HCC-Related lncRNAs: Roles and Mechanisms
Abstract
:1. Introduction
2. Epigenetic Regulation
2.1. DNA Methylation: Beyond the Methyl Mark
2.2. Histone Modifications: The Chromatin Code
2.3. Noncoding RNAs: Epigenetic Mediators
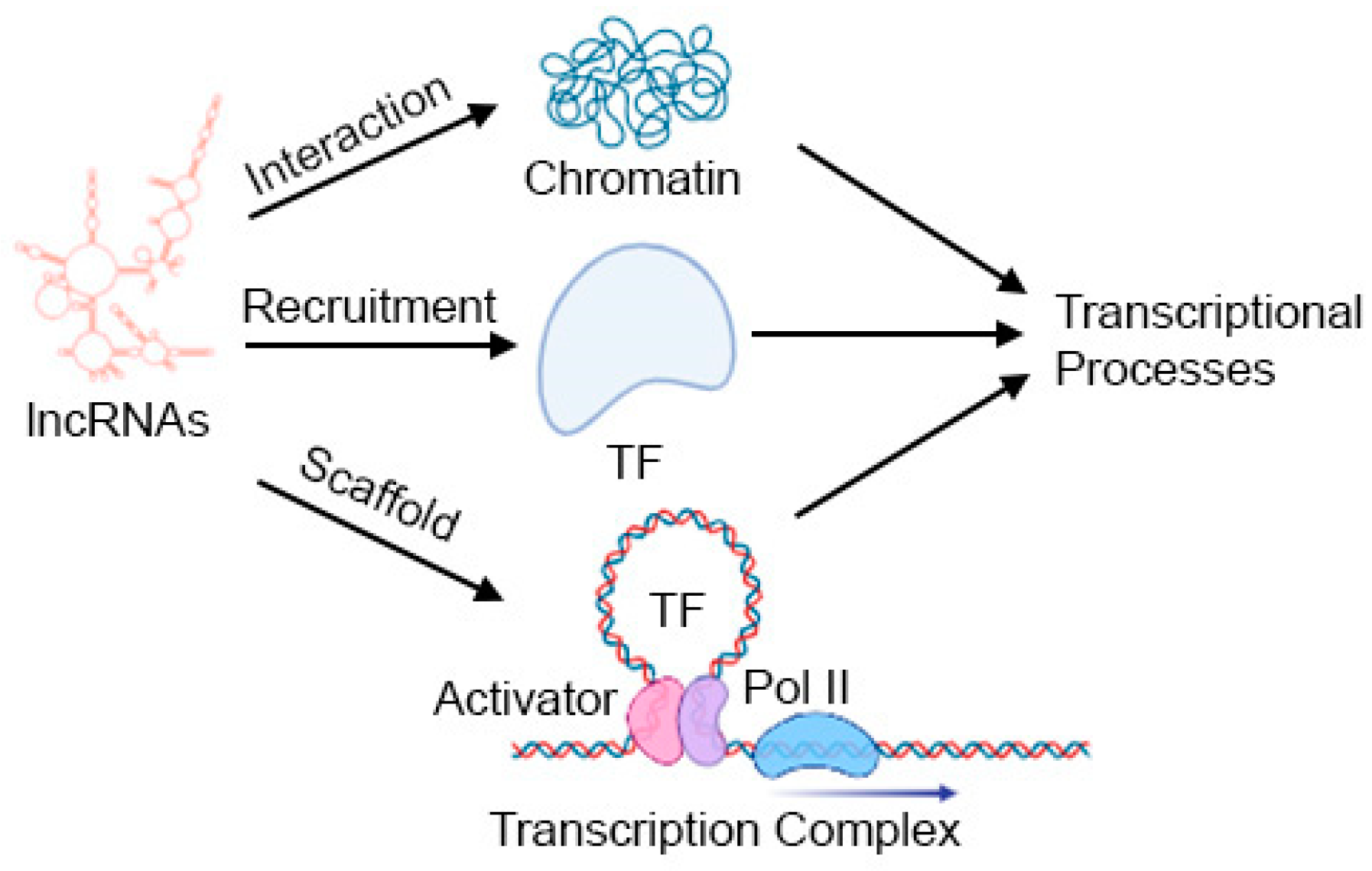
3. Introduction to lncRNAs: Mechanisms and Modes of Action
3.1. Gene Expression Regulation
3.1.1. Regulation of Chromatin
LncRNA–Protein Interaction Mediating Chromatin Regulation
LncRNA-DNA Interaction Mediating Chromatin Regulation
3.1.2. Regulation of Transcription
3.2. Post-Transcriptional Regulation
3.3. Liquid–Liquid Phase Separation (LLPS)
3.4. lncRNAs Coding Micropeptides
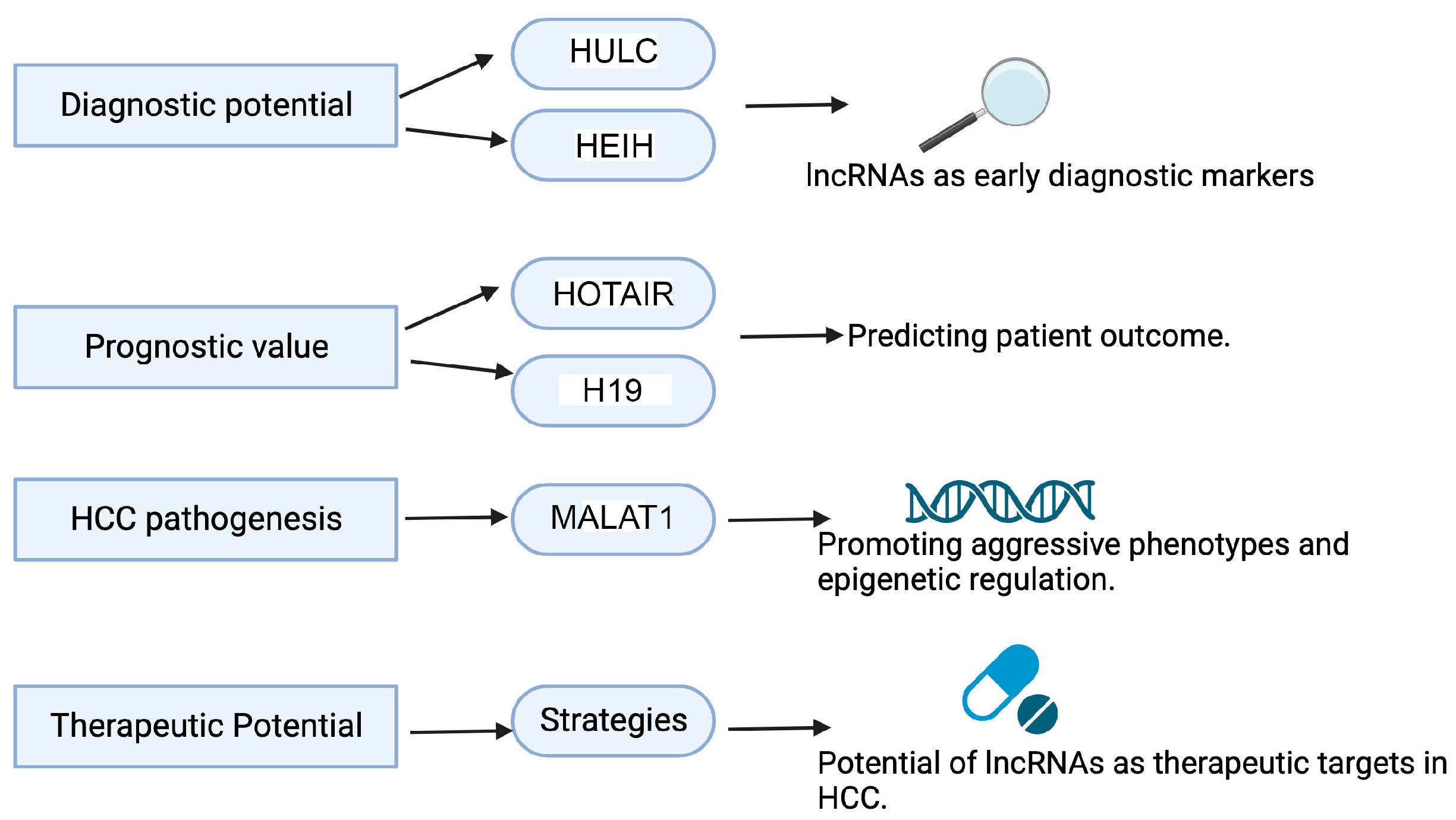
4. Significance of Studying lncRNAs in HCC
5. LncRNAs in NASH and Viral Hepatitis, Risk Factors Contributing to HCC
5.1. NASH
5.2. Viral Hepatitis
6. LncRNAs Regulating HCC
6.1. LncRNAs Functioning as Oncogenes
6.1.1. HOTAIR
6.1.2. MALAT1
6.1.3. HULC
6.1.4. PVT1
6.1.5. HOTTIP
6.1.6. H19
6.1.7. NEAT1
6.1.8. HEIH
6.1.9. SNHG6
6.1.10. Additional lncRNAs Functioning as Oncogenes
LncRNAs Modulating Wnt/β-Catenin Signaling
LncRNAs Promoting HCC by Regulating Metabolism
lncRNAs Regulating EMT and Metastasis
lncRNAs Regulating Vascular Invasion
lncRNAs Regulating Oncogenic AKT Pathway
Miscellaneous Functions of lncRNAs in HCC
6.2. lncRNAs Functioning as Tumor-Suppressor Genes
6.2.1. MEG3
6.2.2. GAS5
6.2.3. FENDRR
6.2.4. Additional lncRNAs Functioning as Tumor Suppressors
7. lncRNAs Modulating Current HCC Treatment
8. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 53BP1 | p53 binding protein 1 |
| AAV | Adeno-associated virus |
| ABCG2 | ATP binding cassette subfamily G member 2 |
| ACSL1 | Acyl-CoA synthetase long-chain family member 1 |
| AEG-1 | Astrocyte elevated gene-1 |
| AFP | Alpha-fetoprotein |
| AKR1C2 | Aldo-keto reductase family 1 member C2 |
| AKT | Akt kinase |
| ALS | Amyotrophic lateral sclerosis |
| AMPKβ2 | Adenosine monophosphate-activated protein kinase subunit beta 2 |
| ANRIL/CDKN2B-AS1 | Antisense noncoding RNA in the INK4 locus/CDKN2B antisense RNA 1 |
| ANXA8 | Annexin A8 |
| APC | APC regulator of WNT signaling pathway |
| ASO | Antisense oligonucleotide |
| ASRPS | A small regulatory peptide of STAT3 |
| ASS1 | Arginosuccinate synthase 1 |
| ATG3 | Autophagy related 3 |
| ATG14 | Autophagy related 14 |
| ATP | Adenosine triphosphate |
| BBOX1-AS1 | BBOX1 antisense RNA 1 |
| BMI1 | BMI1 proto-oncogene, polycomb ring finer |
| B-NSG | NOD-Prkdcscid Il2rgtm1/Bcgen |
| BRD3 | Bromodomain containing 3 |
| CAPRIN1 | Cell cycle associated protein 1 |
| CASC9 | Cancer susceptibility 9 |
| CBX7 | Chromobox 7 |
| CCT3 | Chaperonin containing TCP1 subunit 3 |
| CDIPTOSP | CDIP transferase opposite strand, pseudogene |
| CDKN2A | Cyclin-dependent kinase inhibitor 2A (p16) |
| CDKN2B | Cyclin-dependent kinase inhibitor 2B (p15) |
| CELF1 | CUGBP Elav-like family member 1 |
| ceRNA | Competitive endogenous RNA |
| CHASERR | CHD2 adjacent suppressive regulatory RNA |
| CHD2 | Chromodomain helicase DNA binding protein 2 |
| CIP2A-BP | CIP2A binding peptide |
| CLOCK | Clock circadian regulator |
| COX2 | Cytochrome c oxidase subunit II |
| CREB | cAMP-response element binding protein |
| CSC | Cancer stem cells |
| CT | Computed tomography |
| ctDNA | Circulating tumor deoxyribonucleic acid |
| CRISPR | Clustered regularly interspaced short palindromic repeats |
| CTNNB1 | Catenin beta 1 (β-catenin) |
| CUL4A | Cullin 4A |
| CYTB | Cytochrome b |
| DANCR | Differentiation antagonizing non-protein coding RNA |
| DDR | DNA damage response |
| DDX5 | DEAD-box helicase 5 |
| DDX17 | DEAD-box RNA helicase 17 |
| DDX21 | DExD-box helicase 21 |
| DEN | Diethylnitrosamine |
| DFC | Dense fibrillar component |
| DIGIT | Divergent to GSC, induced by TGF-β family signaling |
| DILC | Downregulated in liver cancer stem cells |
| dilncRNA | Damage-induced lncRNA |
| DNA | Deoxyribonucleic acid |
| DNMT1 | DNA methyltransferase 1 |
| DNMT2 | DNA methyltransferase 2 |
| DPP4 | Dipeptidyl peptidase 4 |
| DSB | Double-strand break repair |
| E2F1 | E2F transcription factor 1 |
| EEF1E1 | Eukaryotic translation elongation factor 1 epsilon 1 |
| EGFR | Epidermal growth factor receptor |
| EMT | Epithelial-mesenchymal transition |
| EPCAM | Epithelial cell adhesion molecule |
| ER | Endoplasmic reticulum |
| ESC | Embryonic stem cells |
| EV | Extracellular vesicle |
| EZH2 | Enhancer of zeste 2 polycomb repressive complex 2 subunit |
| FAF2 | Fas associated factor family member 2 |
| FC | Fibrillar center |
| FDA | Food and drug administration |
| FENDRR | FOXF1 adjacent noncoding developmental regulatory RNA |
| FOXC1 | Forkhead box C1 |
| FOXM1 | Forkhead box M1 |
| FUS | FUS RNA-binding protein |
| GADD45A | Growth arrest and DNA damage-inducible alpha |
| GADD45B | Growth arrest and DNA damage-inducible beta |
| GAS5 | Growth arrest specific 5 |
| GIRGL | glutamine insufficiency regulator of glutaminase lncRNA |
| GLS1 | Glutaminase 1 |
| GPC3 | Glypican-3 |
| GR | Glucocorticoid receptor |
| GRP78 | Glucose regulated protein 78 |
| H19 | H19 imprinted maternally expressed transcript |
| H3K4me3 | Trimethylation of histone H3 at lysine 4 |
| H3K27me3 | Trimethylation of histone H3 at lysine 27 |
| H3K9ac | Acetylation of histone H3 at lysine 9 |
| H3K18ac | Acetylation of histone H3 at lysine 18 |
| H3K56ac | Acetylation of histone H3 at lysine 56 |
| HAND2-AS1 | HAND2 antisense RNA 1 |
| HBV | Hepatitis B virus |
| HCC | Hepatocellular carcinoma |
| HCV | Hepatitis C virus |
| HEIH | Hepatocellular carcinoma upregulated EZH2-associated long noncoding RNA |
| HIF1A | Hypoxia-inducible factor 1 subunit alpha |
| HIG2 | Hypoxia-inducible protein 2 |
| HK2 | Hexokinase 2 |
| HMGA2 | High mobility group AT-hook 2 |
| HNRNPK | Heterogenous nuclear ribonucleoprotein K |
| HNRNPL | Heterogenous nuclear ribonucleoprotein L |
| HNRNPU | Heterogenous nuclear ribonucleoprotein U |
| HOTAIR | HOX transcript antisense RNA |
| HOTTIP | HOXA distal transcript antisense RNA |
| HSP90 | Heat shock protein 90 |
| HULC | Highly upregulated in liver cancer |
| HuR | Human antigen R |
| ICAM1 | Intercellular adhesion molecule 1 |
| ICR | ICAM-1-related |
| IDR | Intrinsically disordered region |
| IGF2 | Insulin-like growth factor 2 |
| IL-6 | Interleukin 6 |
| IL-11 | Interleukin 11 |
| JAK | Janus kinase |
| KLF2 | KLF transcription factor 2 |
| KMT2A/MLL | Lysine methyltransferase 2A |
| LATS1 | Large tumor suppressor kinase 1 |
| LINC-ROR | Long intergenic non-protein coding RNA, regulator of reprogramming |
| LINP1 | LncRNA in non-homologous end joining pathway 1 |
| LLPS | Liquid-liquid phase separation |
| LncRNA | Long noncoding ribonucleic acid |
| lncRNA-ATB | LncRNA-activated by TGF-β |
| LncPRESS1 | LncRNA p53-regulated and ESC-associated |
| LSD1 | Lysine-specific histone demethylase 1 |
| M6A | N6-methyladenosine |
| MAFLD | Metabolic dysfunction-related fatty liver disease |
| MALAT1 | Metastasis associated lung adenocarcinoma transcript 1 |
| MAPK | Mitogen-activated protein kinase |
| MATR3 | Matrin 3 |
| MBNL3 | Muscleblind like splicing regulator 3 |
| MCM2 | Minichromosome maintenance complex component 2 |
| MEG3 | Maternally expressed 3 |
| MEK | MAP kinase-ERK kinase |
| MELTF-AS1 | MELTF antisense RNA 1 |
| MEX3B | Mex-3 RNA-binding family member B |
| miRNA | microRNA |
| MMP | Matrix metallopeptidase |
| MMTV-PyMT | mouse mammary tumor virus (MMTV) long terminal repeat upstream of a cDNA sequence encoding the Polyoma Virus middle T antigen (PyMT) |
| MRI | Magnetic resonance imaging |
| MTCH2 | Mitochondrial carrier 2 |
| mTOR | Mammalian target of rapamycin |
| MVIH | LncRNA associated with microvascular invasion in HCC |
| MYC | MYC proto-oncogene, bHLH transcription factor |
| NAFLD | Non-alcoholic fatty liver disease |
| NASH | Non-alcoholic steatohepatitis |
| NcRNA | Noncoding ribonucleic acid |
| ND3 | NADH dehydrogenase subunit 3 |
| NEAT1 | Nuclear paraspeckle assembly transcript 1 |
| NF-κB | Nuclear factor kappa-light-chain-enhancer of activated B cells |
| NHEJ | Non-homologous end joining |
| NIHCOLE | Noncoding RNA induced in hepatocellular carcinoma with an oncogenic role in ligation efficiency |
| NONO | Non-POU domain-containing octamer binding protein |
| NOP2 | NOP2 nucleolar protein |
| NORAD | Noncoding RNA activated by DNA damage |
| NRAS | NRAS proto-oncogene, GTPase |
| NSCLC | Non-small cell lung cancer |
| PAARH | Progression and angiogenesis associated RNA in HCC |
| PD-1 | Programmed cell death 1 |
| PD-L1 | Programmed cell death 1 ligand 1 |
| PDX | Patient-derived xenograft |
| PGK1 | Phosphoglycerate kinase 1 |
| PHAROH | Pluripotency and hepatocyte associated RNA overexpressed in HCC |
| PHF8 | PHD finger protein 8 |
| PI3K | Phosphatidylinositol 3-kinase |
| PNCTR | Pyrimidine-rich noncoding transcript |
| PNUTS | Phosphatase 1 nuclear targeting subunit |
| Pol I | RNA polymerase I |
| Pol II | RNA polymerase II |
| Pol III | RNA polymerase III |
| PPARA | Peroxisome proliferator–activated receptor alpha |
| PPP1CA | Protein phosphatase 1 catalytic subunit alpha |
| PRAL | p53 regulation-associated lncRNA |
| PRC2 | Polycomb Repressive Complex 2 |
| PRR34-AS1 | PRR34 antisense RNA 1 |
| PTBP1 | Pyrimidine tract binding protein 1 |
| PTEN | Phosphatase and tensin homolog |
| PUM1 | Pumilio RNA-binding family member 1 |
| PUM2 | Pumilio RNA-binding family member 2 |
| PVT1 | Pvt1 oncogene |
| PVTT | Portal vein tumor thrombus |
| PXN-AS1 | Paxillin antisense RNA 1 |
| RAB35 | RAB35, member RAS oncogene family |
| RBP | RNA-binding protein |
| RFA | Radiofrequency ablation |
| RMST | Rhabdomyosarcoma 2 associated transcript |
| RNA | Ribonucleic acid |
| RNase | Ribonuclease |
| RNAi | RNA interference |
| RNP | Ribonucleoprotein |
| RPS6KB1 | Ribosomal protein S6 kinase B1 |
| RXRA | Retinoid X receptor alpha |
| SAT1 | Spermidine/spermine N1-acetyltransferase 1 |
| SCARNA10 | Small cajal body specific RNA 10 |
| SERPINH1 | Serpin family H member 1 |
| SET1 | Histone methyltransferase SET1 |
| SETD2 | SET domain containing 2, histone lysine methyltransferase |
| SF3B3 | Splicing factor 3b subunit 3 |
| SFPQ | Splicing factor proline and glutamine rich |
| shRNA | Short hairpin RNA |
| siRNA | Small interfering RNA |
| SIRT1 | Sirtuin 1 |
| SIRT6 | Sirtuin 6 |
| SLERT | Box H/ACA small nucleolar RNA (snoRNA)–ended lncRNA |
| SNAIL | Zing-finger transcription factor Snail |
| SNAP23 | Synaptosome associated protein 23 |
| SNHG6 | Small nucleolar RNA host gene 6 |
| SNHG9 | Small nucleolar RNA host gene 9 |
| snRNA | Small nuclear RNA |
| snoRNA | Small nucleolar RNA |
| SOCS2 | Suppressor of cytokine signaling 2 |
| SOX2 | SRY-box transcription factor 2 |
| SOX12 | SRY-box transcription factor 12 |
| SPHK1 | Sphingosine kinase 1 |
| SR | Serine/arginine |
| SREBP1 | Sterol regulatory element binding protein 1 |
| STAT1 | Signal transducer and activator of transcription 1 |
| STAT3 | Signal transducer and activator of transcription 3 |
| SUZ12 | SUZ12 polycomb repressive complex 2 subunit |
| TACE | Transarterial chemoembolization |
| TAK1 | Transforming growth factor-β-activated kinase 1 |
| TARID | TCF21 antisense RNA inducing promoter demethylation |
| TCF7L2 | Transcription factor 7 like 2 |
| TCF21 | Transcription factor 21 |
| TDP-43 | Tar DNA binding protein |
| TERT | Telomerase reverse transcriptase |
| TET1 | Tet methylcytosine dioxygenase 1 |
| TEX10 | Testis expressed 10 |
| TGF-β | Transforming growth factor-β |
| TGFBR2 | Transforming growth factor-β receptor 2 |
| TIAR | TIA1 cytotoxic granule associated RNA-binding protein like 1 |
| TKI | Tyrosine kinase inhibitor |
| TNBC | Triple negative breast cancer |
| TP53 | Tumor protein p53 |
| TRAF6 | TNF receptor associated factor 6 |
| Treg | Regulatory T cells |
| TRERNA1 | Translation regulatory lncRNA 1 |
| TUG1 | Taurine upregulated gene 1 |
| URB1-AS1 | URB1-antisense RNA 1 |
| USP22 | Ubiquitin-specific peptidase 22 |
| VAMP3 | Vesicle associated membrane protein 3 |
| VLP | Virus-like particles |
| VEGF | Vascular endothelial growth factor |
| VLDLR | Very low density lipoprotein receptor |
| WDR5 | WD repeat domain 5 |
| WTAP | WT1 associated protein |
| XIST | X inactive specific transcript |
| YAP1 | Yes1 associated transcriptional regulator |
| YBX1 | Y-box binding protein 1 |
| ZBTB7A | Zinc finger and BTB domain containing 7A |
| ZEB1 | Zinc finger E-box binding homeobox 1 |
| ZEB2 | Zinc finger E-box binding homeobox 2 |
| ZFAS1 | ZNFX1 antisense RNA 1 |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Eslam, M.; Sanyal, A.J.; George, J.; International Consensus, P. MAFLD: A Consensus-Driven Proposed Nomenclature for Metabolic Associated Fatty Liver Disease. Gastroenterology 2020, 158, 1999–2014.e1991. [Google Scholar] [CrossRef] [PubMed]
- Pennisi, G.; Celsa, C.; Giammanco, A.; Spatola, F.; Petta, S. The Burden of Hepatocellular Carcinoma in Non-Alcoholic Fatty Liver Disease: Screening Issue and Future Perspectives. Int. J. Mol. Sci. 2019, 20, 5613. [Google Scholar] [CrossRef] [PubMed]
- Rebouissou, S.; Nault, J.C. Advances in molecular classification and precision oncology in hepatocellular carcinoma. J. Hepatol. 2020, 72, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, A. Hepatocellular Carcinoma. N. Engl. J. Med. 2019, 380, 1450–1462. [Google Scholar] [CrossRef]
- Mahpour, A.; Mullen, A.C. Our emerging understanding of the roles of long non-coding RNAs in normal liver function, disease, and malignancy. JHEP Rep. 2021, 3, 100177. [Google Scholar] [CrossRef]
- Nagaraju, G.P.; Dariya, B.; Kasa, P.; Peela, S.; El-Rayes, B.F. Epigenetics in hepatocellular carcinoma. Semin. Cancer Biol. 2022, 86, 622–632. [Google Scholar] [CrossRef]
- Zhang, G.; Li, R.; Zhao, X.; Meng, S.; Ye, J.; Zhao, L. Validation of the American Joint Committee on Cancer eighth edition staging system in patients undergoing hepatectomy for hepatocellular carcinoma: A US population-based study. J. Surg. Res. 2018, 222, 55–68. [Google Scholar] [CrossRef]
- Labgaa, I.; Villanueva, A.; Dormond, O.; Demartines, N.; Melloul, E. The Role of Liquid Biopsy in Hepatocellular Carcinoma Prognostication. Cancers 2021, 13, 659. [Google Scholar] [CrossRef]
- Kulik, L.; El-Serag, H.B. Epidemiology and Management of Hepatocellular Carcinoma. Gastroenterology 2019, 156, 477–491.e471. [Google Scholar] [CrossRef]
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2021, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- El-Khoueiry, A.B.; Sangro, B.; Yau, T.; Crocenzi, T.S.; Kudo, M.; Hsu, C.; Kim, T.Y.; Choo, S.P.; Trojan, J.; Welling, T.H.R.; et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): An open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet 2017, 389, 2492–2502. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.S.; Ryoo, B.Y.; Hsu, C.H.; Numata, K.; Stein, S.; Verret, W.; Hack, S.P.; Spahn, J.; Liu, B.; Abdullah, H.; et al. Atezolizumab with or without bevacizumab in unresectable hepatocellular carcinoma (GO30140): An open-label, multicentre, phase 1b study. Lancet Oncol. 2020, 21, 808–820. [Google Scholar] [CrossRef] [PubMed]
- Casak, S.J.; Donoghue, M.; Fashoyin-Aje, L.; Jiang, X.; Rodriguez, L.; Shen, Y.L.; Xu, Y.; Jiang, X.; Liu, J.; Zhao, H.; et al. FDA Approval Summary: Atezolizumab Plus Bevacizumab for the Treatment of Patients with Advanced Unresectable or Metastatic Hepatocellular Carcinoma. Clin. Cancer Res. 2021, 27, 1836–1841. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.S.; Qin, S.; Ikeda, M.; Galle, P.R.; Ducreux, M.; Kim, T.Y.; Kudo, M.; Breder, V.; Merle, P.; Kaseb, A.O.; et al. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N. Engl. J. Med. 2020, 382, 1894–1905. [Google Scholar] [CrossRef]
- Reig, M.; Forner, A.; Rimola, J.; Ferrer-Fabrega, J.; Burrel, M.; Garcia-Criado, A.; Kelley, R.K.; Galle, P.R.; Mazzaferro, V.; Salem, R.; et al. BCLC strategy for prognosis prediction and treatment recommendation: The 2022 update. J. Hepatol. 2022, 76, 681–693. [Google Scholar] [CrossRef] [PubMed]
- Anstee, Q.M.; Reeves, H.L.; Kotsiliti, E.; Govaere, O.; Heikenwalder, M. From NASH to HCC: Current concepts and future challenges. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 411–428. [Google Scholar] [CrossRef]
- Bird, A. Perceptions of epigenetics. Nature 2007, 447, 396–398. [Google Scholar] [CrossRef]
- Hon, G.C.; Rajagopal, N.; Shen, Y.; McCleary, D.F.; Yue, F.; Dang, M.D.; Ren, B. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat. Genet. 2013, 45, 1198–1206. [Google Scholar] [CrossRef]
- Jones, P.A.; Takai, D. The role of DNA methylation in mammalian epigenetics. Science 2001, 293, 1068–1070. [Google Scholar] [CrossRef]
- Esteller, M. Epigenetics in cancer. N. Engl. J. Med. 2008, 358, 1148–1159. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, A.; Portela, A.; Sayols, S.; Battiston, C.; Hoshida, Y.; Mendez-Gonzalez, J.; Imbeaud, S.; Letouze, E.; Hernandez-Gea, V.; Cornella, H.; et al. DNA methylation-based prognosis and epidrivers in hepatocellular carcinoma. Hepatology 2015, 61, 1945–1956. [Google Scholar] [CrossRef] [PubMed]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Arrowsmith, C.H.; Bountra, C.; Fish, P.V.; Lee, K.; Schapira, M. Epigenetic protein families: A new frontier for drug discovery. Nat. Rev. Drug Discov. 2012, 11, 384–400. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Mu, H.; Xiao, Y.; Zhao, Z.; Cui, X.; Wu, D. Comprehensive Analysis of Histone Modifications in Hepatocellular Carcinoma Reveals Different Subtypes and Key Prognostic Models. J. Oncol. 2022, 2022, 5961603. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef] [PubMed]
- Gebert, L.F.R.; MacRae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Henson, R.; Wehbe-Janek, H.; Ghoshal, K.; Jacob, S.T.; Patel, T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 2007, 133, 647–658. [Google Scholar] [CrossRef]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef]
- Hartford, C.C.R.; Lal, A. When Long Noncoding Becomes Protein Coding. Mol. Cell Biol. 2020, 40, e00528-19. [Google Scholar] [CrossRef]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 925–933. [Google Scholar] [CrossRef] [PubMed]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Cao, J. The functional role of long non-coding RNAs and epigenetics. Biol. Proced. Online 2014, 16, 11. [Google Scholar] [CrossRef] [PubMed]
- Dueva, R.; Akopyan, K.; Pederiva, C.; Trevisan, D.; Dhanjal, S.; Lindqvist, A.; Farnebo, M. Neutralization of the Positive Charges on Histone Tails by RNA Promotes an Open Chromatin Structure. Cell Chem. Biol. 2019, 26, 1436–1449.e1435. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Yang, Y.W.; Liu, B.; Sanyal, A.; Corces-Zimmerman, R.; Chen, Y.; Lajoie, B.R.; Protacio, A.; Flynn, R.A.; Gupta, R.A.; et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011, 472, 120–124. [Google Scholar] [CrossRef]
- Jain, A.K.; Xi, Y.; McCarthy, R.; Allton, K.; Akdemir, K.C.; Patel, L.R.; Aronow, B.; Lin, C.; Li, W.; Yang, L.; et al. LncPRESS1 Is a p53-Regulated LncRNA that Safeguards Pluripotency by Disrupting SIRT6-Mediated De-acetylation of Histone H3K56. Mol. Cell 2016, 64, 967–981. [Google Scholar] [CrossRef]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef]
- Yap, K.L.; Li, S.; Munoz-Cabello, A.M.; Raguz, S.; Zeng, L.; Mujtaba, S.; Gil, J.; Walsh, M.J.; Zhou, M.M. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell 2010, 38, 662–674. [Google Scholar] [CrossRef]
- Arab, K.; Karaulanov, E.; Musheev, M.; Trnka, P.; Schafer, A.; Grummt, I.; Niehrs, C. GADD45A binds R-loops and recruits TET1 to CpG island promoters. Nat. Genet. 2019, 51, 217–223. [Google Scholar] [CrossRef]
- Arab, K.; Park, Y.J.; Lindroth, A.M.; Schafer, A.; Oakes, C.; Weichenhan, D.; Lukanova, A.; Lundin, E.; Risch, A.; Meister, M.; et al. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol. Cell 2014, 55, 604–614. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.; Wang, X.; Youmans, D.T.; Cech, T.R. How do lncRNAs regulate transcription? Sci. Adv. 2017, 3, eaao2110. [Google Scholar] [CrossRef]
- Kino, T.; Hurt, D.E.; Ichijo, T.; Nader, N.; Chrousos, G.P. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal 2010, 3, ra8. [Google Scholar] [CrossRef]
- Ng, S.Y.; Bogu, G.K.; Soh, B.S.; Stanton, L.W. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol. Cell 2013, 51, 349–359. [Google Scholar] [CrossRef]
- Rom, A.; Melamed, L.; Gil, N.; Goldrich, M.J.; Kadir, R.; Golan, M.; Biton, I.; Perry, R.B.; Ulitsky, I. Regulation of CHD2 expression by the Chaserr long noncoding RNA gene is essential for viability. Nat. Commun. 2019, 10, 5092. [Google Scholar] [CrossRef]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar] [CrossRef]
- Yap, K.; Mukhina, S.; Zhang, G.; Tan, J.S.C.; Ong, H.S.; Makeyev, E.V. A Short Tandem Repeat-Enriched RNA Assembles a Nuclear Compartment to Control Alternative Splicing and Promote Cell Survival. Mol. Cell 2018, 72, 525–540 e513. [Google Scholar] [CrossRef]
- Tay, Y.; Rinn, J.; Pandolfi, P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014, 505, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Grelet, S.; Link, L.A.; Howley, B.; Obellianne, C.; Palanisamy, V.; Gangaraju, V.K.; Diehl, J.A.; Howe, P.H. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat. Cell Biol. 2017, 19, 1105–1115. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.; Brangwynne, C.P. Liquid phase condensation in cell physiology and disease. Science 2017, 357, eaaf4382. [Google Scholar] [CrossRef]
- Lin, Y.; Protter, D.S.; Rosen, M.K.; Parker, R. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol. Cell 2015, 60, 208–219. [Google Scholar] [CrossRef]
- Adnane, S.; Marino, A.; Leucci, E. LncRNAs in human cancers: Signal from noise. Trends Cell Biol. 2022, 32, 565–573. [Google Scholar] [CrossRef] [PubMed]
- Clemson, C.M.; Hutchinson, J.N.; Sara, S.A.; Ensminger, A.W.; Fox, A.H.; Chess, A.; Lawrence, J.B. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 2009, 33, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Takakuwa, H.; Yamazaki, T.; Souquere, S.; Adachi, S.; Yoshino, H.; Fujiwara, N.; Yamamoto, T.; Natsume, T.; Nakagawa, S.; Pierron, G.; et al. Shell protein composition specified by the lncRNA NEAT1 domains dictates the formation of paraspeckles as distinct membraneless organelles. Nat. Cell Biol. 2023, 25, 1664–1675. [Google Scholar] [CrossRef]
- Nakagawa, S.; Shimada, M.; Yanaka, K.; Mito, M.; Arai, T.; Takahashi, E.; Fujita, Y.; Fujimori, T.; Standaert, L.; Marine, J.C.; et al. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development 2014, 141, 4618–4627. [Google Scholar] [CrossRef]
- Adriaens, C.; Standaert, L.; Barra, J.; Latil, M.; Verfaillie, A.; Kalev, P.; Boeckx, B.; Wijnhoven, P.W.; Radaelli, E.; Vermi, W.; et al. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat. Med. 2016, 22, 861–868. [Google Scholar] [CrossRef]
- Daneshvar, K.; Ardehali, M.B.; Klein, I.A.; Hsieh, F.K.; Kratkiewicz, A.J.; Mahpour, A.; Cancelliere, S.O.L.; Zhou, C.; Cook, B.M.; Li, W.; et al. lncRNA DIGIT and BRD3 protein form phase-separated condensates to regulate endoderm differentiation. Nat. Cell Biol. 2020, 22, 1211–1222. [Google Scholar] [CrossRef]
- Wu, M.; Xu, G.; Han, C.; Luan, P.F.; Xing, Y.H.; Nan, F.; Yang, L.Z.; Huang, Y.; Yang, Z.H.; Shan, L.; et al. lncRNA SLERT controls phase separation of FC/DFCs to facilitate Pol I transcription. Science 2021, 373, 547–555. [Google Scholar] [CrossRef]
- Xing, Y.H.; Yao, R.W.; Zhang, Y.; Guo, C.J.; Jiang, S.; Xu, G.; Dong, R.; Yang, L.; Chen, L.L. SLERT Regulates DDX21 Rings Associated with Pol I Transcription. Cell 2017, 169, 664–678 e616. [Google Scholar] [CrossRef] [PubMed]
- Elguindy, M.M.; Mendell, J.T. NORAD-induced Pumilio phase separation is required for genome stability. Nature 2021, 595, 303–308. [Google Scholar] [CrossRef]
- Li, R.H.; Tian, T.; Ge, Q.W.; He, X.Y.; Shi, C.Y.; Li, J.H.; Zhang, Z.; Liu, F.Z.; Sang, L.J.; Yang, Z.Z.; et al. A phosphatidic acid-binding lncRNA SNHG9 facilitates LATS1 liquid-liquid phase separation to promote oncogenic YAP signaling. Cell Res. 2021, 31, 1088–1105. [Google Scholar] [CrossRef]
- Lu, X.; Wang, J.; Wang, W.; Lu, C.; Qu, T.; He, X.; Liu, X.; Guo, R.; Zhang, E. Copy number amplification and SP1-activated lncRNA MELTF-AS1 regulates tumorigenesis by driving phase separation of YBX1 to activate ANXA8 in non-small cell lung cancer. Oncogene 2022, 41, 3222–3238. [Google Scholar] [CrossRef]
- Thapar, R.; Wang, J.L.; Hammel, M.; Ye, R.; Liang, K.; Sun, C.; Hnizda, A.; Liang, S.; Maw, S.S.; Lee, L.; et al. Mechanism of efficient double-strand break repair by a long non-coding RNA. Nucleic Acids Res. 2021, 49, 1199–1200. [Google Scholar] [CrossRef]
- Pessina, F.; Giavazzi, F.; Yin, Y.; Gioia, U.; Vitelli, V.; Galbiati, A.; Barozzi, S.; Garre, M.; Oldani, A.; Flaus, A.; et al. Functional transcription promoters at DNA double-strand breaks mediate RNA-driven phase separation of damage-response factors. Nat. Cell Biol. 2019, 21, 1286–1299. [Google Scholar] [CrossRef]
- Pandya-Jones, A.; Markaki, Y.; Serizay, J.; Chitiashvili, T.; Mancia Leon, W.R.; Damianov, A.; Chronis, C.; Papp, B.; Chen, C.K.; McKee, R.; et al. A protein assembly mediates Xist localization and gene silencing. Nature 2020, 587, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Cao, L.; Thorne, R.F.; Zhang, X.D.; Li, J.; Shao, F.; Zhang, L.; Wu, M. LncRNA GIRGL drives CAPRIN1-mediated phase separation to suppress glutaminase-1 translation under glutamine deprivation. Sci. Adv. 2021, 7, eabe5708. [Google Scholar] [CrossRef]
- Wang, C.; Duan, Y.; Duan, G.; Wang, Q.; Zhang, K.; Deng, X.; Qian, B.; Gu, J.; Ma, Z.; Zhang, S.; et al. Stress Induces Dynamic, Cytotoxicity-Antagonizing TDP-43 Nuclear Bodies via Paraspeckle LncRNA NEAT1-Mediated Liquid-Liquid Phase Separation. Mol. Cell 2020, 79, 443–458.e447. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, S.; Zhu, X.; Zhang, L.; Deng, J.; Li, F.; Guo, B.; Zhang, S.; Wu, R.; Zhang, Z.; et al. LncRNA-encoded polypeptide ASRPS inhibits triple-negative breast cancer angiogenesis. J. Exp. Med. 2020, 217, jem.20190950. [Google Scholar] [CrossRef]
- Guo, B.; Wu, S.; Zhu, X.; Zhang, L.; Deng, J.; Li, F.; Wang, Y.; Zhang, S.; Wu, R.; Lu, J.; et al. Micropeptide CIP2A-BP encoded by LINC00665 inhibits triple-negative breast cancer progression. EMBO J. 2020, 39, e102190. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.G.; Yang, J.; Zhu, Y.; Zhu, Q.; Pan, W.; Deng, S.; He, Y.; Zuo, D.; Wang, P.; Han, Y.; et al. The microprotein encoded by exosomal lncAKR1C2 promotes gastric cancer lymph node metastasis by regulating fatty acid metabolism. Cell Death Dis. 2023, 14, 708. [Google Scholar] [CrossRef]
- Zhang, Q.; Wei, T.; Yan, L.; Zhu, S.; Jin, W.; Bai, Y.; Zeng, Y.; Zhang, X.; Yin, Z.; Yang, J.; et al. Hypoxia-Responsive lncRNA AC115619 Encodes a Micropeptide That Suppresses m6A Modifications and Hepatocellular Carcinoma Progression. Cancer Res. 2023, 83, 2496–2512. [Google Scholar] [CrossRef] [PubMed]
- Tornini, V.A.; Miao, L.; Lee, H.J.; Gerson, T.; Dube, S.E.; Schmidt, V.; Kroll, F.; Tang, Y.; Du, K.; Kuchroo, M.; et al. linc-mipep and linc-wrb encode micropeptides that regulate chromatin accessibility in vertebrate-specific neural cells. eLife 2023, 12, e82249. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liao, Z.; Liu, F.; Su, C.; Zhu, H.; Li, Y.; Tao, R.; Liang, H.; Zhang, B.; Zhang, X. Long noncoding RNA HULC promotes hepatocellular carcinoma progression. Aging 2019, 11, 9111–9127. [Google Scholar] [CrossRef] [PubMed]
- Wan, T.; Wang, H.; Gou, M.; Si, H.; Wang, Z.; Yan, H.; Liu, T.; Chen, S.; Fan, R.; Qian, N.; et al. LncRNA HEIH promotes cell proliferation, migration and invasion in cholangiocarcinoma by modulating miR-98-5p/HECTD4. Biomed. Pharmacother. 2020, 125, 109916. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Chen, S.; Yang, G.; Gu, F.; Li, M.; Zhong, B.; Hu, J.; Hoffman, A.; Chen, M. Long noncoding RNA HOTAIR as an independent prognostic marker in cancer: A meta-analysis. PLoS ONE 2014, 9, e105538. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zeng, J.; Chen, W.; Fan, J.; Hylemon, P.B.; Zhou, H. Long Noncoding RNA H19: A Novel Oncogene in Liver Cancer. Noncoding RNA 2023, 9, 19. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Guo, J.; Liu, J.; Mao, X.; Xu, K. Long Non-coding RNA MALAT1: A Key Player in Liver Diseases. Front. Med. 2021, 8, 734643. [Google Scholar] [CrossRef]
- Wong, L.S.; Wong, C.M. Decoding the Roles of Long Noncoding RNAs in Hepatocellular Carcinoma. Int. J. Mol. Sci. 2021, 22, 3137. [Google Scholar] [CrossRef]
- He, Z.; Yang, D.; Fan, X.; Zhang, M.; Li, Y.; Gu, X.; Yang, M. The Roles and Mechanisms of lncRNAs in Liver Fibrosis. Int. J. Mol. Sci. 2020, 21, 1482. [Google Scholar] [CrossRef]
- Yan, C.; Chen, J.; Chen, N. Long noncoding RNA MALAT1 promotes hepatic steatosis and insulin resistance by increasing nuclear SREBP-1c protein stability. Sci. Rep. 2016, 6, 22640. [Google Scholar] [CrossRef]
- Xiong, H.; Ni, Z.; He, J.; Jiang, S.; Li, X.; He, J.; Gong, W.; Zheng, L.; Chen, S.; Li, B.; et al. LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene 2017, 36, 3528–3540. [Google Scholar] [CrossRef] [PubMed]
- Unfried, J.P.; Fortes, P. LncRNAs in HCV Infection and HCV-Related Liver Disease. Int. J. Mol. Sci. 2020, 21, 2255. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xu, L.; Lu, B.; Zhao, M.; Li, L.; Sun, W.; Qiu, Z.; Zhang, B. LncRNA H19/microRNA-675/PPARalpha axis regulates liver cell injury and energy metabolism remodelling induced by hepatitis B X protein via Akt/mTOR signalling. Mol. Immunol. 2019, 116, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Kogo, R.; Shimamura, T.; Mimori, K.; Kawahara, K.; Imoto, S.; Sudo, T.; Tanaka, F.; Shibata, K.; Suzuki, A.; Komune, S.; et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011, 71, 6320–6326. [Google Scholar] [CrossRef]
- Verma, S.; Sahu, B.D.; Mugale, M.N. Role of lncRNAs in hepatocellular carcinoma. Life Sci. 2023, 325, 121751. [Google Scholar] [CrossRef] [PubMed]
- Ji, P.; Diederichs, S.; Wang, W.; Boing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003, 22, 8031–8041. [Google Scholar] [CrossRef]
- Gutschner, T.; Hammerle, M.; Eissmann, M.; Hsu, J.; Kim, Y.; Hung, G.; Revenko, A.; Arun, G.; Stentrup, M.; Gross, M.; et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013, 73, 1180–1189. [Google Scholar] [CrossRef]
- Huang, Z.; Zhou, J.K.; Peng, Y.; He, W.; Huang, C. The role of long noncoding RNAs in hepatocellular carcinoma. Mol. Cancer 2020, 19, 77. [Google Scholar] [CrossRef]
- Panzitt, K.; Tschernatsch, M.M.; Guelly, C.; Moustafa, T.; Stradner, M.; Strohmaier, H.M.; Buck, C.R.; Denk, H.; Schroeder, R.; Trauner, M.; et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology 2007, 132, 330–342. [Google Scholar] [CrossRef]
- Xie, H.; Ma, H.; Zhou, D. Plasma HULC as a promising novel biomarker for the detection of hepatocellular carcinoma. BioMed Res. Int. 2013, 2013, 136106. [Google Scholar] [CrossRef]
- Tu, Z.Q.; Li, R.J.; Mei, J.Z.; Li, X.H. Down-regulation of long non-coding RNA GAS5 is associated with the prognosis of hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 2014, 7, 4303–4309. [Google Scholar] [PubMed]
- Wang, W.; Zhou, R.; Wu, Y.; Liu, Y.; Su, W.; Xiong, W.; Zeng, Z. PVT1 Promotes Cancer Progression via MicroRNAs. Front. Oncol. 2019, 9, 609. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Yu, Y.; Li, H.; Hu, Q.; Chen, X.; He, Y.; Xue, C.; Ren, F.; Ren, Z.; Li, J.; et al. Long non-coding RNA PVT1 promotes tumor progression by regulating the miR-143/HK2 axis in gallbladder cancer. Mol. Cancer 2019, 18, 33. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ren, H. Long noncoding RNA PVT1 promotes tumor cell proliferation, invasion, migration and inhibits apoptosis in oral squamous cell carcinoma by regulating miR-150-5p/GLUT-1. Oncol. Rep. 2020, 44, 1524–1538. [Google Scholar] [CrossRef] [PubMed]
- Quagliata, L.; Matter, M.S.; Piscuoglio, S.; Arabi, L.; Ruiz, C.; Procino, A.; Kovac, M.; Moretti, F.; Makowska, Z.; Boldanova, T.; et al. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology 2014, 59, 911–923. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Unfried, J.P.; Sangro, P.; Prats-Mari, L.; Sangro, B.; Fortes, P. The Landscape of lncRNAs in Hepatocellular Carcinoma: A Translational Perspective. Cancers 2021, 13, 2651. [Google Scholar] [CrossRef]
- Zhang, Y.; Lin, S.; Yang, X.; Zhang, X. Prognostic and Clinicopathological Significance of lncRNA MVIH in Cancer Patients. J. Cancer 2019, 10, 1503–1510. [Google Scholar] [CrossRef]
- Abbastabar, M.; Sarfi, M.; Golestani, A.; Khalili, E. lncRNA involvement in hepatocellular carcinoma metastasis and prognosis. EXCLI J. 2018, 17, 900–913. [Google Scholar] [CrossRef]
- Matouk, I.J.; DeGroot, N.; Mezan, S.; Ayesh, S.; Abu-lail, R.; Hochberg, A.; Galun, E. The H19 non-coding RNA is essential for human tumor growth. PLoS ONE 2007, 2, e845. [Google Scholar] [CrossRef]
- Zhao, H.; Peng, R.; Liu, Q.; Liu, D.; Du, P.; Yuan, J.; Peng, G.; Liao, Y. The lncRNA H19 interacts with miR-140 to modulate glioma growth by targeting iASPP. Arch. Biochem. Biophys. 2016, 610, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Liu, S.; Ye, F.; Shen, Y.; Tie, Y.; Zhu, J.; Wei, L.; Jin, Y.; Fu, H.; Wu, Y.; et al. Long Noncoding RNA MEG3 Interacts with p53 Protein and Regulates Partial p53 Target Genes in Hepatoma Cells. PLoS ONE 2015, 10, e0139790. [Google Scholar] [CrossRef] [PubMed]
- He, J.H.; Han, Z.P.; Liu, J.M.; Zhou, J.B.; Zou, M.X.; Lv, Y.B.; Li, Y.G.; Cao, M.R. Overexpression of Long Non-Coding RNA MEG3 Inhibits Proliferation of Hepatocellular Carcinoma Huh7 Cells via Negative Modulation of miRNA-664. J. Cell Biochem. 2017, 118, 3713–3721. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.J.; Xie, S.L.; Li, Q.; Ma, J.; Wang, G.Y. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J. Int. Med. Res. 2011, 39, 2119–2128. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.M.; Zhu, X.; Wang, W.M.; Lu, Y.F.; Hu, B.G.; Wang, H.; Liang, W.C.; Wang, S.S.; Ko, C.H.; Waye, M.M.; et al. Hotair mediates hepatocarcinogenesis through suppressing miRNA-218 expression and activating P14 and P16 signaling. J. Hepatol. 2015, 63, 886–895. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Xing, Z.; Mani, S.K.; Bancel, B.; Durantel, D.; Zoulim, F.; Tran, E.J.; Merle, P.; Andrisani, O. RNA helicase DEAD box protein 5 regulates Polycomb repressive complex 2/Hox transcript antisense intergenic RNA function in hepatitis B virus infection and hepatocarcinogenesis. Hepatology 2016, 64, 1033–1048. [Google Scholar] [CrossRef]
- Yang, L.; Peng, X.; Li, Y.; Zhang, X.; Ma, Y.; Wu, C.; Fan, Q.; Wei, S.; Li, H.; Liu, J. Long non-coding RNA HOTAIR promotes exosome secretion by regulating RAB35 and SNAP23 in hepatocellular carcinoma. Mol. Cancer 2019, 18, 78. [Google Scholar] [CrossRef]
- Li, H.; An, J.; Wu, M.; Zheng, Q.; Gui, X.; Li, T.; Pu, H.; Lu, D. LncRNA HOTAIR promotes human liver cancer stem cell malignant growth through downregulation of SETD2. Oncotarget 2015, 6, 27847–27864. [Google Scholar] [CrossRef]
- Su, D.N.; Wu, S.P.; Chen, H.T.; He, J.H. HOTAIR, a long non-coding RNA driver of malignancy whose expression is activated by FOXC1, negatively regulates miRNA-1 in hepatocellular carcinoma. Oncol. Lett. 2016, 12, 4061–4067. [Google Scholar] [CrossRef]
- Kong, J.H.; Qiu, Y.J.; Li, Y.; Zhang, H.J.; Wang, W.P. TGF-β1 elevates P-gp and BCRP in hepatocellular carcinoma through HOTAIR/miR-145 axis. Biopharm. Drug Dispos. 2019, 40, 70–80. [Google Scholar] [CrossRef]
- Cheng, D.; Deng, J.G.; Zhang, B.; He, X.Y.; Meng, Z.; Li, G.L.; Ye, H.L.; Zheng, S.Y.; Wei, L.S.; Deng, X.G.; et al. LncRNA HOTAIR epigenetically suppresses miR-122 expression in hepatocellular carcinoma via DNA methylation. EBioMedicine 2018, 36, 159–170. [Google Scholar] [CrossRef]
- Ding, C.; Cheng, S.; Yang, Z.; Lv, Z.; Xiao, H.; Du, C.; Peng, C.; Xie, H.; Zhou, L.; Wu, J.; et al. Long non-coding RNA HOTAIR promotes cell migration and invasion via down-regulation of RNA binding motif protein 38 in hepatocellular carcinoma cells. Int. J. Mol. Sci. 2014, 15, 4060–4076. [Google Scholar] [CrossRef]
- Zhou, D.; Wang, Y.; Hu, H.; Liu, H.; Deng, J.; Li, L.; Zheng, C. lncRNA MALAT1 promotes HCC metastasis through the peripheral vascular infiltration via miRNA-613: A primary study using contrast ultrasound. World J. Surg. Oncol. 2022, 20, 203. [Google Scholar] [CrossRef]
- Sasaki, Y.T.; Ideue, T.; Sano, M.; Mituyama, T.; Hirose, T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc. Natl. Acad. Sci. USA 2009, 106, 2525–2530. [Google Scholar] [CrossRef]
- Malakar, P.; Shilo, A.; Mogilevsky, A.; Stein, I.; Pikarsky, E.; Nevo, Y.; Benyamini, H.; Elgavish, S.; Zong, X.; Prasanth, K.V.; et al. Long Noncoding RNA MALAT1 Promotes Hepatocellular Carcinoma Development by SRSF1 Upregulation and mTOR Activation. Cancer Res. 2017, 77, 1155–1167. [Google Scholar] [CrossRef]
- Malakar, P.; Stein, I.; Saragovi, A.; Winkler, R.; Stern-Ginossar, N.; Berger, M.; Pikarsky, E.; Karni, R. Long Noncoding RNA MALAT1 Regulates Cancer Glucose Metabolism by Enhancing mTOR-Mediated Translation of TCF7L2. Cancer Res. 2019, 79, 2480–2493. [Google Scholar] [CrossRef]
- Liu, D.; Zhu, Y.; Pang, J.; Weng, X.; Feng, X.; Guo, Y. Knockdown of long non-coding RNA MALAT1 inhibits growth and motility of human hepatoma cells via modulation of miR-195. J. Cell Biochem. 2018, 119, 1368–1380. [Google Scholar] [CrossRef]
- Chen, L.; Yao, H.; Wang, K.; Liu, X. Long Non-Coding RNA MALAT1 Regulates ZEB1 Expression by Sponging miR-143-3p and Promotes Hepatocellular Carcinoma Progression. J. Cell Biochem. 2017, 118, 4836–4843. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Miao, R.; Liu, S.; Wan, Y.; Zhang, S.; Deng, Y.; Bi, J.; Qu, K.; Zhang, J.; Liu, C. Down-regulation of miR-146b-5p by long noncoding RNA MALAT1 in hepatocellular carcinoma promotes cancer growth and metastasis. Oncotarget 2017, 8, 28683–28695. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wang, G.; Tao, K.; Cai, K.; Wu, K.; Ye, L.; Bai, J.; Yin, Y.; Wang, J.; Shuai, X.; et al. Long noncoding RNA metastasis-associated lung adenocarcinoma transcript 1 cooperates with enhancer of zeste homolog 2 to promote hepatocellular carcinoma development by modulating the microRNA-22/Snail family transcriptional repressor 1 axis. Cancer Sci. 2020, 111, 1582–1595. [Google Scholar] [CrossRef]
- Pan, Y.; Tong, S.; Cui, R.; Fan, J.; Liu, C.; Lin, Y.; Tang, J.; Xie, H.; Lin, P.; Zheng, T.; et al. Long Non-Coding MALAT1 Functions as a Competing Endogenous RNA to Regulate Vimentin Expression by Sponging miR-30a-5p in Hepatocellular Carcinoma. Cell Physiol. Biochem. 2018, 50, 108–120. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhou, L.; Li, H.; Sun, T.; Wen, X.; Li, X.; Meng, Y.; Li, Y.; Liu, M.; Liu, S.; et al. Nuclear-Encoded lncRNA MALAT1 Epigenetically Controls Metabolic Reprogramming in HCC Cells through the Mitophagy Pathway. Mol. Ther. Nucleic Acids 2021, 23, 264–276. [Google Scholar] [CrossRef]
- Li, S.P.; Xu, H.X.; Yu, Y.; He, J.D.; Wang, Z.; Xu, Y.J.; Wang, C.Y.; Zhang, H.M.; Zhang, R.X.; Zhang, J.J.; et al. LncRNA HULC enhances epithelial-mesenchymal transition to promote tumorigenesis and metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1 signaling pathway. Oncotarget 2016, 7, 42431–42446. [Google Scholar] [CrossRef]
- Lu, Z.; Xiao, Z.; Liu, F.; Cui, M.; Li, W.; Yang, Z.; Li, J.; Ye, L.; Zhang, X. Long non-coding RNA HULC promotes tumor angiogenesis in liver cancer by up-regulating sphingosine kinase 1 (SPHK1). Oncotarget 2016, 7, 241–254. [Google Scholar] [CrossRef]
- Xin, X.; Wu, M.; Meng, Q.; Wang, C.; Lu, Y.; Yang, Y.; Li, X.; Zheng, Q.; Pu, H.; Gui, X.; et al. Long noncoding RNA HULC accelerates liver cancer by inhibiting PTEN via autophagy cooperation to miR15a. Mol. Cancer 2018, 17, 94. [Google Scholar] [CrossRef]
- Du, Y.; Kong, G.; You, X.; Zhang, S.; Zhang, T.; Gao, Y.; Ye, L.; Zhang, X. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J. Biol. Chem. 2012, 287, 26302–26311. [Google Scholar] [CrossRef]
- Cui, M.; Xiao, Z.; Wang, Y.; Zheng, M.; Song, T.; Cai, X.; Sun, B.; Ye, L.; Zhang, X. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res. 2015, 75, 846–857. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.; Zheng, M.; Sun, B.; Wang, Y.; Ye, L.; Zhang, X. A long noncoding RNA perturbs the circadian rhythm of hepatoma cells to facilitate hepatocarcinogenesis. Neoplasia 2015, 17, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Tseng, Y.Y.; Moriarity, B.S.; Gong, W.; Akiyama, R.; Tiwari, A.; Kawakami, H.; Ronning, P.; Reuland, B.; Guenther, K.; Beadnell, T.C.; et al. PVT1 dependence in cancer with MYC copy-number increase. Nature 2014, 512, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; Yang, Z.; Lv, Z.; Du, C.; Xiao, H.; Peng, C.; Cheng, S.; Xie, H.; Zhou, L.; Wu, J.; et al. Long non-coding RNA PVT1 is associated with tumor progression and predicts recurrence in hepatocellular carcinoma patients. Oncol. Lett. 2015, 9, 955–963. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Yuan, J.H.; Wang, S.B.; Yang, F.; Yuan, S.X.; Ye, C.; Yang, N.; Zhou, W.P.; Li, W.L.; Li, W.; et al. Oncofetal long noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. Hepatology 2014, 60, 1278–1290. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Peng, X.; Jin, H.; Liu, J. Long non-coding RNA PVT1 promotes autophagy as ceRNA to target ATG3 by sponging microRNA-365 in hepatocellular carcinoma. Gene 2019, 697, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Luo, X.; He, W.; Chen, G.; Li, Y.; Li, W.; Wang, X.; Lai, Y.; Ye, Y. Long Non-Coding RNA PVT1/miR-150/ HIG2 Axis Regulates the Proliferation, Invasion and the Balance of Iron Metabolism of Hepatocellular Carcinoma. Cell Physiol. Biochem. 2018, 49, 1403–1419. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.; Liu, J.; Liu, B.; Zeng, Y.; Chen, P.; Su, Y. Long noncoding RNA PVT1 inhibits interferon-alpha mediated therapy for hepatocellular carcinoma cells by interacting with signal transducer and activator of transcription 1. Biochem. Biophys. Res. Commun. 2018, 500, 973–980. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Yan, X.; Jin, Y.; Yang, X.; Yu, X.; Zhou, L.; Han, S.; Yuan, Q.; Yang, M. MiRNA-192 [corrected] and miRNA-204 Directly Suppress lncRNA HOTTIP and Interrupt GLS1-Mediated Glutaminolysis in Hepatocellular Carcinoma. PLoS Genet. 2015, 11, e1005726. [Google Scholar] [CrossRef]
- Wei, H.; Xu, Z.; Chen, L.; Wei, Q.; Huang, Z.; Liu, G.; Li, W.; Wang, J.; Tang, Q.; Pu, J. Long non-coding RNA PAARH promotes hepatocellular carcinoma progression and angiogenesis via upregulating HOTTIP and activating HIF-1alpha/VEGF signaling. Cell Death Dis. 2022, 13, 102. [Google Scholar] [CrossRef]
- Wang, Y.; Hylemon, P.B.; Zhou, H. Long Noncoding RNA H19: A Key Player in Liver Diseases. Hepatology 2021, 74, 1652–1659. [Google Scholar] [CrossRef]
- Ye, Y.; Guo, J.; Xiao, P.; Ning, J.; Zhang, R.; Liu, P.; Yu, W.; Xu, L.; Zhao, Y.; Yu, J. Macrophages-induced long noncoding RNA H19 up-regulation triggers and activates the miR-193b/MAPK1 axis and promotes cell aggressiveness in hepatocellular carcinoma. Cancer Lett. 2020, 469, 310–322. [Google Scholar] [CrossRef]
- Zhang, J.; Han, C.; Ungerleider, N.; Chen, W.; Song, K.; Wang, Y.; Kwon, H.; Ma, W.; Wu, T. A Transforming Growth Factor-beta and H19 Signaling Axis in Tumor-Initiating Hepatocytes That Regulates Hepatic Carcinogenesis. Hepatology 2019, 69, 1549–1563. [Google Scholar] [CrossRef]
- Huang, Z.; Chu, L.; Liang, J.; Tan, X.; Wang, Y.; Wen, J.; Chen, J.; Wu, Y.; Liu, S.; Liao, J.; et al. H19 Promotes HCC Bone Metastasis Through Reducing Osteoprotegerin Expression in a Protein Phosphatase 1 Catalytic Subunit Alpha/p38 Mitogen-Activated Protein Kinase-Dependent Manner and Sponging microRNA 200b-3p. Hepatology 2021, 74, 214–232. [Google Scholar] [CrossRef]
- Souquere, S.; Beauclair, G.; Harper, F.; Fox, A.; Pierron, G. Highly ordered spatial organization of the structural long noncoding NEAT1 RNAs within paraspeckle nuclear bodies. Mol. Biol. Cell 2010, 21, 4020–4027. [Google Scholar] [CrossRef]
- West, J.A.; Davis, C.P.; Sunwoo, H.; Simon, M.D.; Sadreyev, R.I.; Wang, P.I.; Tolstorukov, M.Y.; Kingston, R.E. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell 2014, 55, 791–802. [Google Scholar] [CrossRef]
- Slack, F.J.; Chinnaiyan, A.M. The Role of Non-coding RNAs in Oncology. Cell 2019, 179, 1033–1055. [Google Scholar] [CrossRef]
- Zhang, X.N.; Zhou, J.; Lu, X.J. The long noncoding RNA NEAT1 contributes to hepatocellular carcinoma development by sponging miR-485 and enhancing the expression of the STAT3. J. Cell Physiol. 2018, 233, 6733–6741. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhou, Y.; Yang, L.; Ma, Y.; Peng, X.; Yang, S.; Li, H.; Liu, J. LncRNA NEAT1 promotes autophagy via regulating miR-204/ATG3 and enhanced cell resistance to sorafenib in hepatocellular carcinoma. J. Cell Physiol. 2020, 235, 3402–3413. [Google Scholar] [CrossRef]
- Guo, S.; Chen, W.; Luo, Y.; Ren, F.; Zhong, T.; Rong, M.; Dang, Y.; Feng, Z.; Chen, G. Clinical implication of long non-coding RNA NEAT1 expression in hepatocellular carcinoma patients. Int. J. Clin. Exp. Pathol. 2015, 8, 5395–5402. [Google Scholar]
- Tu, J.; Zhao, Z.; Xu, M.; Lu, X.; Chang, L.; Ji, J. NEAT1 upregulates TGF-beta1 to induce hepatocellular carcinoma progression by sponging hsa-mir-139-5p. J. Cell Physiol. 2018, 233, 8578–8587. [Google Scholar] [CrossRef]
- Zhang, H.; Su, X.; Burley, S.K.; Zheng, X.F.S. mTOR regulates aerobic glycolysis through NEAT1 and nuclear paraspeckle-mediated mechanism in hepatocellular carcinoma. Theranostics 2022, 12, 3518–3533. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Zhang, L.; Huo, X.S.; Yuan, J.H.; Xu, D.; Yuan, S.X.; Zhu, N.; Zhou, W.P.; Yang, G.S.; Wang, Y.Z.; et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology 2011, 54, 1679–1689. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Yang, X.; Qi, Q.; Gao, Y.; Wei, Q.; Han, S. lncRNA-HEIH in serum and exosomes as a potential biomarker in the HCV-related hepatocellular carcinoma. Cancer Biomark. 2018, 21, 651–659. [Google Scholar] [CrossRef]
- Shen, Q.; Jiang, S.; Wu, M.; Zhang, L.; Su, X.; Zhao, D. LncRNA HEIH Confers Cell Sorafenib Resistance in Hepatocellular Carcinoma by Regulating miR-98-5p/PI3K/AKT Pathway. Cancer Manag. Res. 2020, 12, 6585–6595. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Zhang, T.; Zhang, D.; Xie, L.; Zou, X.; Lei, L.; Wu, D.; Liu, L. The long non-coding RNA, SNHG6-003, functions as a competing endogenous RNA to promote the progression of hepatocellular carcinoma. Oncogene 2017, 36, 1112–1122. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Yuan, Y.; Li, C.; Guo, T.; Qi, H.; Xiao, Y.; Dong, X.; Liu, Z.; Liu, Q. Upregulation of SNHG6 regulates ZEB1 expression by competitively binding miR-101-3p and interacting with UPF1 in hepatocellular carcinoma. Cancer Lett. 2016, 383, 183–194. [Google Scholar] [CrossRef]
- Wu, G.; Ju, X.; Wang, Y.; Li, Z.; Gan, X. Up-regulation of SNHG6 activates SERPINH1 expression by competitive binding to miR-139-5p to promote hepatocellular carcinoma progression. Cell Cycle 2019, 18, 1849–1867. [Google Scholar] [CrossRef]
- Chen, S.; Xie, C.; Hu, X. lncRNA SNHG6 functions as a ceRNA to up-regulate c-Myc expression via sponging let-7c-5p in hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2019, 519, 901–908. [Google Scholar] [CrossRef]
- Fan, X.; Zhao, Z.; Song, J.; Zhang, D.; Wu, F.; Tu, J.; Xu, M.; Ji, J. LncRNA-SNHG6 promotes the progression of hepatocellular carcinoma by targeting miR-6509-5p and HIF1A. Cancer Cell Int. 2021, 21, 150. [Google Scholar] [CrossRef]
- Liu, F.; Tian, T.; Zhang, Z.; Xie, S.; Yang, J.; Zhu, L.; Wang, W.; Shi, C.; Sang, L.; Guo, K.; et al. Long non-coding RNA SNHG6 couples cholesterol sensing with mTORC1 activation in hepatocellular carcinoma. Nat. Metab. 2022, 4, 1022–1040. [Google Scholar] [CrossRef]
- Cao, C.; Sun, J.; Zhang, D.; Guo, X.; Xie, L.; Li, X.; Wu, D.; Liu, L. The long intergenic noncoding RNA UFC1, a target of MicroRNA 34a, interacts with the mRNA stabilizing protein HuR to increase levels of beta-catenin in HCC cells. Gastroenterology 2015, 148, 415–426 e418. [Google Scholar] [CrossRef]
- Zhu, P.; Wang, Y.; Huang, G.; Ye, B.; Liu, B.; Wu, J.; Du, Y.; He, L.; Fan, Z. lnc-beta-Catm elicits EZH2-dependent beta-catenin stabilization and sustains liver CSC self-renewal. Nat. Struct. Mol. Biol. 2016, 23, 631–639. [Google Scholar] [CrossRef]
- Yuan, S.X.; Wang, J.; Yang, F.; Tao, Q.F.; Zhang, J.; Wang, L.L.; Yang, Y.; Liu, H.; Wang, Z.G.; Xu, Q.G.; et al. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology 2016, 63, 499–511. [Google Scholar] [CrossRef]
- Chen, M.; Zhang, C.; Liu, W.; Du, X.; Liu, X.; Xing, B. Long noncoding RNA LINC01234 promotes hepatocellular carcinoma progression through orchestrating aspartate metabolic reprogramming. Mol. Ther. 2022, 30, 2354–2369. [Google Scholar] [CrossRef]
- Xu, K.; Xia, P.; Gongye, X.; Zhang, X.; Ma, S.; Chen, Z.; Zhang, H.; Liu, J.; Liu, Y.; Guo, Y.; et al. A novel lncRNA RP11-386G11.10 reprograms lipid metabolism to promote hepatocellular carcinoma progression. Mol. Metab. 2022, 63, 101540. [Google Scholar] [CrossRef]
- Sondergaard, J.N.; Sommerauer, C.; Atanasoai, I.; Hinte, L.C.; Geng, K.; Guiducci, G.; Brautigam, L.; Aouadi, M.; Stojic, L.; Barragan, I.; et al. CCT3-LINC00326 axis regulates hepatocarcinogenic lipid metabolism. Gut 2022, 71, 2081–2092. [Google Scholar] [CrossRef]
- Lin, Y.H.; Wu, M.H.; Huang, Y.H.; Yeh, C.T.; Cheng, M.L.; Chi, H.C.; Tsai, C.Y.; Chung, I.H.; Chen, C.Y.; Lin, K.H. Taurine up-regulated gene 1 functions as a master regulator to coordinate glycolysis and metastasis in hepatocellular carcinoma. Hepatology 2018, 67, 188–203. [Google Scholar] [CrossRef]
- Huang, M.D.; Chen, W.M.; Qi, F.Z.; Sun, M.; Xu, T.P.; Ma, P.; Shu, Y.Q. Long non-coding RNA TUG1 is up-regulated in hepatocellular carcinoma and promotes cell growth and apoptosis by epigenetically silencing of KLF2. Mol. Cancer 2015, 14, 165. [Google Scholar] [CrossRef]
- Huang, M.D.; Chen, W.M.; Qi, F.Z.; Xia, R.; Sun, M.; Xu, T.P.; Yin, L.; Zhang, E.B.; De, W.; Shu, Y.Q. Long non-coding RNA ANRIL is upregulated in hepatocellular carcinoma and regulates cell apoptosis by epigenetic silencing of KLF2. J. Hematol. Oncol. 2015, 8, 50. [Google Scholar] [CrossRef]
- Yuan, J.H.; Yang, F.; Wang, F.; Ma, J.Z.; Guo, Y.J.; Tao, Q.F.; Liu, F.; Pan, W.; Wang, T.T.; Zhou, C.C.; et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 2014, 25, 666–681. [Google Scholar] [CrossRef]
- Qin, M.; Meng, Y.; Luo, C.; He, S.; Qin, F.; Yin, Y.; Huang, J.; Zhao, H.; Hu, J.; Deng, Z.; et al. lncRNA PRR34-AS1 promotes HCC development via modulating Wnt/beta-catenin pathway by absorbing miR-296-5p and upregulating E2F2 and SOX12. Mol. Ther. Nucleic Acids 2021, 25, 37–52. [Google Scholar] [CrossRef]
- Li, T.; Xie, J.; Shen, C.; Cheng, D.; Shi, Y.; Wu, Z.; Deng, X.; Chen, H.; Shen, B.; Peng, C.; et al. Amplification of Long Noncoding RNA ZFAS1 Promotes Metastasis in Hepatocellular Carcinoma. Cancer Res. 2015, 75, 3181–3191. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.Z.; Li, F.; Cheng, S.T.; Xu, Y.; Deng, H.J.; Gu, D.Y.; Wang, J.; Chen, W.X.; Zhou, Y.J.; Yang, M.L.; et al. DDX17-regulated alternative splicing that produced an oncogenic isoform of PXN-AS1 to promote HCC metastasis. Hepatology 2022, 75, 847–865. [Google Scholar] [CrossRef]
- Yuan, J.H.; Liu, X.N.; Wang, T.T.; Pan, W.; Tao, Q.F.; Zhou, W.P.; Wang, F.; Sun, S.H. The MBNL3 splicing factor promotes hepatocellular carcinoma by increasing PXN expression through the alternative splicing of lncRNA-PXN-AS1. Nat. Cell Biol. 2017, 19, 820–832. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.X.; Yang, F.; Yang, Y.; Tao, Q.F.; Zhang, J.; Huang, G.; Yang, Y.; Wang, R.Y.; Yang, S.; Huo, X.S.; et al. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients’ poor recurrence-free survival after hepatectomy. Hepatology 2012, 56, 2231–2241. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Liu, S.; Cheng, Y.; Lu, L.; Shi, J.; Xu, G.; Li, N.; Cheng, K.; Wu, M.; Cheng, S.; et al. ICAM-1-Related Noncoding RNA in Cancer Stem Cells Maintains ICAM-1 Expression in Hepatocellular Carcinoma. Clin. Cancer Res. 2016, 22, 2041–2050. [Google Scholar] [CrossRef] [PubMed]
- Klingenberg, M.; Gross, M.; Goyal, A.; Polycarpou-Schwarz, M.; Miersch, T.; Ernst, A.S.; Leupold, J.; Patil, N.; Warnken, U.; Allgayer, H.; et al. The Long Noncoding RNA Cancer Susceptibility 9 and RNA Binding Protein Heterogeneous Nuclear Ribonucleoprotein L Form a Complex and Coregulate Genes Linked to AKT Signaling. Hepatology 2018, 68, 1817–1832. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Huan, L.; Wang, J.; Wu, Y.; Huang, S.; He, X. LncRNA RP11-295G20.2 regulates hepatocellular carcinoma cell growth and autophagy by targeting PTEN to lysosomal degradation. Cell Discov. 2021, 7, 118. [Google Scholar] [CrossRef]
- Unfried, J.P.; Marin-Baquero, M.; Rivera-Calzada, A.; Razquin, N.; Martin-Cuevas, E.M.; de Braganca, S.; Aicart-Ramos, C.; McCoy, C.; Prats-Mari, L.; Arribas-Bosacoma, R.; et al. Long Noncoding RNA NIHCOLE Promotes Ligation Efficiency of DNA Double-Strand Breaks in Hepatocellular Carcinoma. Cancer Res. 2021, 81, 4910–4925. [Google Scholar] [CrossRef]
- Xia, A.; Yuan, W.; Wang, Q.; Xu, J.; Gu, Y.; Zhang, L.; Chen, C.; Wang, Z.; Wu, D.; He, Q.; et al. The cancer-testis lncRNA lnc-CTHCC promotes hepatocellular carcinogenesis by binding hnRNP K and activating YAP1 transcription. Nat. Cancer 2022, 3, 203–218. [Google Scholar] [CrossRef]
- Yu, A.T.; Berasain, C.; Bhatia, S.; Rivera, K.; Liu, B.; Rigo, F.; Pappin, D.J.; Spector, D.L. PHAROH lncRNA regulates Myc translation in hepatocellular carcinoma via sequestering TIAR. eLife 2021, 10, e68263. [Google Scholar] [CrossRef]
- Tang, X.; Feng, D.; Li, M.; Zhou, J.; Li, X.; Zhao, D.; Hao, B.; Li, D.; Ding, K. Transcriptomic Analysis of mRNA-lncRNA-miRNA Interactions in Hepatocellular Carcinoma. Sci. Rep. 2019, 9, 16096. [Google Scholar] [CrossRef]
- Anwar, S.L.; Krech, T.; Hasemeier, B.; Schipper, E.; Schweitzer, N.; Vogel, A.; Kreipe, H.; Lehmann, U. Loss of imprinting and allelic switching at the DLK1-MEG3 locus in human hepatocellular carcinoma. PLoS ONE 2012, 7, e49462. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, J.; Lv, Y.; Zhang, C.; Guo, S. LncRNA meg3 suppresses hepatocellular carcinoma in vitro and vivo studies. Am. J. Transl. Res. 2019, 11, 4089–4099. [Google Scholar]
- Chang, L.; Wang, G.; Jia, T.; Zhang, L.; Li, Y.; Han, Y.; Zhang, K.; Lin, G.; Zhang, R.; Li, J.; et al. Armored long non-coding RNA MEG3 targeting EGFR based on recombinant MS2 bacteriophage virus-like particles against hepatocellular carcinoma. Oncotarget 2016, 7, 23988–24004. [Google Scholar] [CrossRef]
- Chang, L.; Li, C.; Lan, T.; Wu, L.; Yuan, Y.; Liu, Q.; Liu, Z. Decreased expression of long non-coding RNA GAS5 indicates a poor prognosis and promotes cell proliferation and invasion in hepatocellular carcinoma by regulating vimentin. Mol. Med. Rep. 2016, 13, 1541–1550. [Google Scholar] [CrossRef]
- Gao, X.; Lu, C.; Liu, Z.; Lin, Y.; Huang, J.; Lu, L.; Li, S.; Huang, X.; Tang, M.; Huang, S.; et al. RBM38 Reverses Sorafenib Resistance in Hepatocellular Carcinoma Cells by Combining and Promoting lncRNA-GAS5. Cancers 2023, 15, 2897. [Google Scholar] [CrossRef]
- Zhang, W.Y.; Zhan, H.L.; Li, M.K.; Wu, G.D.; Liu, Z.; Wu, L.F. Long noncoding RNA Gas5 induces cell apoptosis and inhibits tumor growth via activating the CHOP-dependent endoplasmic reticulum stress pathway in human hepatoblastoma HepG2 cells. J. Cell Biochem. 2022, 123, 231–247. [Google Scholar] [CrossRef]
- Zhao, P.; Cui, X.; Zhao, L.; Liu, L.; Wang, D. Overexpression of Growth-Arrest-Specific Transcript 5 Improved Cisplatin Sensitivity in Hepatocellular Carcinoma Through Sponging miR-222. DNA Cell Biol. 2020, 39, 724–732. [Google Scholar] [CrossRef]
- Wang, B.; Xian, J.; Zang, J.; Xiao, L.; Li, Y.; Sha, M.; Shen, M. Long non-coding RNA FENDRR inhibits proliferation and invasion of hepatocellular carcinoma by down-regulating glypican-3 expression. Biochem. Biophys. Res. Commun. 2019, 509, 143–147. [Google Scholar] [CrossRef]
- Grote, P.; Wittler, L.; Hendrix, D.; Koch, F.; Wahrisch, S.; Beisaw, A.; Macura, K.; Blass, G.; Kellis, M.; Werber, M.; et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell 2013, 24, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Zhao, H.; Feng, X.; Li, H.; Qiu, C.; Yi, X.; Tang, H.; Zhang, J. Long Non-coding RNA FENDRR Acts as a miR-423-5p Sponge to Suppress the Treg-Mediated Immune Escape of Hepatocellular Carcinoma Cells. Mol. Ther. Nucleic Acids 2019, 17, 516–529. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Sun, W.; Shen, W.; Xia, M.; Chen, C.; Xiang, D.; Ning, B.; Cui, X.; Li, H.; Li, X.; et al. Long non-coding RNA DILC regulates liver cancer stem cells via IL-6/STAT3 axis. J. Hepatol. 2016, 64, 1283–1294. [Google Scholar] [CrossRef]
- Zhou, C.C.; Yang, F.; Yuan, S.X.; Ma, J.Z.; Liu, F.; Yuan, J.H.; Bi, F.R.; Lin, K.Y.; Yin, J.H.; Cao, G.W.; et al. Systemic genome screening identifies the outcome associated focal loss of long noncoding RNA PRAL in hepatocellular carcinoma. Hepatology 2016, 63, 850–863. [Google Scholar] [CrossRef]
- Lin, Y.H.; Wu, M.H.; Liu, Y.C.; Lyu, P.C.; Yeh, C.T.; Lin, K.H. LINC01348 suppresses hepatocellular carcinoma metastasis through inhibition of SF3B3-mediated EZH2 pre-mRNA splicing. Oncogene 2021, 40, 4675–4685. [Google Scholar] [CrossRef]
- Ni, W.; Zhang, Y.; Zhan, Z.; Ye, F.; Liang, Y.; Huang, J.; Chen, K.; Chen, L.; Ding, Y. A novel lncRNA uc.134 represses hepatocellular carcinoma progression by inhibiting CUL4A-mediated ubiquitination of LATS1. J. Hematol. Oncol. 2017, 10, 91. [Google Scholar] [CrossRef]
- Liu, F.; Yuan, J.H.; Huang, J.F.; Yang, F.; Wang, T.T.; Ma, J.Z.; Zhang, L.; Zhou, C.C.; Wang, F.; Yu, J.; et al. Long noncoding RNA FTX inhibits hepatocellular carcinoma proliferation and metastasis by binding MCM2 and miR-374a. Oncogene 2016, 35, 5422–5434. [Google Scholar] [CrossRef]
- Song, W.; Zheng, C.; Liu, M.; Xu, Y.; Qian, Y.; Zhang, Z.; Su, H.; Li, X.; Wu, H.; Gong, P.; et al. TRERNA1 upregulation mediated by HBx promotes sorafenib resistance and cell proliferation in HCC via targeting NRAS by sponging miR-22-3p. Mol. Ther. 2021, 29, 2601–2616. [Google Scholar] [CrossRef]
- Fan, L.; Huang, X.; Chen, J.; Zhang, K.; Gu, Y.H.; Sun, J.; Cui, S.Y. Long Noncoding RNA MALAT1 Contributes to Sorafenib Resistance by Targeting miR-140-5p/Aurora-A Signaling in Hepatocellular Carcinoma. Mol. Cancer Ther. 2020, 19, 1197–1209. [Google Scholar] [CrossRef]
- Zhi, Y.; Abudoureyimu, M.; Zhou, H.; Wang, T.; Feng, B.; Wang, R.; Chu, X. FOXM1-Mediated LINC-ROR Regulates the Proliferation and Sensitivity to Sorafenib in Hepatocellular Carcinoma. Mol. Ther. Nucleic Acids 2019, 16, 576–588. [Google Scholar] [CrossRef]
- Zhou, K.; Nguyen, R.; Qiao, L.; George, J. Single cell RNA-seq analysis identifies a noncoding RNA mediating resistance to sorafenib treatment in HCC. Mol. Cancer 2022, 21, 6. [Google Scholar] [CrossRef]
- Takahashi, K.; Yan, I.K.; Wood, J.; Haga, H.; Patel, T. Involvement of extracellular vesicle long noncoding RNA (linc-VLDLR) in tumor cell responses to chemotherapy. Mol. Cancer Res. 2014, 12, 1377–1387. [Google Scholar] [CrossRef]
- Tao, H.; Zhang, Y.; Li, J.; Liu, J.; Yuan, T.; Wang, W.; Liang, H.; Zhang, E.; Huang, Z. Oncogenic lncRNA BBOX1-AS1 promotes PHF8-mediated autophagy and elicits sorafenib resistance in hepatocellular carcinoma. Mol. Ther. Oncolytics 2023, 28, 88–103. [Google Scholar] [CrossRef]
- Gao, Y.; Tong, M.; Wong, T.L.; Ng, K.Y.; Xie, Y.N.; Wang, Z.; Yu, H.; Loh, J.J.; Li, M.; Ma, S. Long Noncoding RNA URB1-Antisense RNA 1 (AS1) Suppresses Sorafenib-Induced Ferroptosis in Hepatocellular Carcinoma by Driving Ferritin Phase Separation. ACS Nano 2023, 17, 22240–22258. [Google Scholar] [CrossRef]
- Zhang, J.; Pan, T.; Zhou, W.; Zhang, Y.; Xu, G.; Xu, Q.; Li, S.; Gao, Y.; Wang, Z.; Xu, J.; et al. Long noncoding RNA LINC01132 enhances immunosuppression and therapy resistance via NRF1/DPP4 axis in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2022, 41, 270. [Google Scholar] [CrossRef]
- Han, Y.; Jiang, W.; Wang, Y.; Zhao, M.; Li, Y.; Ren, L. Serum long non-coding RNA SCARNA10 serves as a potential diagnostic biomarker for hepatocellular carcinoma. BMC Cancer 2022, 22, 431. [Google Scholar] [CrossRef]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics—Challenges and potential solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef]
- Rebouissou, S.; Zucman-Rossi, J.; Moreau, R.; Qiu, Z.; Hui, L. Note of caution: Contaminations of hepatocellular cell lines. J. Hepatol. 2017, 67, 896–897. [Google Scholar] [CrossRef]
- DiStefano, J.K.; Gerhard, G.S. Long Noncoding RNAs and Human Liver Disease. Annu. Rev. Pathol. 2022, 17, 1–21. [Google Scholar] [CrossRef]
- Wong, L.S.; Wei, L.; Wang, G.; Law, C.T.; Tsang, F.H.; Chin, W.C.; Ng, I.O.; Wong, C.M. In Vivo Genome-Wide CRISPR Activation Screening Identifies Functionally Important Long Noncoding RNAs in Hepatocellular Carcinoma. Cell Mol. Gastroenterol. Hepatol. 2022, 14, 1053–1076. [Google Scholar] [CrossRef]
- Li, G.Z.; Meng, G.X.; Pan, G.Q.; Zhang, X.; Yan, L.J.; Li, R.Z.; Ding, Z.N.; Tan, S.Y.; Wang, D.X.; Tian, B.W.; et al. MALAT1/ mir-1-3p mediated BRF2 expression promotes HCC progression via inhibiting the LKB1/AMPK signaling pathway. Cancer Cell Int. 2023, 23, 188. [Google Scholar] [CrossRef]
- Wang, Y.; Zhu, P.; Luo, J.; Wang, J.; Liu, Z.; Wu, W.; Du, Y.; Ye, B.; Wang, D.; He, L.; et al. LncRNA HAND2-AS1 promotes liver cancer stem cell self-renewal via BMP signaling. EMBO J. 2019, 38, e101110. [Google Scholar] [CrossRef]
- Chakraborty, E.; Sarkar, D. Emerging Therapies for Hepatocellular Carcinoma (HCC). Cancers 2022, 14, 2798. [Google Scholar] [CrossRef]
- Rajasekaran, D.; Srivastava, J.; Ebeid, K.; Gredler, R.; Akiel, M.; Jariwala, N.; Robertson, C.L.; Shen, X.N.; Siddiq, A.; Fisher, P.B.; et al. Combination of Nanoparticle-Delivered siRNA for Astrocyte Elevated Gene-1 (AEG-1) and All-trans Retinoic Acid (ATRA): An Effective Therapeutic Strategy for Hepatocellular Carcinoma (HCC). Bioconjug. Chem. 2015, 26, 1651–1661. [Google Scholar] [CrossRef]
- Srivastava, J.; Robertson, C.L.; Ebeid, K.; Dozmorov, M.; Rajasekaran, D.; Mendoza, R.; Siddiq, A.; Akiel, M.A.; Jariwala, N.; Shen, X.N.; et al. A novel role of astrocyte elevated gene-1 (AEG-1) in regulating nonalcoholic steatohepatitis (NASH). Hepatology 2017, 66, 466–480. [Google Scholar] [CrossRef]

| lncRNAs | Associated Protein | Mechanism | Phenotypes/Outcomes | References |
|---|---|---|---|---|
| DIGIT | BRD3 | Forms phase-separated condensates of BRD3, facilitating the histone H3 acetylation of lysine 18 (H3K18ac) of enhancers of endoderm transcription factors | Endoderm differentiation | [57] |
| SLERT | DDX21 | Formation of fibrillar center (FC) and dense fibrillar component (DFC) in the nucleolus | RNA polymerase I (Pol I) transcription and ribosomal RNA production | [58,59] |
| NORAD | PUM1 and PUM2 | Sequestration of Pumilio proteins in PUM condensates termed NP bodies | Inhibition of Pumilio function, preventing aberrant mitosis and maintaining genomic stability | [60] |
| SNHG9 | LATS1 | LATS1 phase separation inhibits LATS1-mediated YAP phosphorylation | Activates YAP-driven gene transcription, thus promoting breast cancer | [61] |
| MELTF-AS1 | YBX1 | Phase separation of oncogenic RBP YBX1 | Activation of ANXA8 transcription, leading to promotion of non-small-cell lung cancer (NSCLC) | [62] |
| LINP1 | Ku70/Ku80 | Multimerization of Ku to form filamentous Ku-containing aggregates | Facilitates Ku-mediated DNA repair by non-homologous end joining (NHEJ) | [63] |
| dilncRNA | DNA-damage-response (DDR) proteins, such as 53BP1 | Molecular crowding of DDR proteins in LLPS condensates | Facilitates DNA double-strand break (DSB) repair | [64] |
| XiST | PTBP1, MATR3, TDP-43 and CELF1 RBPs | Forms a condensate in the inactive X (Xi)-compartment | X-chromosome inactivation | [65] |
| GIRGL | CAPRIN1 | Sequesters CAPRIN1 and GLS1 mRNA in stress granules | Inhibition of GLS1 mRNA translation, facilitating the survival of cancer cells under glutamine-deprived conditions | [66] |
| NEAT1 | TDP-43 | Formation of nuclear bodies in response to stress | Mitigation of stress, the dysfunction of which might be a cause of ALS | [67] |
| lncRNAs | Expression | Role in HCC | Outcome in HCC | References |
|---|---|---|---|---|
| HOTAIR | Upregulated | Oncogene | Promotes metastasis | [38,84,85] |
| MALAT1 | Upregulated | Oncogene | Promotes proliferation, migration, invasion, and metastasis | [86,87,88] |
| HULC | Upregulated | Oncogene | Promotes proliferation and inhibits apoptosis | [85,89,90] |
| GAS5 | Downregulated | Tumor Suppressor | Suppresses proliferation and invasion | [88,91] |
| PVT1 | Upregulated | Oncogene | Promotes proliferation, migration, and invasion | [92,93,94] |
| HOTTIP | Upregulated | Oncogene | Promotes proliferation and angiogenesis | [95,96,97] |
| MVIH | Upregulated | Oncogene | Promotes proliferation, invasion, metastasis and angiogenesis | [98,99] |
| H19 | Upregulated | Oncogene | Promotes proliferation, metastasis and angiogenesis | [99,100,101] |
| MEG3 | Downregulated | Tumor suppressor | Inhibits proliferation, migration and invasion | [102,103] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shah, M.; Sarkar, D. HCC-Related lncRNAs: Roles and Mechanisms. Int. J. Mol. Sci. 2024, 25, 597. https://doi.org/10.3390/ijms25010597
Shah M, Sarkar D. HCC-Related lncRNAs: Roles and Mechanisms. International Journal of Molecular Sciences. 2024; 25(1):597. https://doi.org/10.3390/ijms25010597
Chicago/Turabian StyleShah, Mimansha, and Devanand Sarkar. 2024. "HCC-Related lncRNAs: Roles and Mechanisms" International Journal of Molecular Sciences 25, no. 1: 597. https://doi.org/10.3390/ijms25010597





