Analysis of MicroRNA-Transcription Factors Co-Regulatory Network Linking Depression and Vitamin D Deficiency
Abstract
:1. Introduction
2. Results
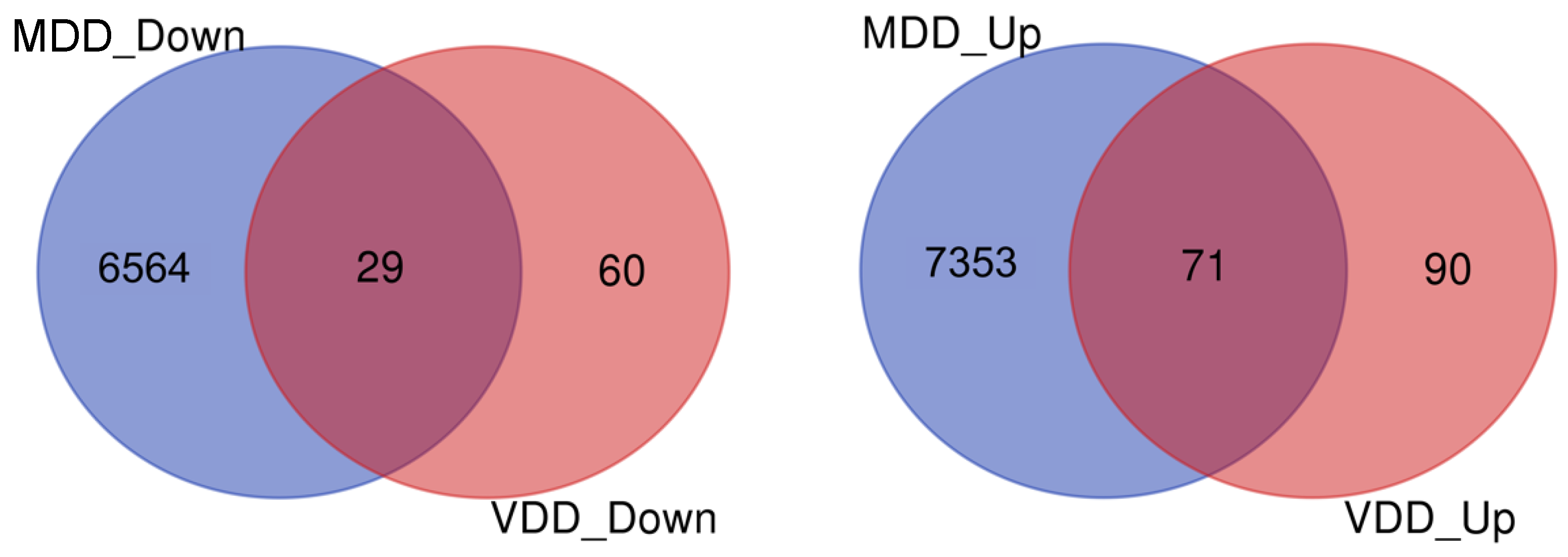
2.1. Identification of DEGs
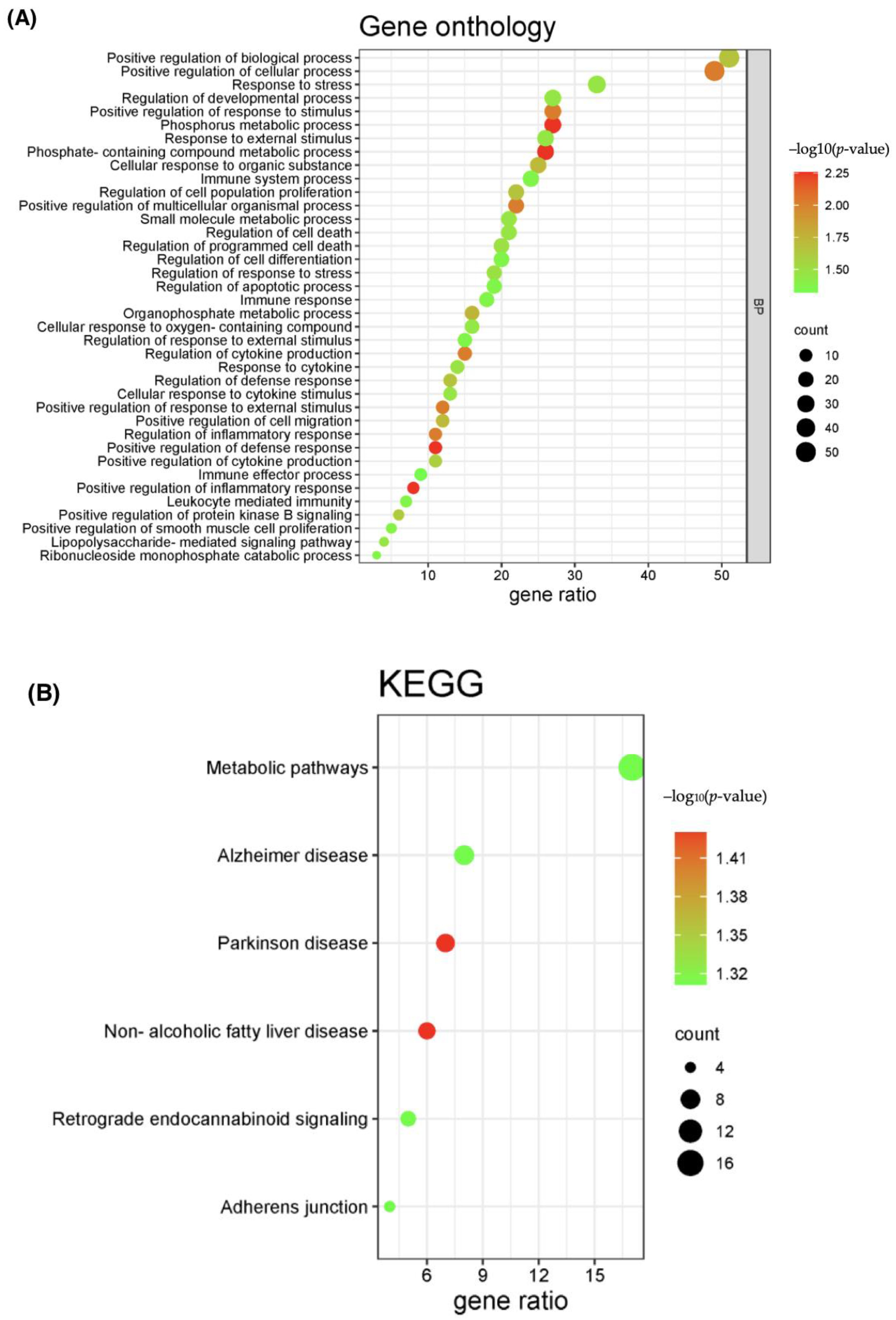
2.2. PPI Network and Functional Enrichment Analysis of Common DEGs
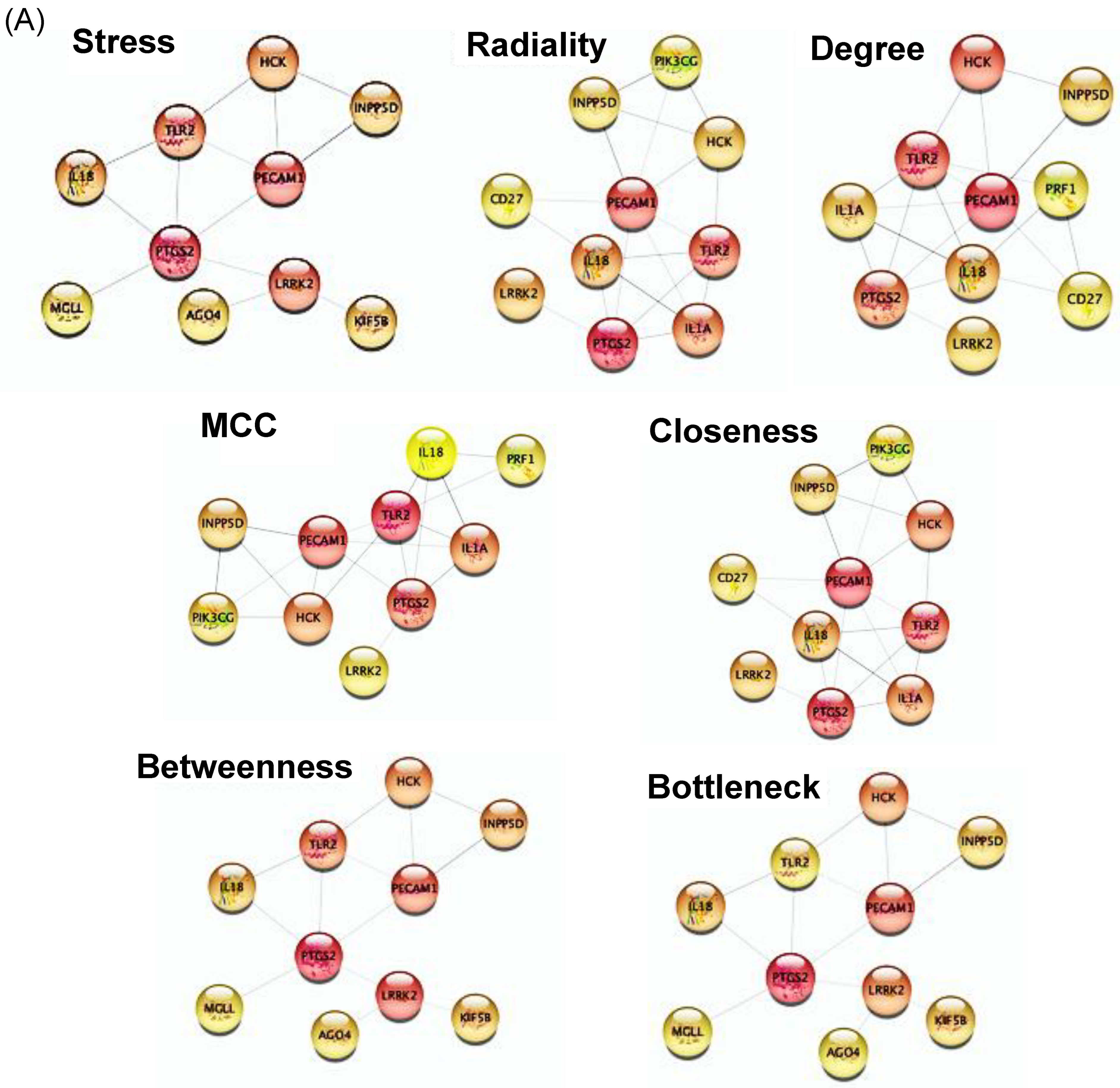
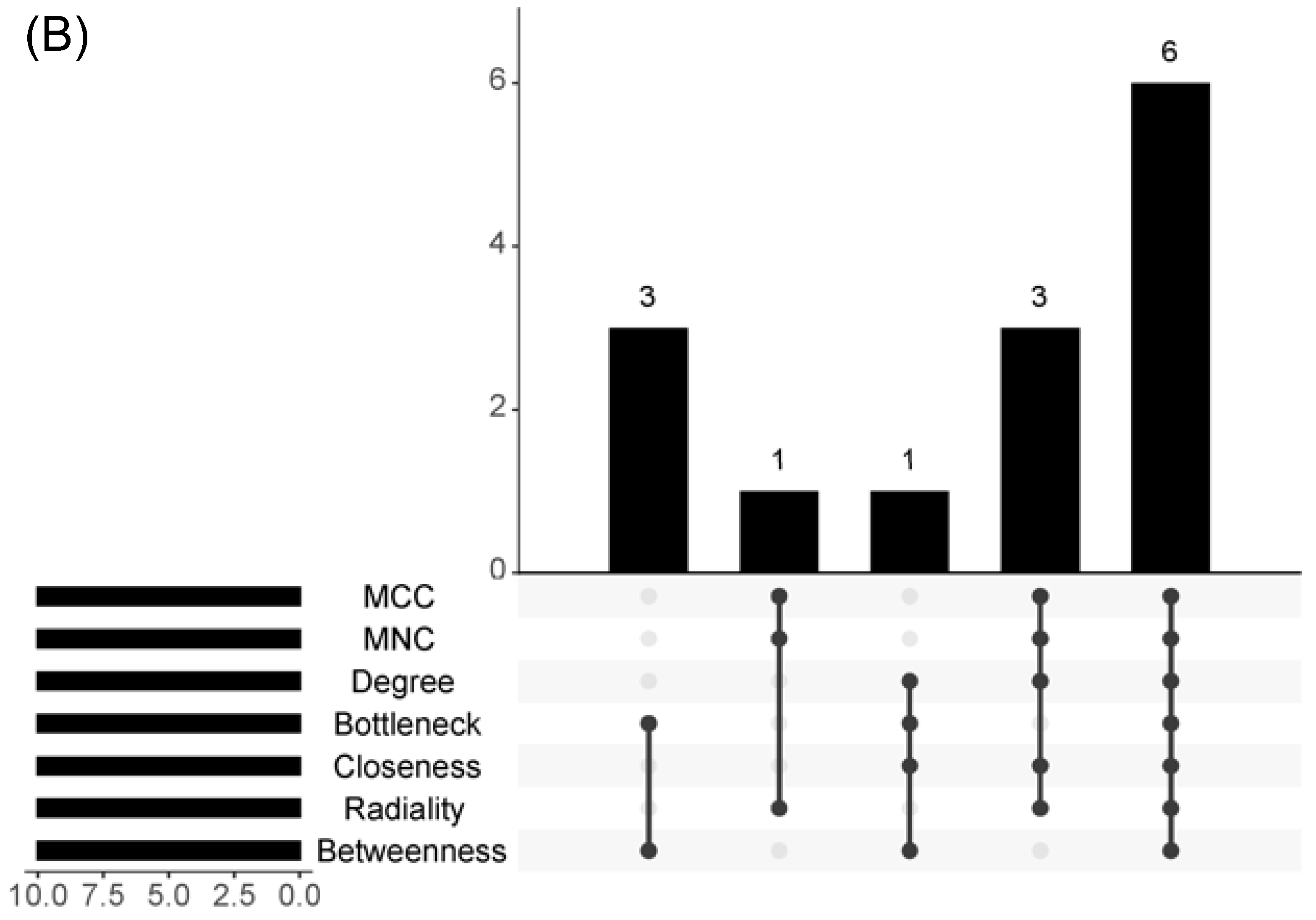
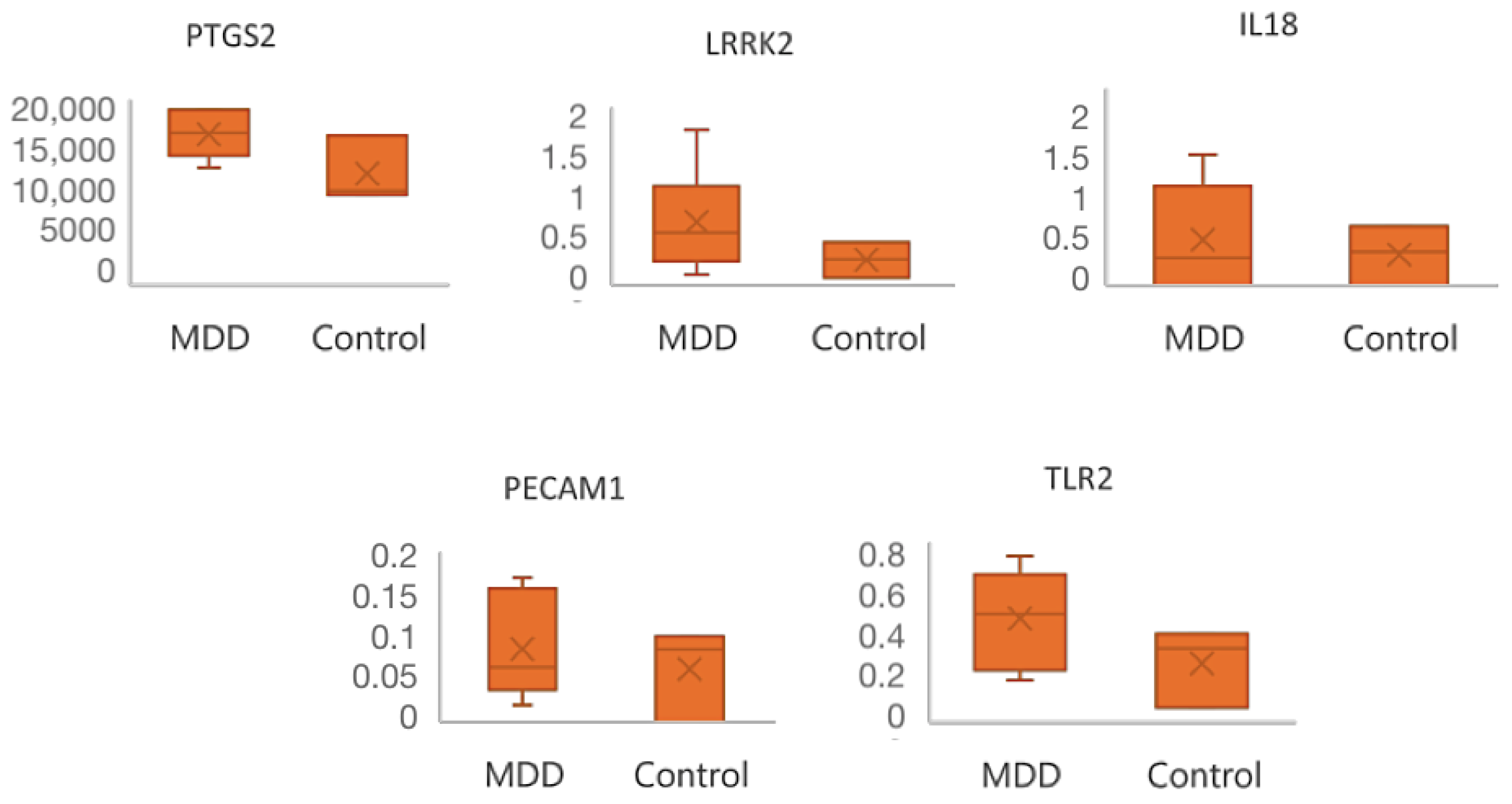
2.3. Hub Genes Identification
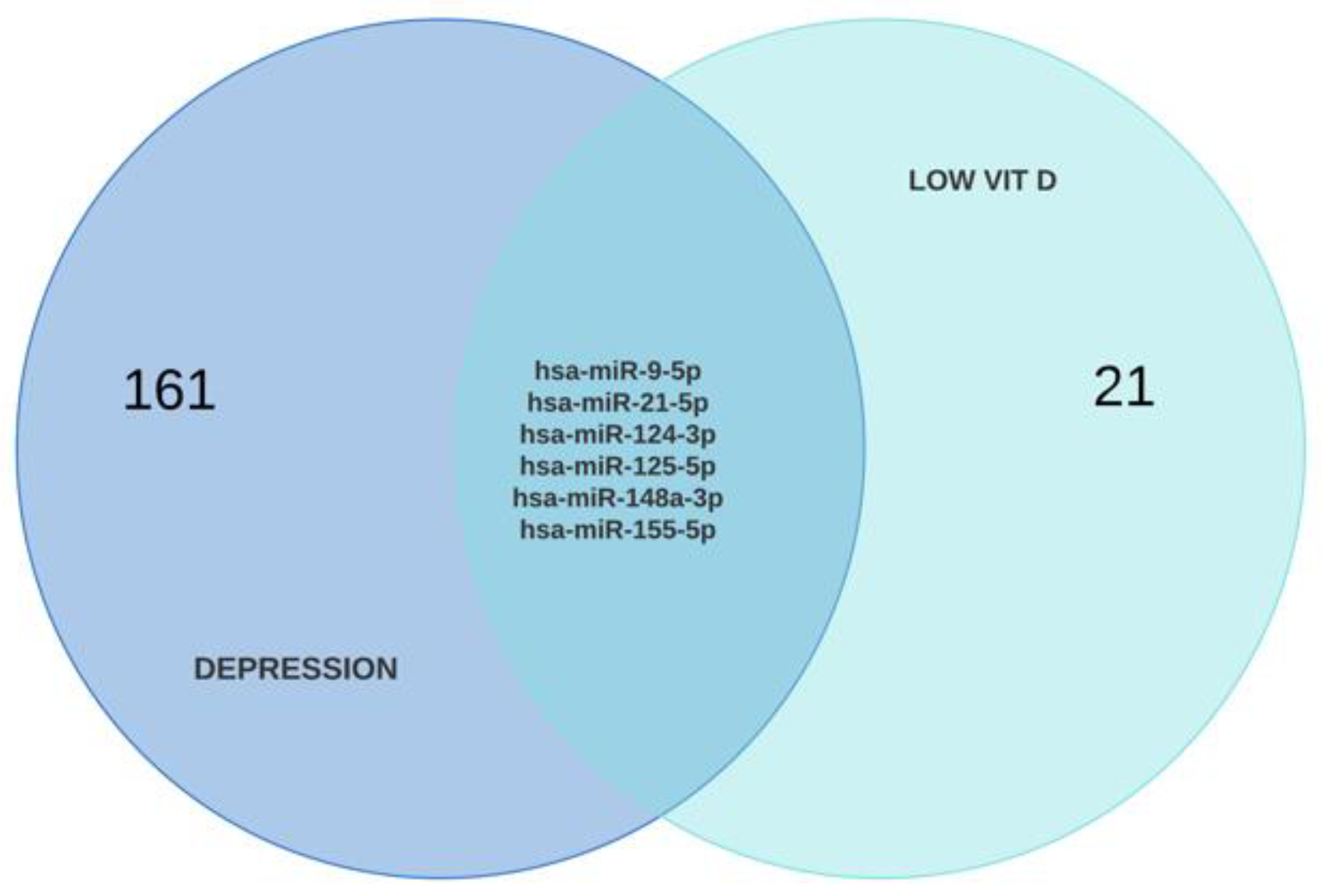
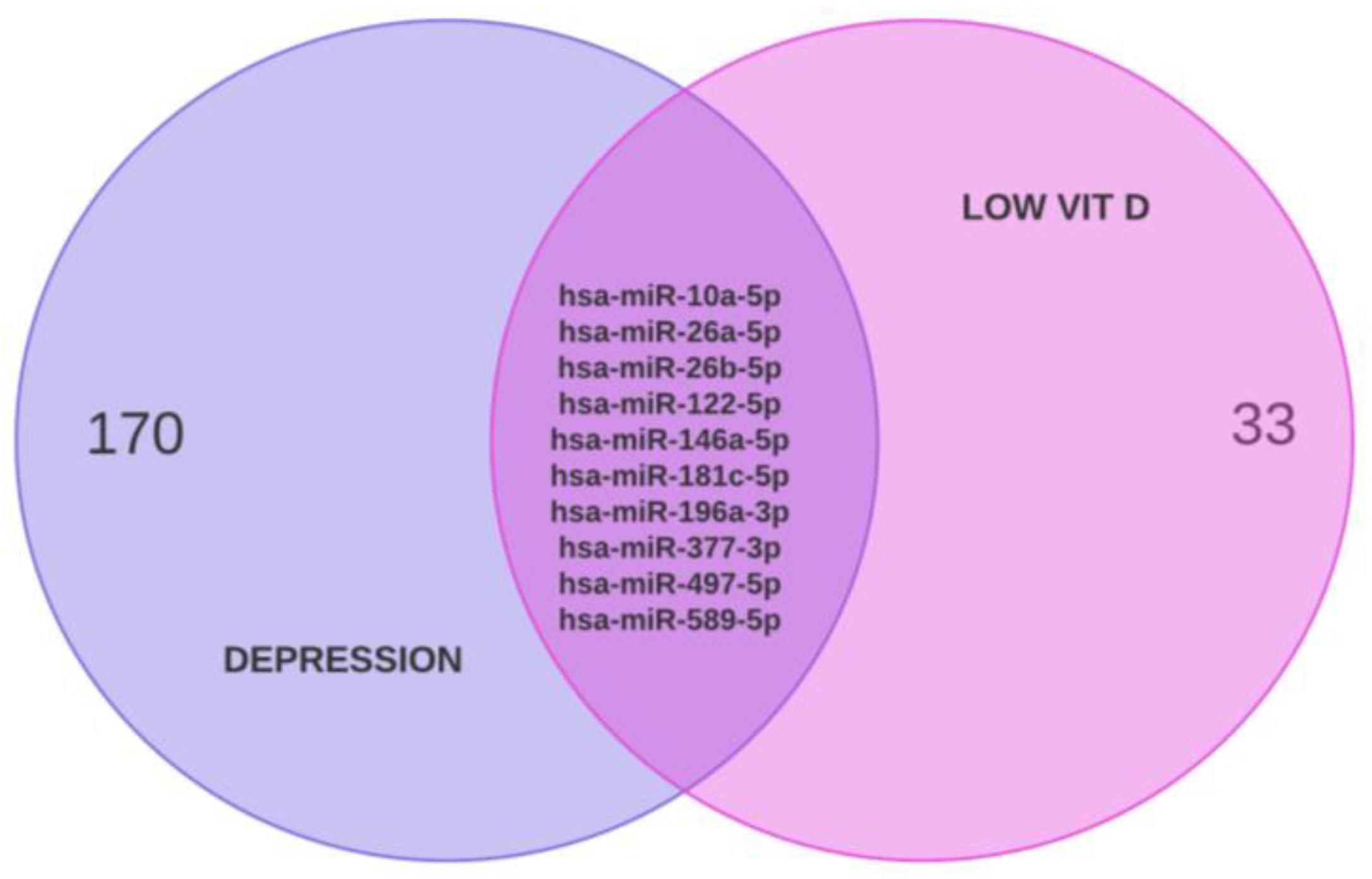
2.4. Construction of TF-Hub Genes-MicroRNA Regulatory Networks
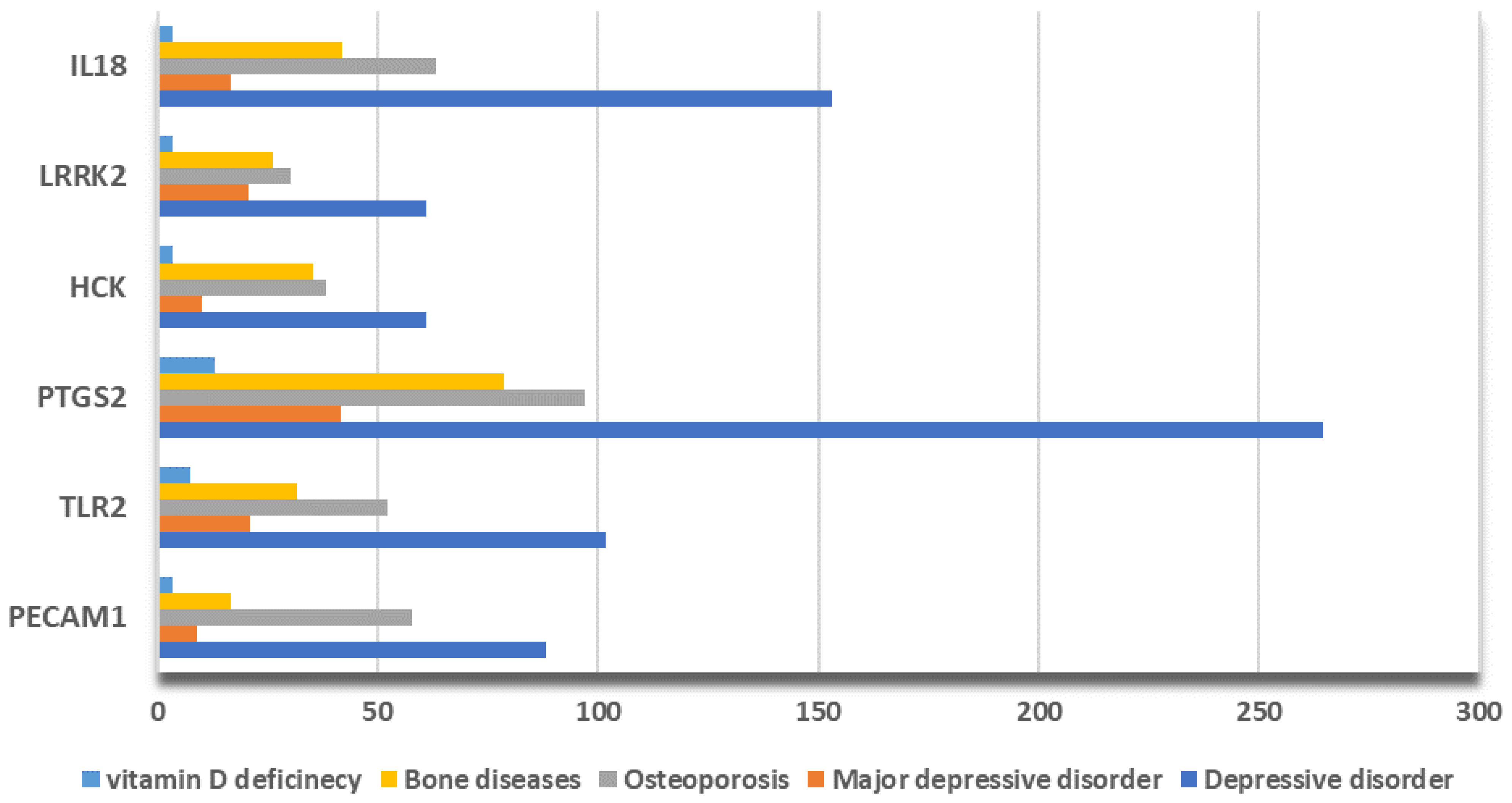
2.5. Data Validation
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Data Collection and Identification of DEGs
5.2. Construction of Protein–Protein Interaction Network and Functional Enrichment Pathway Analysis
5.3. Hub Genes Selection
5.4. Construction of Transcription Factors (TFs) and MicroRNAs (miRNAs) Regulatory Networks
5.5. Data Validation
- Case–control, human studies on patients diagnosed with any form of depression or low (non-drug-related) vitamin D;
- Research articles reporting correlations between depression/vitamin D deficiency and differentially expressed microRNAs (with an adjusted p-value < 0.05);
- Full-text articles in English.
- Studies evaluating the effect of certain antidepressants or vitamin D supplementation on microRNA expression;
- Cancer-related research articles;
- Articles with insufficient data for subsequent analysis.
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Institute of Health Metrics and Evaluation. Global Health Data Exchange (GHDx). Available online: https://vizhub.healthdata.org/gbd-results/ (accessed on 4 March 2023).
- Ma, Y.; Xiang, Q.; Yan, C.; Liao, H.; Wang, J. Relationship between chronic diseases and depression: The mediating effect of pain. BMC Psychiatry 2021, 21, 436. [Google Scholar] [CrossRef]
- Fortin, M.; Stewart, M.; Poitras, M.-E.; Almirall, J.; Maddocks, H. A systematic review of prevalence studies on multimorbidity: Toward a more uniform methodology. Ann. Fam. Med. 2012, 10, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Read, J.R.; Sharpe, L.; Modini, M.; Dear, B.F. Multimorbidity and depression: A systematic review and meta-analysis. J. Affect. Disord. 2017, 221, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.; Lou, P.; Shang, Y.; Zhang, P.; Wang, J.; Chang, G.; Shi, C. Prevalence and determinants of depressive and anxiety symptoms in adults with type 2 diabetes in China: A cross-sectional study. BMJ Open 2016, 6, e012540. [Google Scholar] [CrossRef]
- Liu, B.; Liu, J.; Wang, M.; Zhang, Y.; Li, L. From Serotonin to Neuroplasticity: Evolvement of Theories for Major Depressive Disorder. Front. Cell. Neurosci. 2017, 11, 305. [Google Scholar] [CrossRef] [PubMed]
- Fries, G.R.; Saldana, V.A.; Finnstein, J.; Rein, T. Molecular pathways of major depressive disorder converge on the synapse. Mol. Psychiatry 2023, 28, 284–297. [Google Scholar] [CrossRef] [PubMed]
- Mariani, N.; Cattane, N.; Pariante, C.; Cattaneo, A. Gene expression studies in Depression development and treatment: An overview of the underlying molecular mechanisms and biological processes to identify biomarkers. Transl. Psychiatry 2021, 11, 354. [Google Scholar] [CrossRef]
- Gold, P.W. The PPARg System in Major Depression: Pathophysiologic and Therapeutic Implications. Int. J. Mol. Sci. 2021, 22, 9248. [Google Scholar] [CrossRef]
- Kim, I.-B.; Lee, J.-H.; Park, S.-C. The Relationship between Stress, Inflammation, and Depression. Biomedicines 2022, 10, 1929. [Google Scholar] [CrossRef]
- Harsanyi, S.; Kupcova, I.; Danisovic, L.; Klein, M. Selected Biomarkers of Depression: What Are the Effects of Cytokines and Inflammation? Int. J. Mol. Sci. 2022, 24, 578. [Google Scholar] [CrossRef]
- Chand, S.P.; Arif, H. Depression. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2023. Available online: http://www.ncbi.nlm.nih.gov/books/NBK430847/ (accessed on 17 July 2023).
- Nuñez, N.A.; Joseph, B.; Pahwa, M.; Kumar, R.; Resendez, M.G.; Prokop, L.J.; Veldic, M.; Seshadri, A.; Biernacka, J.M.; Frye, M.A.; et al. Augmentation strategies for treatment resistant major depression: A systematic review and network meta-analysis. J. Affect. Disord. 2022, 302, 385–400. [Google Scholar] [CrossRef] [PubMed]
- Kishi, T.; Ikuta, T.; Sakuma, K.; Okuya, M.; Hatano, M.; Matsuda, Y.; Iwata, N. Antidepressants for the treatment of adults with major depressive disorder in the maintenance phase: A systematic review and network meta-analysis. Mol. Psychiatry 2023, 28, 402–409. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.; Sportelli, V.; Ziller, M.; Spengler, D. Epigenomics of Major Depressive Disorders and Schizophrenia: Early Life Decides. Int. J. Mol. Sci. 2017, 18, 1711. [Google Scholar] [CrossRef]
- Etchecopar-Etchart, D.; Korchia, T.; Loundou, A.; Llorca, P.-M.; Auquier, P.; Lançon, C.; Boyer, L.; Fond, G. Comorbid Major Depressive Disorder in Schizophrenia: A Systematic Review and Meta-Analysis. Schizophr. Bull. 2021, 47, 298–308. [Google Scholar] [CrossRef]
- Haller, H.; Anheyer, D.; Cramer, H.; Dobos, G. Complementary therapies for clinical depression: An overview of systematic reviews. BMJ Open 2019, 9, e028527. [Google Scholar] [CrossRef] [PubMed]
- Kris-Etherton, P.M.; Petersen, K.S.; Hibbeln, J.R.; Hurley, D.; Kolick, V.; Peoples, S.; Rodriguez, N.; Woodward-Lopez, G. Nutrition and behavioral health disorders: Depression and anxiety. Nutr. Rev. 2021, 79, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Quan, Z.; Li, H.; Quan, Z.; Qing, H. Appropriate Macronutrients or Mineral Elements Are Beneficial to Improve Depression and Reduce the Risk of Depression. Int. J. Mol. Sci. 2023, 24, 7098. [Google Scholar] [CrossRef]
- Appleton, K.M.; Sallis, H.M.; Perry, R.; Ness, A.R.; Churchill, R. Omega-3 fatty acids for depression in adults. Cochrane Database Syst. Rev. 2015, 2015, CD004692. [Google Scholar] [CrossRef]
- Park, Y.; Ah, Y.-M.; Yu, Y.M. Vitamin D supplementation for depression in older adults: A meta-analysis of randomized controlled trials. Front. Nutr. 2023, 10, 1169436. [Google Scholar] [CrossRef]
- Menon, V.; Kar, S.K.; Suthar, N.; Nebhinani, N. Vitamin D and Depression: A Critical Appraisal of the Evidence and Future Directions. Ind. J. Psychol. Med. 2020, 42, 11–21. [Google Scholar] [CrossRef]
- Somoza-Moncada, M.M.; Turrubiates-Hernández, F.J.; Muñoz-Valle, J.F.; Gutiérrez-Brito, J.A.; Díaz-Pérez, S.A.; Aguayo-Arelis, A.; Hernández-Bello, J. Vitamin D in Depression: A Potential Bioactive Agent to Reduce Suicide and Suicide Attempt Risk. Nutrients 2023, 15, 1765. [Google Scholar] [CrossRef]
- Spedding, S. Vitamin D and Depression: A Systematic Review and Meta-Analysis Comparing Studies with and without Biological Flaws. Nutrients 2014, 6, 1501–1518. [Google Scholar] [CrossRef] [PubMed]
- Nair, R.; Maseeh, A. Vitamin D: The “sunshine” vitamin. J. Pharmacol. Pharmacother. 2012, 3, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Geng, C.; Shaikh, A.S.; Han, W.; Chen, D.; Guo, Y.; Jiang, P. Vitamin D and depression: Mechanisms, determination and application. Asia Pac. J. Clin. Nutr. 2019, 28, 689–694. [Google Scholar] [CrossRef]
- Hassamal, S. Chronic stress, neuroinflammation, and depression: An overview of pathophysiological mechanisms and emerging anti-inflammatories. Front. Psychiatry 2023, 14, 1130989. [Google Scholar] [CrossRef] [PubMed]
- Parel, N.S.; Krishna, P.V.; Gupta, A.; Uthayaseelan, K.; Uthayaseelan, K.; Kadari, M.; Subhan, M.; Kasire, S.P. Depression and Vitamin D: A Peculiar Relationship. Cureus 2022, 14, e24363. [Google Scholar] [CrossRef]
- Dasgupta, B.; Dufour, E.; Mamdouh, Z.; Muller, W.A. A Novel and Critical Role for Tyrosine 663 in Platelet Endothelial Cell Adhesion Molecule-1 Trafficking and Transendothelial Migration. J. Immunol. 2009, 182, 5041–5051. [Google Scholar] [CrossRef]
- Bulut, Y.; Michelsen, K.S.; Hayrapetian, L.; Naiki, Y.; Spallek, R.; Singh, M.; Arditi, M. Mycobacterium tuberculosis heat shock proteins use diverse toll-like receptor pathways to activate pro-inflammatory signals. J. Biol. Chem. 2005, 280, 20961–20967. [Google Scholar] [CrossRef]
- Le, H.D.; Meisel, J.A.; de Meijer, V.E.; Gura, K.M.; Puder, M. The essentiality of arachidonic acid and docosahexaenoic acid. Prostagland. Leukot. Essent. Fat. Acids 2009, 81, 165–170. [Google Scholar] [CrossRef]
- Zach, S.; Felk, S.; Gillardon, F. Signal Transduction Protein Array Analysis Links LRRK2 to Ste20 Kinases and PKC Zeta That Modulate Neuronal Plasticity. PLoS ONE 2010, 5, e13191. [Google Scholar] [CrossRef]
- Guiet, R.; Poincloux, R.; Castandet, J.; Marois, L.; Labrousse, A.; Le Cabec, V.; Maridonneau-Parini, I. Hematopoietic cell kinase (Hck) isoforms and phagocyte duties—From signaling and actin reorganization to migration and phagocytosis. Eur. J. Cell Biol. 2008, 87, 527–542. [Google Scholar] [CrossRef]
- Tsutsumi, N.; Kimura, T.; Arita, K.; Ariyoshi, M.; Ohnishi, H.; Yamamoto, T.; Zuo, X.; Maenaka, K.; Park, E.Y.; Kondo, N.; et al. The structural basis for receptor recognition of human interleukin-18. Nat. Commun. 2014, 5, 5340. [Google Scholar] [CrossRef]
- Dantzer, R.; O’Connor, J.C.; Freund, G.G.; Johnson, R.W.; Kelley, K.W. From inflammation to sickness and depression: When the immune system subjugates the brain. Nat. Rev. Neurosci. 2008, 9, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Berk, M.; Williams, L.J.; Jacka, F.N.; O’neil, A.; Pasco, J.A.; Moylan, S.; Allen, N.B.; Stuart, A.L.; Hayley, A.C.; Byrne, M.L.; et al. So depression is an inflammatory disease, but where does the inflammation come from? BMC Med. 2013, 11, 200. [Google Scholar] [CrossRef] [PubMed]
- Jellinger, K.A. The pathobiological basis of depression in Parkinson disease: Challenges and outlooks. J. Neural Transm. 2022, 129, 1397–1418. [Google Scholar] [CrossRef] [PubMed]
- Linnemann, C.; Lang, U.E. Pathways Connecting Late-Life Depression and Dementia. Front. Pharmacol. 2020, 11, 279. [Google Scholar] [CrossRef]
- Yarar, E. Role and Function of Endocannabinoid System in Major Depressive Disease. Med. Cannabis Cannabinoids 2021, 4, 1–12. [Google Scholar] [CrossRef]
- Xu, H.; Li, T.; Gong, Q.; Xu, H.; Hu, Y.; Lü, W.; Yang, X.; Li, J.; Xu, W.; Kuang, W. Genetic variations in the retrograde endocannabinoid signaling pathway in Chinese patients with major depressive disorder. Front. Neurol. 2023, 14, 1153509. [Google Scholar] [CrossRef]
- Akil, H.; Gordon, J.; Hen, R.; Javitch, J.; Mayberg, H.; McEwen, B.; Meaney, M.J.; Nestler, E.J.; Akil, H.; Gordon, J.; et al. Treatment resistant depression: A multi-scale, systems biology approach. Neurosci. Biobehav. Rev. 2018, 84, 272–288. [Google Scholar] [CrossRef]
- Banafshe, H.R.; Khoshnoud, M.J.; Abed, A.; Saghazadeh, M.; Mesdaghinia, A. Vitamin D supplementation attenuates the be-havioral scores of neuropathic pain in rats. Nutr. Neurosci. 2019, 22, 700–705. [Google Scholar] [CrossRef]
- Song, Q.; Feng, Y.-B.; Wang, L.; Shen, J.; Li, Y.; Fan, C.; Wang, P.; Yu, S.Y. COX-2 inhibition rescues depression-like behaviors via suppressing glial activation, oxidative stress and neuronal apoptosis in rats. Neuropharmacology 2019, 160, 107779. [Google Scholar] [CrossRef]
- Beurel, E.; Toups, M.; Nemeroff, C.B. The Bidirectional Relationship of Depression and Inflammation: Double Trouble. Neuron 2020, 107, 234–256. [Google Scholar] [CrossRef] [PubMed]
- Mehdi, S.; Wani, S.U.D.; Krishna, K.; Kinattingal, N.; Roohi, T.F. A review on linking stress, depression, and insulin resistance via low-grade chronic inflammation. Biochem. Biophys. Rep. 2023, 36, 101571. [Google Scholar] [CrossRef] [PubMed]
- Almeida Moreira, L.L.K.; Lima, L.A.; Alexandre de Aquino, P.E.; de Sousa, J.A.C.; Gadelha, C.V.J.; Calou, I.B.F.; Lopes, M.J.P.; Lima, F.A.V.; Neves, K.R.T.; de Andrade, G.M.; et al. Vitamin D (VD3) antioxidative and anti-inflammatory activities: Peripheral and central effects. Eur. J. Pharmacol. 2020, 879, 173099. [Google Scholar] [CrossRef] [PubMed]
- Rui, Q.; Ni, H.; Li, D.; Gao, R.; Chen, G. The Role of LRRK2 in Neurodegeneration of Parkinson Disease. Curr. Neuropharmacol. 2018, 16, 1348–1357. [Google Scholar] [CrossRef] [PubMed]
- Mutti, V.; Carini, G.; Filippini, A.; Castrezzati, S.; Giugno, L.; Gennarelli, M.; Russo, I. LRRK2 Kinase Inhibition Attenuates Neuroinflammation and Cytotoxicity in Animal Models of Alzheimer’s and Parkinson’s Disease-Related Neuroinflammation. Cells 2023, 12, 1799. [Google Scholar] [CrossRef]
- Wile, D.J.; Agarwal, P.A.; Schulzer, M.; Mak, E.; Dinelle, K.; Shahinfard, E.; Vafai, N.; Hasegawa, K.; Zhang, J.; McKenzie, J.; et al. Serotonin and dopamine transporter PET changes in the premotor phase of LRRK2 parkinsonism: Cross-sectional studies. Lancet Neurol. 2017, 16, 351–359. [Google Scholar] [CrossRef]
- Filippone, A.; Cucinotta, L.; Bova, V.; Lanza, M.; Casili, G.; Paterniti, I.; Campolo, M.; Cuzzocrea, S.; Esposito, E. Inhibition of LRRK2 Attenuates Depression-Related Symptoms in Mice with Moderate Traumatic Brain Injury. Cells 2023, 12, 1040. [Google Scholar] [CrossRef]
- Anwar, J.; Alenezi, S.K.; Alhowail, A.H. Molecular insights into the pathogenic impact of vitamin D deficiency in neurological disorders. Biomed. Pharmacother. 2023, 162, 114718. [Google Scholar] [CrossRef]
- Mullany, L.E.; Herrick, J.S.; Wolff, R.K.; Stevens, J.R.; Samowitz, W.; Slattery, M.L. MicroRNA-transcription factor interactions and their combined effect on target gene expression in colon cancer cases. Genes Chromosom. Cancer 2018, 57, 192–202. [Google Scholar] [CrossRef]
- Luo, H.; Liu, Z.; Liu, B.; Li, H.; Yang, Y.; Xu, Z.-Q.D. Virus-Mediated Overexpression of ETS-1 in the Ventral Hippocampus Counteracts Depression-Like Behaviors in Rats. Neurosci. Bull. 2019, 35, 1035–1044. [Google Scholar] [CrossRef]
- Shi, W.; Zhang, X.; Xu, C.; Pang, R.; Fan, Z.; Wan, X.; Jiang, Z.; Li, H.; Li, Z.; Zhang, H. Identification of Hub Genes and Pathways Associated with Oxidative Stress of Cartilage in Osteonecrosis of Femoral Head Using Bioinformatics Analysis. CARTILAGE 2022, 13, 19476035221074000. [Google Scholar] [CrossRef]
- Roberson-Nay, R.; Wolen, A.R.; Lapato, D.M.; Lancaster, E.E.; Webb, B.T.; Verhulst, B.; Hettema, J.M.; York, T.P. Twin study of early-onset major depres-sionfinds DNA methylation enrichment for neuro developmental genes. bioRxiv 2018. bioRxiv:422345. [Google Scholar]
- Arasappan, D.; Eickhoff, S.B.; Nemeroff, C.B.; Hofmann, H.A.; Jabbi, M. Transcription Factor Motifs Associated with Anterior Insula Gene Expression Underlying Mood Disorder Phenotypes. Mol. Neurobiol. 2021, 58, 1978–1989. [Google Scholar] [CrossRef] [PubMed]
- Koo, J.W.; Russo, S.J.; Ferguson, D.; Nestler, E.J.; Duman, R.S. Nuclear factor-kappaB is a critical mediator of stress-impaired neurogenesis and depressive behavior. Proc. Natl. Acad. Sci. USA 2010, 107, 2669–2674. [Google Scholar] [CrossRef] [PubMed]
- Caviedes, A.; Lafourcade, C.; Soto, C.; Wyneken, U. BDNF/NF-κB Signaling in the Neurobiology of Depression. Curr. Pharm. Des. 2017, 23, 3154–3163. [Google Scholar] [CrossRef] [PubMed]
- Karin, M. Nuclear factor-κB in cancer development and progression. Nature 2006, 441, 431–436. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, J.; Ge, X.; Du, J.; Deb, D.K.; Li, Y.C. Vitamin D Receptor Inhibits Nuclear Factor κB Activation by Interacting with IκB Kinase β Protein. J. Biol. Chem. 2013, 288, 19450–19458. [Google Scholar] [CrossRef]
- Bayer, S.J.B.; Yang, G.S.; Lyon, D.E.P. Genetic Variation Associated with Depressive Symptoms in Breast Cancer Patients: A Systematic Review. Cancer Nurs. 2022, 45, E197–E205. [Google Scholar] [CrossRef]
- Charalambous, M.P.; The Colorectal Cancer Study Group; Maihöfner, C.; Bhambra, U.; Lightfoot, T.; Gooderham, N.J. Upregulation of cyclooxygenase-2 is accompanied by increased expression of nuclear factor-κB and IκB kinase-α in human colorectal cancer epithelial cells. Br. J. Cancer 2003, 88, 1598–1604. [Google Scholar] [CrossRef]
- Vergani, E.; Dugo, M.; Cossa, M.; Frigerio, S.; Di Guardo, L.; Gallino, G.; Mattavelli, I.; Vergani, B.; Lalli, L.; Tamborini, E.; et al. miR-146a-5p impairs melanoma resistance to kinase inhibitors by targeting COX2 and regulating NFkB-mediated inflammatory mediators. Cell Commun. Signal. 2020, 18, 156. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.H.; Oh, J.Y. The Effect of miR-146a on the Gene Expression of Immunoregulatory Cytokines in Human Mesenchymal Stromal Cells. Int. J. Mol. Sci. 2020, 21, 6809. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.-J.; Wu, D.; Yang, X.-F.; Li, T. miR-146a-5p Modulates Adult Hippocampal Neurogenesis Deficits Through Klf4/p-Stat3 Signaling in APP/PS1 Mice. Neuroscience 2023, 526, 314–325. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-P.; Zhong, M.; Sun, J.-X.; He, J.; Gao, Y.; Qin, F.-X. miR-146a reduces depressive behavior by inhibiting microglial activation. Mol. Med. Rep. 2021, 23, 463. [Google Scholar] [CrossRef] [PubMed]
- Hung, Y.Y.; Wu, M.K.; Tsai, M.C.; Huang, Y.L.; Kang, H.Y. Aberrant Expression of Intracellular let-7e, miR-146a, and miR-155 Correlates with Severity of Depression in Patients with Major Depressive Disorder and Is Ameliorated after Antidepressant Treatment. Cells 2019, 8, 647. [Google Scholar] [CrossRef]
- Enatescu, V.R.; Papava, I.; Enatescu, I.; Antonescu, M.; Anghel, A.; Seclaman, E.; Sirbu, I.O.; Marian, C. Circulating Plasma Micro RNAs in Patients with Major Depressive Disorder Treated with Antidepressants: A Pilot Study. Psychiatry Investig. 2016, 13, 549–557. [Google Scholar] [CrossRef]
- Hutchison, E.R.; Kawamoto, E.M.; Taub, D.D.; Lal, A.; Abdelmohsen, K.; Zhang, Y.; Wood, W.H.; Lehrmann, E.; Camandola, S.; Becker, K.; et al. Evidence for miR-181 involvement in neuroinflammatory responses of astrocytes. Glia 2013, 61, 1018–1028. [Google Scholar] [CrossRef]
- Sun, X.; Sit, A.; Feinberg, M.W. Role of miR-181 family in regulating vascular inflammation and immunity. Trends Cardiovasc. Med. 2014, 24, 105–112. [Google Scholar] [CrossRef]
- Manzano-Crespo, M.; Atienza, M.; Cantero, J.L. Lower serum expression of miR-181c-5p is associated with increased plasma levels of amyloid-beta 1–40 and cerebral vulnerability in normal aging. Transl. Neurodegener. 2019, 8, 34. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucl. Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucl. Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Conway, J.R.; Lex, A.; Gehlenborg, N. UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics 2017, 33, 2938–2940. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Soufan, O.; Ewald, J.; Hancock, R.E.; Basu, N.; Xia, J. NetworkAnalyst 3.0: A visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucl. Acids Res. 2019, 47, W234–W241. [Google Scholar] [CrossRef]



| GSE Number | Participants (Patients/Controls) | Biological Sample | Pathological Condition | Analytical Platform |
|---|---|---|---|---|
| GSE190518 | 3/3 | Blood | Depression | Illumina HiSeq 4000 |
| GSE98793 | 128/64 | Blood | Depression | Affymetrix GeneChip system |
| GSE76826 | 10/12 | Blood | Depression | Agilent microarray |
| GSE217811 | 10/10 | Plasma | Depression | Agilent microarray |
| GSE101521 | 18/38 | Brain | Depression | Illumina MiSeq 2500 |
| GSE23848 | 20/15 | Blood | Depression | Sentrix Human-6 v2 Expression BeadChip |
| GSE80655 | 69/70 | Brain | Depression | Illumina HiSeq 2000 |
| GSE169459 | 3/3 | Blood | Depression | Agilent microarray |
| GSE157939 | 80/80 | Blood | Hypovitaminosis D | Fluidigm BioMark |
| GSE22523 | 2/2 | Blood | Hypovitaminosis D | Affymetrix GeneChip system |
| Gene Symbol | Full Name | Function |
|---|---|---|
| PECAM1 | Platelet And Endothelial Cell Adhesion Molecule 1 | Cell adhesion molecule, which is required for leukocyte transendothelial migration (TEM) under most inflammatory conditions [29]. |
| TLR2 | Toll Like Receptor 2 | Modulates the host’s inflammatory response and has been implicated in the pathogenesis of several autoimmune diseases [30]. |
| PTGS2 | Prostaglandin-Endoperoxide Synthase 2 | With a particular role in the inflammatory response, during neuroinflammation, plays a role in neuronal secretion of specialized pre-resolving mediator [31]. |
| LRRK2 | Leucine Rich Repeat Kinase 2 | Regulates neuronal process morphology in the intact central nervous system [32]. |
| HCK | HCK Proto-Oncogene, Src Family Tyrosine Kinase | Plays an important role in the regulation of innate immune responses, including neutrophiles, monocytes, and macrophages [33]. |
| IL18 | Interleukin 18 | Pro-inflammatory cytokine primarily involved in epithelial barrier repair [34]. |
| adj. p-Value | logFC | TF | GEO Dataset |
|---|---|---|---|
| 2.31 × 10−2 | −1.18 | ETS1 | GSE217811 |
| 2.97 × 10−2 | 0.62 | TFAP2A | |
| 4.69 × 10−2 | 0.75 | NFKB2 | |
| 2.91 × 10−2 | 0.55 | CTCF | |
| 8.78 × 10−3 | 0.58 | RELA | GSE23848 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sala-Cirtog, M.; Sirbu, I.-O. Analysis of MicroRNA-Transcription Factors Co-Regulatory Network Linking Depression and Vitamin D Deficiency. Int. J. Mol. Sci. 2024, 25, 1114. https://doi.org/10.3390/ijms25021114
Sala-Cirtog M, Sirbu I-O. Analysis of MicroRNA-Transcription Factors Co-Regulatory Network Linking Depression and Vitamin D Deficiency. International Journal of Molecular Sciences. 2024; 25(2):1114. https://doi.org/10.3390/ijms25021114
Chicago/Turabian StyleSala-Cirtog, Maria, and Ioan-Ovidiu Sirbu. 2024. "Analysis of MicroRNA-Transcription Factors Co-Regulatory Network Linking Depression and Vitamin D Deficiency" International Journal of Molecular Sciences 25, no. 2: 1114. https://doi.org/10.3390/ijms25021114





