircSV2b participates in oxidative stress regulation through miR-5107-5p-Foxk1-Akt1 axis in Parkinson's disease. Redox Biol. 2022, 56, 102430. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Li, M.; Wang, S.; Li, Z.; Li, H.; Li, S. CircRNA SRRM4 affects glucose metabolism by regulating PKM alternative splicing via SRSF3 deubiquitination in epilepsy. Neuropathol. Appl. Neurobiol. 2023, 49, e12850. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wilusz, J.E.; Chen, L.L. Biogenesis and Regulatory Roles of Circular RNAs. Annu. Rev. Cell Dev. Biol. 2022, 38, 263–289. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Li, C.; Li, X.; Li, Y.; Zhong, Y.; Ling, L. Altered circular RNA expression profiles in the non-ischemic thalamus in focal cortical infarction mice. Aging 2020, 12, 13206–13219. [Google Scholar] [CrossRef]
- Ren, L.; Jiang, Q.; Mo, L.; Tan, L.; Dong, Q.; Meng, L.; Yang, N.; Li, G. Mechanisms of circular RNA degradation. Commun. Biol. 2022, 5, 1355. [Google Scholar] [CrossRef]
- Xu, B.; Huang, X.; Yan, Y.; Zhao, Z.; Yang, J.; Zhu, L.; Yang, Y.; Liang, B.; Gu, L.; Su, L. Analysis of expression profiles and bioinformatics suggests that plasma exosomal circular RNAs may be involved in ischemic stroke in the Chinese Han population. Metab. Brain. Dis. 2022, 37, 665–676. [Google Scholar] [CrossRef]
- Li, L.; Si, X.; Ruan, J.; Ni, Z.; Li, X.; Sang, H.; Xia, W.; Huang, J.; Liu, K.; Lu, S.; et al. Circular RNA hsa_circ_0003574 as a biomarker for prediction and diagnosis of ischemic stroke caused by intracranial atherosclerotic stenosis. Front. Pharmacol. 2022, 13, 961866. [Google Scholar] [CrossRef]
- Xiao, Q.; Hou, R.; Li, H.; Zhang, S.; Zhang, F.; Zhu, X.; Pan, X. Circulating Exosomal circRNAs Contribute to Potential Diagnostic Value of Large Artery Atherosclerotic Stroke. Front. Immunol. 2021, 12, 830018. [Google Scholar] [CrossRef] [PubMed]
- Zu, J.; Zuo, L.; Zhang, L.; Wang, Z.; Shi, Y.; Gu, L.; Zhang, Z. Circular RNA FUNDC1 for Prediction of Acute Phase Outcome and Long-Term Survival of Acute Ischemic Stroke. Front. Neurol. 2022, 13, 846198. [Google Scholar] [CrossRef] [PubMed]
- Zuo, L.; Zhang, L.; Zu, J.; Wang, Z.; Han, B.; Chen, B.; Cheng, M.; Ju, M.; Li, M.; Shu, G.; et al. Circulating Circular RNAs as Biomarkers for the Diagnosis and Prediction of Outcomes in Acute Ischemic Stroke. Stroke 2020, 51, 319–323. [Google Scholar] [CrossRef]
- Chen, B.; Yi, J.; Xu, Y.; Zheng, P.; Tang, R.; Liu, B. Construction of a circRNA-miRNA-mRNA network revealed the potential mechanism of Buyang Huanwu Decoction in the treatment of cerebral ischemia. Biomed. Pharmacother. 2022, 145, 112445. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.G.; Buller, B.; Chopp, M. Exosomes—beyond stem cells for restorative therapy in stroke and neurological injury. Nat. Rev. Neurol. 2019, 15, 193–203. [Google Scholar] [CrossRef]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef]
- Liang, Y.; Duan, L.; Lu, J.; Xia, J. Engineering exosomes for targeted drug delivery. Theranostics 2021, 11, 3183–3195. [Google Scholar] [CrossRef]
- Yang, L.; Han, B.; Zhang, Z.; Wang, S.; Bai, Y.; Zhang, Y.; Tang, Y.; Du, L.; Xu, L.; Wu, F.; et al. Extracellular Vesicle-Mediated Delivery of Circular RNA SCMH1 Promotes Functional Recovery in Rodent and Nonhuman Primate Ischemic Stroke Models. Circulation 2020, 142, 556–574. [Google Scholar] [CrossRef]

| Phenotype | CircRNA | Animals and/or Cells | Model | Expression | Targets | The Roles in Stroke | References |
|---|---|---|---|---|---|---|---|
| Apoptosis | CircFOXP1 | Mice, A172 cells | tMCAO, OGD/R | Decreased | STAT3 | Alleviation | Yang et al. [30] |
| CircCDC14A | Mice, HT22 cells | MCAO/R, OGD/R | Increased | miR-23a-3p | Aggravation | Huo et al. [31] | |
| CircOGDH | Mice | MCAO | Increased | miR-5112 | Aggravation | Liu et al. [32] | |
| CircPRDX3 | Mice, N2a cells | MCAO/R, OGD/R | Decreased | miR-641 | Alleviation | Chen et al. [33] | |
| Circ_0000566 | HBMECs | OGD/R | Increased | miR-18a-5p | Aggravation | Liu et al. [34] | |
| Circ_0000647 | SK-N-SH cells | OGD/R | Increased | miR-126-5p | Aggravation | Dai et al. [35] | |
| CircFUNDC1 | HCN-2 cell, HBMECs | OGD | Increased | miR-496 | Aggravation | Wang et al. [36] | |
| CircTLK1 | Mice, N2a cells | MCAO/R, OGD/R | Increased | miR-26a-5p | Aggravation | Wu et al. [37] | |
| CircUSP36 | Mice, A172 cells | MCAO/R, OGD/R | Decreased | miR-139-3p | Alleviation | Yang et al. [38] | |
| CircHIPK3 | Mice, bEnd.3 cells | tMCAO, OGD/R | Decreased | miR-148b-3p | Aggravation | Chen et al. [39] | |
| Circ_0010729 | HBMECs | OGD/R | Increased | miR-665 | Aggravation | Ouyang et al. [40] | |
| CircPUM1 | Mice, SK-N-SH cells | tMCAO, OGD/R | Decreased | miR-340-5p | Alleviation | Hu et al. [41] | |
| CircHECTD1 | Mice, HT22 cells | MCAO, OGD | Increased | miR-133b, miR-27a-3p | Aggravation | Zhang et al. [42], Dai et al. [43] | |
| CircPHC3 | HBMECs | OGD | Increased | miR-455-5p | Aggravation | Xu et al. [44] | |
| CircTTC3 | Mice, Primary astrocytes | MCAO/R, OGD/R | Increased | miR-372-3p | Aggravation | Yang et al. [45] | |
| CircCCDC9 | Mice | tMCAO | Decreased | Notch1, NICD | Alleviation | Wu et al. [46] | |
| CircUCK2 | Mice, HT22 cells | MCAO, OGD | Decreased | miR-125b-5p | Alleviation | Chen et al. [47] | |
| Circ_016719 | Mice, HT22 cells | tMCAO, OGD/R | Increased | miR-29c | Aggravation | Tang et al. [48] | |
| CircLIFR | Mice, bEnd.3 cells | MCAO, OGD | Decreased | miR-100 | Alleviation | Zhao et al. [49] | |
| Circ_0025984 | Rats, A172 cells | MCAO, OGD | Decreased | miR-134-3p | Alleviation | Zhou et al. [50] | |
| CDR1as | Mice | MCAO/R | Decreased | miR-7 | Alleviation | Mehta et al. [51] | |
| Autophagy | Circ-FoxO3 | Mice, bEnd.3 cells and HBMECs | MCAO/R, OGD/R | Increased | mTOR, E2F1 | Alleviation | Yang et al. [52] |
| CircSHOC2 | Primary astrocytes | OGD/R | Increased | miR-7670a-3p | Alleviation | Chen et al. [53] | |
| Inflammation | Circ_0000831 | Mice, BV-2 cells | MCAO, OGD | Decreased | miR-16-5p | Alleviation | Huang et al. [54] |
| CircDLGAP4 | HCN-2 cells | OGD | Decreased | miR-503-3p | Alleviation | Qiu et al. [55] | |
| CircRps5 | Mice, ADSCs | MCAO, OGD | Increased | miR-124-3 | Alleviation | Yang et al. [56] | |
| Oxidative stress | Circ-Memo1 | HBMECs | H/R | Increased | miR-17-5p | Aggravation | Ren et al. [57] |
| CircDLGAP4 | Mice, N2a cells | tMCAO, OGD | Decreased | AUF1 | Alleviation | Liu et al. [58] | |
| CircCTNNB1 | Mice, Primary astrocytes | MCAO/R, OGD/R | Decreased | miR-96-5p | Alleviation | Chen et al. [59] | |
| Angiogenesis | CircPDS5B | Mice, HBMECs | tMCAO, OGD/R | Increased | hnRNPL | Aggravation | Jiang et al. [60] |
| CircHECTD1 | Mice, HBMECs | MCAO, OGD/R | Increased | miR-335 | Aggravation | He et al. [61] | |
| CircFUNDC1 | HBMECs | OGD | Increased | miR-375 | Aggravation | Bai et al. [62] | |
| Circ_0006768 | HBNECs | OGD/R | Decreased | miR-222-3p | Alleviation | Li et al. [63] | |
| CircPHKA2 | HBMECs | OGD | Decreased | miR-574-5p | Alleviation | Yang et al. [64] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, G.; Liu, G.; Wang, Z.; Li, Y.; Fang, W. Circular RNAs: Promising Treatment Targets and Biomarkers of Ischemic Stroke. Int. J. Mol. Sci. 2024, 25, 178. https://doi.org/10.3390/ijms25010178
Xu G, Liu G, Wang Z, Li Y, Fang W. Circular RNAs: Promising Treatment Targets and Biomarkers of Ischemic Stroke. International Journal of Molecular Sciences. 2024; 25(1):178. https://doi.org/10.3390/ijms25010178
Chicago/Turabian StyleXu, Guangchen, Ge Liu, Ziyu Wang, Yunman Li, and Weirong Fang. 2024. "Circular RNAs: Promising Treatment Targets and Biomarkers of Ischemic Stroke" International Journal of Molecular Sciences 25, no. 1: 178. https://doi.org/10.3390/ijms25010178




