BMP9-ID1 Pathway Attenuates N6-Methyladenosine Levels of CyclinD1 to Promote Cell Proliferation in Hepatocellular Carcinoma
Abstract
:1. Introduction
2. Results
2.1. BMP9 Expression Is Related to the Expression of CyclinD1 in HCC Tissues
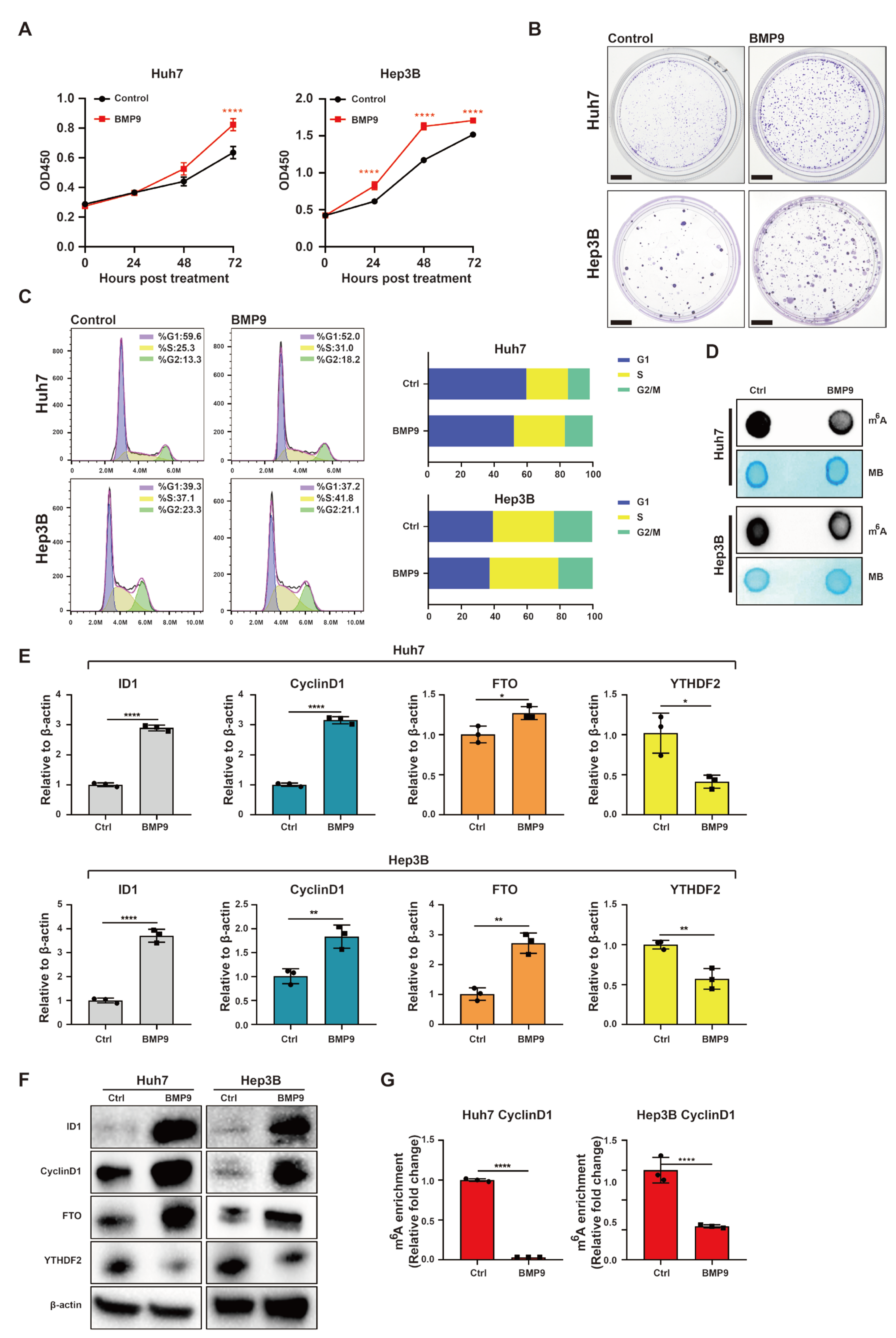
2.2. BMP9 Enhances CyclinD1 Expression in HCC Cells to Facilitate Cell Cycle Progression via Suppressing m6A Methylation within the 5′-UTR of CyclinD1 mRNA
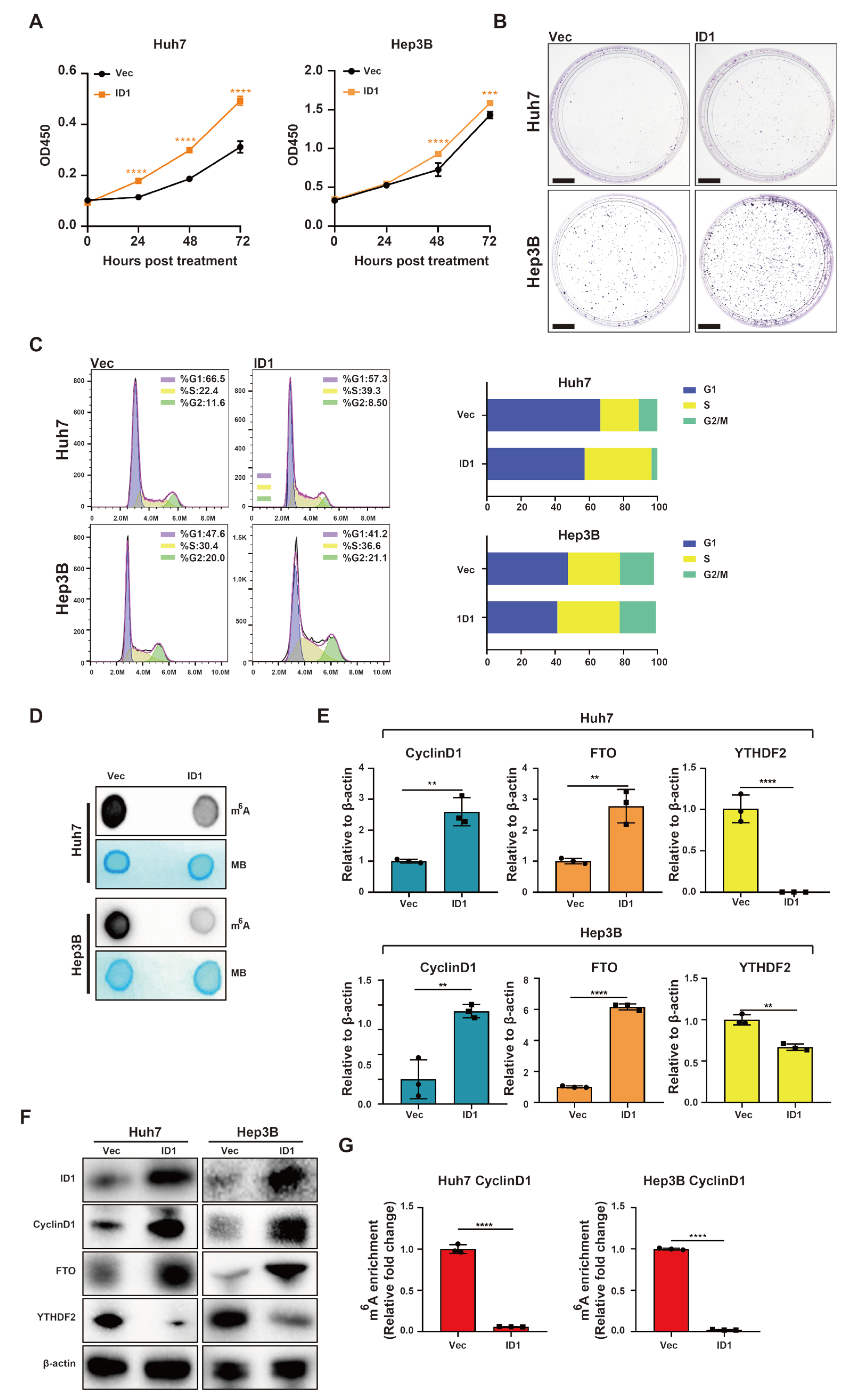
2.3. The BMP9-ID1 Pathway Facilitates HCC Cell Cycle Progression by Suppressing m6A Methylation within the 5′ UTR of CyclinD1 mRNA
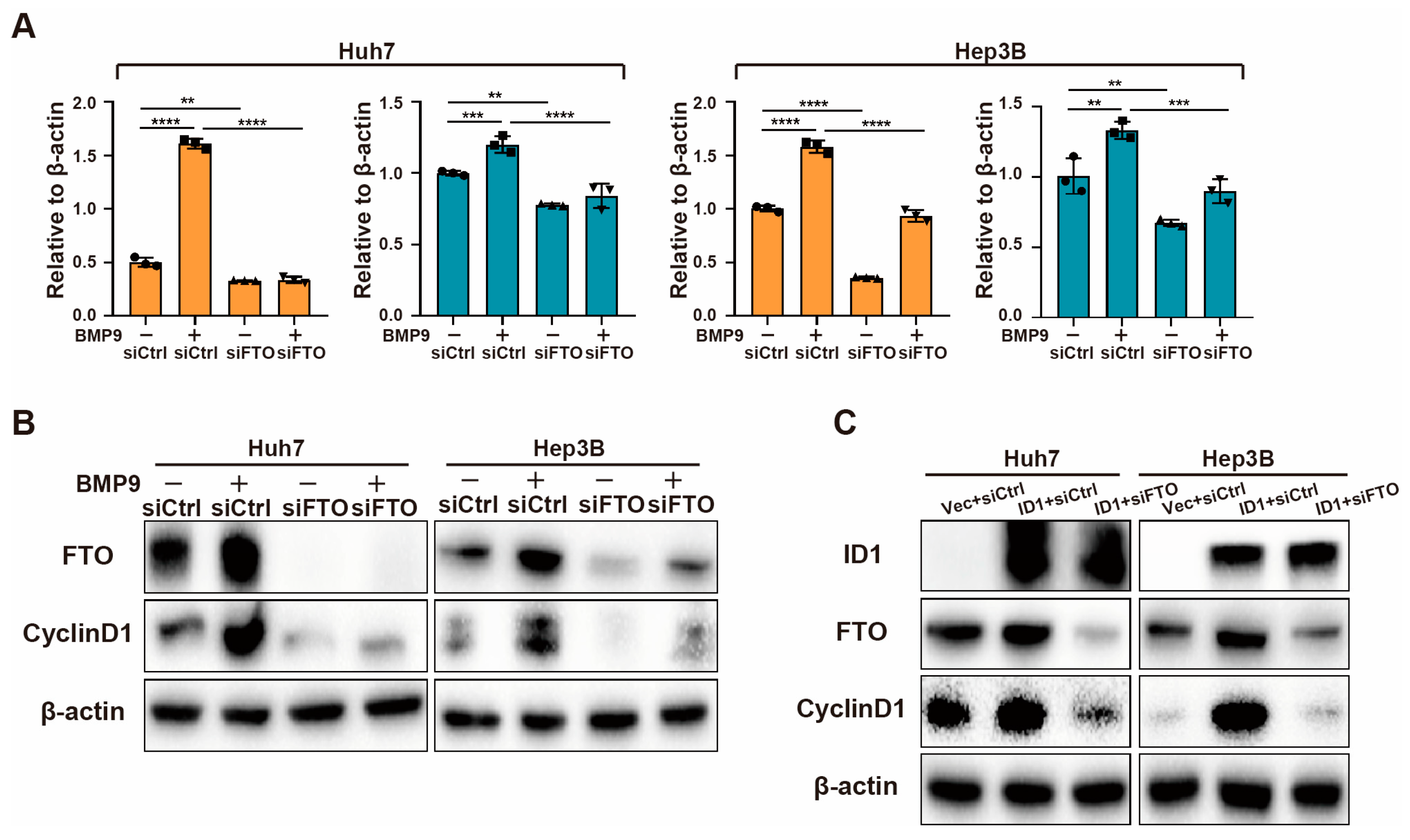
2.4. BMP9-ID1 Pathway Promotes CyclinD1 Expression via FTO
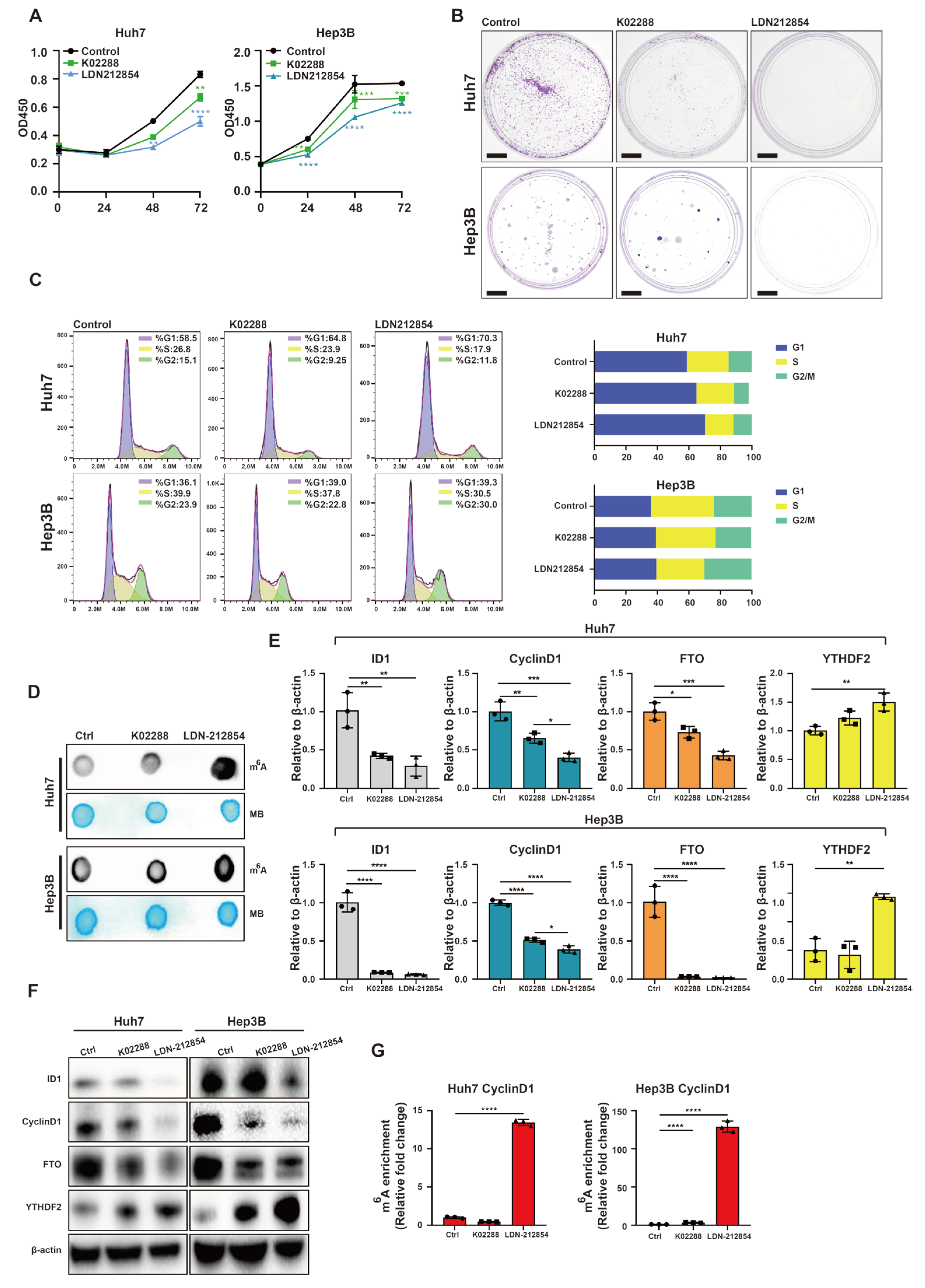
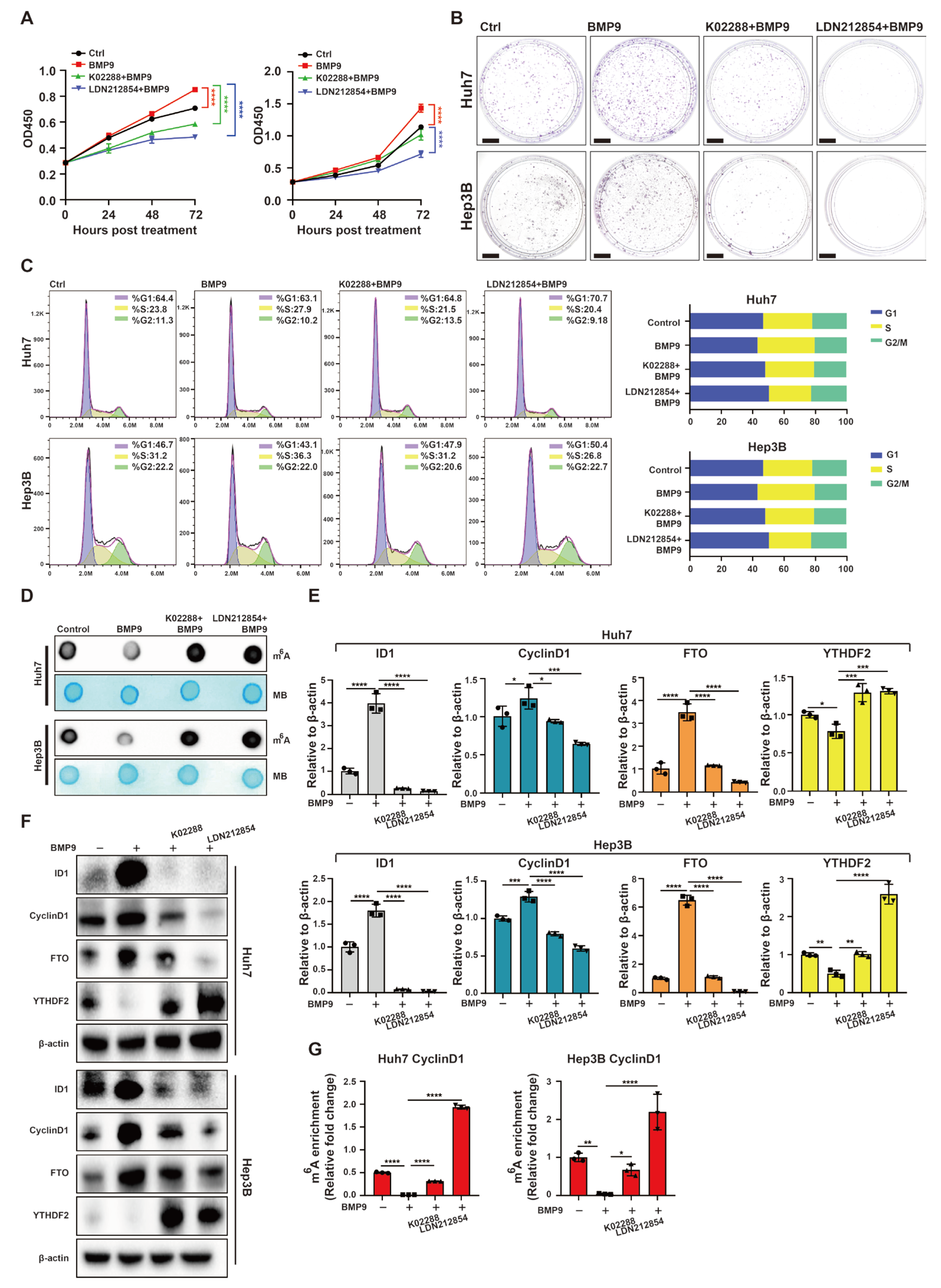
2.5. BMP Receptor Inhibitors Attenuate Upregulation of Progression of Cell Cycle and Downregulate m6A Methylation within the 5′ UTR of CyclinD1 mRNA Induced by BMP9 in HCC Cells
2.6. BMP9 Receptor Inhibitor Suppresses HCC Tumor Growth and CyclinD1 Expression While Inducing Global RNA m6A Methylation In Vivo
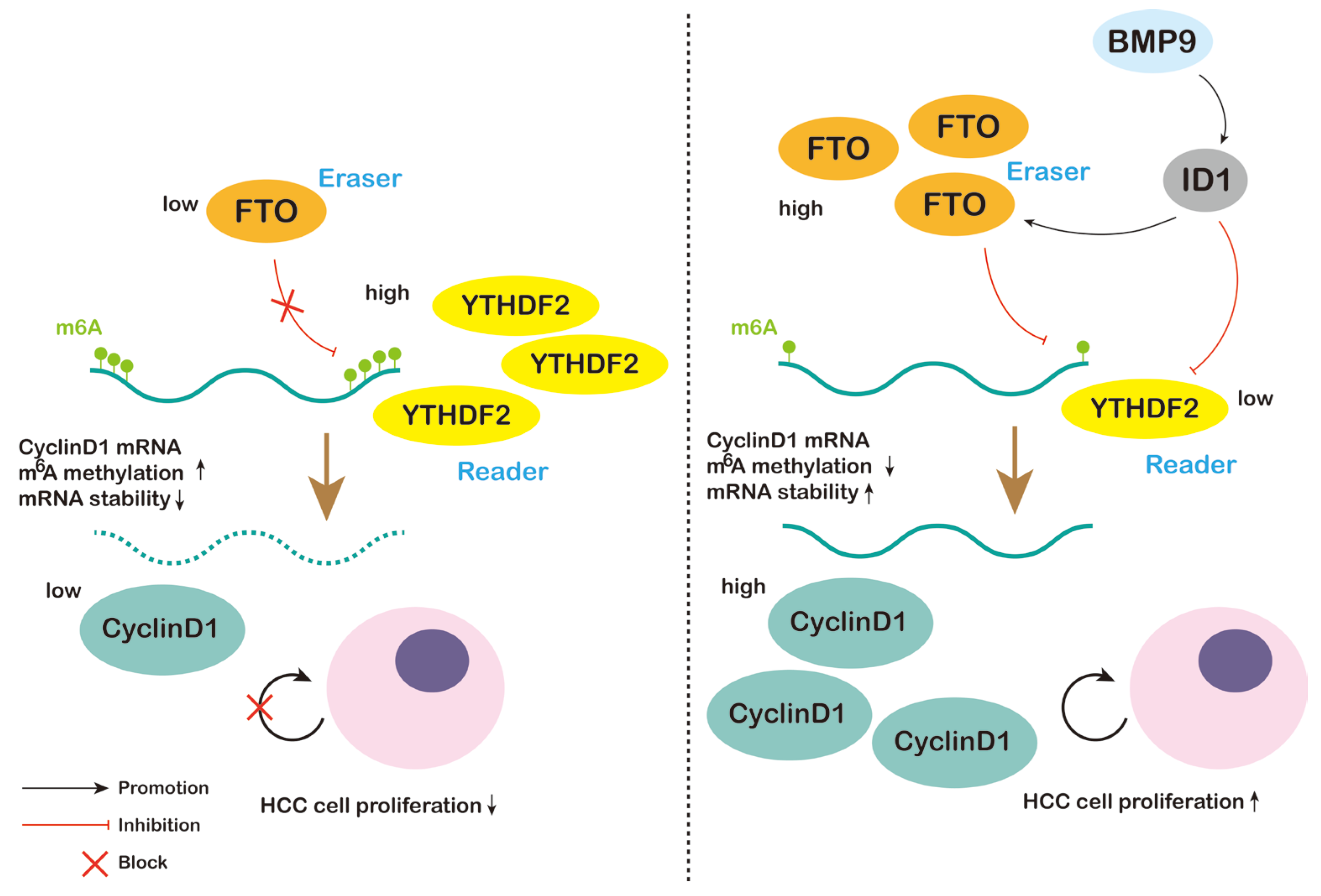
3. Discussion
4. Materials and Methods
4.1. Clinical Samples
4.2. Cell Lines and Reagents
4.3. RNA Interference and Plasmid Transfection
4.4. Cell Proliferation Assay and Colony Formation
4.5. Cell Cycle Analysis
4.6. Real-Time Quantitative PCR
4.7. Western Blotting
4.8. IHC Staining
4.9. m6A Dot Blot Assay
4.10. Methylated RNA Immunoprecipitation (MeRIP)-qPCR
4.11. Animal Studies
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.W.; Talati, C.; Kim, R. Hepatocellular carcinoma (HCC): Beyond sorafenib-chemotherapy. J. Gastrointest. Oncol. 2017, 8, 256–265. [Google Scholar] [CrossRef] [PubMed]
- Medavaram, S.; Zhang, Y. Emerging therapies in advanced hepatocellular carcinoma. Exp. Hematol. Oncol. 2018, 7, 17. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.N.; Green, J.; Wang, Z.; Deng, Y.; Qiao, M.; Peabody, M.; Zhang, Q.; Ye, J.; Yan, Z.; Denduluri, S.; et al. Bone Morphogenetic Protein (BMP) signaling in development and human diseases. Genes Dis. 2014, 1, 87–105. [Google Scholar] [CrossRef] [PubMed]
- Mostafa, S.; Pakvasa, M.; Coalson, E.; Zhu, A.; Alverdy, A.; Castillo, H.; Fan, J.; Li, A.; Feng, Y.; Wu, D.; et al. The wonders of BMP9: From mesenchymal stem cell differentiation, angiogenesis, neurogenesis, tumorigenesis, and metabolism to regenerative medicine. Genes Dis. 2019, 6, 201–223. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Dominissini, D.; Rechavi, G.; He, C. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat. Rev. Genet. 2014, 15, 293–306. [Google Scholar] [CrossRef] [PubMed]
- He, P.C.; He, C. m(6) A RNA methylation: From mechanisms to therapeutic potential. EMBO J. 2021, 40, e105977. [Google Scholar] [CrossRef]
- Song, H.; Zhang, J.; Liu, B.; Xu, J.; Cai, B.; Yang, H.; Straube, J.; Yu, X.; Ma, T. Biological roles of RNA m(5)C modification and its implications in Cancer immunotherapy. Biomark. Res. 2022, 10, 15. [Google Scholar] [CrossRef]
- Li, J.; Zhang, H.; Wang, H. N(1)-methyladenosine modification in cancer biology: Current status and future perspectives. Comput. Struct. Biotechnol. J. 2022, 20, 6578–6585. [Google Scholar] [CrossRef]
- Luo, Y.; Yao, Y.; Wu, P.; Zi, X.; Sun, N.; He, J. The potential role of N(7)-methylguanosine (m7G) in cancer. J. Hematol. Oncol. 2022, 15, 63. [Google Scholar] [CrossRef]
- Zhang, X.; Zhu, W.Y.; Shen, S.Y.; Shen, J.H.; Chen, X.D. Biological roles of RNA m7G modification and its implications in cancer. Biol. Direct 2023, 18, 58. [Google Scholar] [CrossRef]
- Sun, H.; Li, K.; Liu, C.; Yi, C. Regulation and functions of non-m(6)A mRNA modifications. Nat. Rev. Mol. Cell Biol. 2023, 24, 714–731. [Google Scholar] [CrossRef]
- Gilbert, W.V.; Bell, T.A.; Schaening, C. Messenger RNA modifications: Form, distribution, and function. Science 2016, 352, 1408–1412. [Google Scholar] [CrossRef] [PubMed]
- Boo, S.H.; Kim, Y.K. The emerging role of RNA modifications in the regulation of mRNA stability. Exp. Mol. Med. 2020, 52, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Nau, F. The methylation of tRNA. Biochimie 1976, 58, 629–645. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.M.; Masuda, I.; Foster, L.J. tRNA methylation: An unexpected link to bacterial resistance and persistence to antibiotics and beyond. Wiley Interdiscip. Rev. RNA 2020, 11, e1609. [Google Scholar] [CrossRef] [PubMed]
- Bachellerie, J.P.; Cavaille, J. Guiding ribose methylation of rRNA. Trends Biochem. Sci. 1997, 22, 257–261. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Guo, J.; Fan, Z. Interactions between m6A modification and miRNAs in malignant tumors. Cell Death Dis. 2021, 12, 598. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Pan, T. RNA epigenetics. Transl. Res. J. Lab. Clin. Med. 2015, 165, 28–35. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Du, X.; Chen, M.; Han, L.; Sun, J. Novel insight into the functions of N(6)-methyladenosine modified lncRNAs in cancers (Review). Int. J. Oncol. 2022, 61, 152. [Google Scholar] [CrossRef]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef] [PubMed]
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Qian, Y.; Jia, G. The detection and functions of RNA modification m(6)A based on m(6)A writers and erasers. J. Biol. Chem. 2021, 297, 100973. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Wei, J.; He, C. Where, When, and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol. Cell 2019, 74, 640–650. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Jaffrey, S.R. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 2014, 15, 313–326. [Google Scholar] [CrossRef]
- Kostyusheva, A.; Brezgin, S.; Glebe, D.; Kostyushev, D.; Chulanov, V. Host-cell interactions in HBV infection and pathogenesis: The emerging role of m6A modification. Emerg. Microbes Infect. 2021, 10, 2264–2275. [Google Scholar] [CrossRef]
- Yang, L.; Tian, S.; Zheng, X.; Zhang, M.; Zhou, X.; Shang, Y.; Han, Y. N6-methyladenosine RNA methylation in liver diseases: From mechanism to treatment. J. Gastroenterol. 2023, 58, 718–733. [Google Scholar] [CrossRef]
- Pan, X.Y.; Huang, C.; Li, J. The emerging roles of m(6)A modification in liver carcinogenesis. Int. J. Biol. Sci. 2021, 17, 271–284. [Google Scholar] [CrossRef]
- Qie, S.; Diehl, J.A. Cyclin D1, cancer progression, and opportunities in cancer treatment. J. Mol. Med. 2016, 94, 1313–1326. [Google Scholar] [CrossRef]
- Casimiro, M.C.; Velasco-Velazquez, M.; Aguirre-Alvarado, C.; Pestell, R.G. Overview of cyclins D1 function in cancer and the CDK inhibitor landscape: Past and present. Expert Opin. Investig. Drugs 2014, 23, 295–304. [Google Scholar] [CrossRef]
- Kim, J.K.; Diehl, J.A. Nuclear cyclin D1: An oncogenic driver in human cancer. J. Cell. Physiol. 2009, 220, 292–296. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, M.; Wei, F.Y.; Chujo, T.; Oki, S.; Yakita, M.; Kobayashi, D.; Araki, N.; Takahashi, N.; Yoshida, R.; Nakayama, H.; et al. FTO Demethylates Cyclin D1 mRNA and Controls Cell-Cycle Progression. Cell Rep. 2020, 31, 107464. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.Q.; Liu, B.B.; Xu, K.S. New insights into BMP9 signaling in liver diseases. Mol. Cell. Biochem. 2021, 476, 3591–3600. [Google Scholar] [CrossRef]
- Herrera, B.; Dooley, S.; Breitkopf-Heinlein, K. Potential roles of bone morphogenetic protein (BMP)-9 in human liver diseases. Int. J. Mol. Sci. 2014, 15, 5199–5220. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.; Wang, K.; Wang, L. Systematic analysis of the expression profile and prognostic significance of m6A regulators and PD-L1 in hepatocellular carcinoma. Discover. Oncol. 2022, 13, 131. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Chen, H.; Zhang, X.; Liu, W.; Ding, Y.; Huang, D.; Zhai, J.; Wei, W.; Wen, J.; Chen, D.; et al. METTL3 drives NAFLD-related hepatocellular carcinoma and is a therapeutic target for boosting immunotherapy. Cell Rep. Med. 2023, 4, 101144. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Zhuang, Y.; Zhang, J.; Chen, M.; Wu, S. METTL14 Inhibits Hepatocellular Carcinoma Metastasis Through Regulating EGFR/PI3K/AKT Signaling Pathway in an m6A-Dependent Manner. Cancer Manag. Res. 2020, 12, 13173–13184. [Google Scholar] [CrossRef]
- Gan, X.; Dai, Z.; Ge, C.; Yin, H.; Wang, Y.; Tan, J.; Sun, S.; Zhou, W.; Yuan, S.; Yang, F. FTO promotes liver inflammation by suppressing m6A mRNA methylation of IL-17RA. Front. Oncol. 2022, 12, 989353. [Google Scholar] [CrossRef]
- Bian, X.; Shi, D.; Xing, K.; Zhou, H.; Lu, L.; Yu, D.; Wu, W. AMD1 upregulates hepatocellular carcinoma cells stemness by FTO mediated mRNA demethylation. Clin. Transl. Med. 2021, 11, e352. [Google Scholar] [CrossRef]
- Li, J.; Zhu, L.; Shi, Y.; Liu, J.; Lin, L.; Chen, X. m6A demethylase FTO promotes hepatocellular carcinoma tumorigenesis via mediating PKM2 demethylation. Am. J. Transl. Res. 2019, 11, 6084–6092. [Google Scholar]
- Mittenbuhler, M.J.; Saedler, K.; Nolte, H.; Kern, L.; Zhou, J.; Qian, S.B.; Meder, L.; Ullrich, R.T.; Bruning, J.C.; Wunderlich, F.T. Hepatic FTO is dispensable for the regulation of metabolism but counteracts HCC development in vivo. Mol. Metab. 2020, 42, 101085. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; You, S.; Yu, Y.Q.; Zhang, S.; Li, P.T.; Ye, Y.H.; Zhao, W.X.; Li, J.; Li, Q.; Jiao, H.; et al. Decreased nuclear expression of FTO in human primary hepatocellular carcinoma is associated with poor prognosis. Int. J. Clin. Exp. Pathol. 2019, 12, 3376–3383. [Google Scholar] [PubMed]
- Liu, L.; Gu, M.; Ma, J.; Wang, Y.; Li, M.; Wang, H.; Yin, X.; Li, X. CircGPR137B/miR-4739/FTO feedback loop suppresses tumorigenesis and metastasis of hepatocellular carcinoma. Mol. Cancer 2022, 21, 149. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Chen, Y.; Mao, Q.; Jiang, X.; Jiang, W.; Chen, J.; Xu, W.; Zhong, L.; Sun, X. Overexpression of YTHDF1 is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Biomark. Sect. A Dis. Markers 2018, 21, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Cao, M.; Gao, F.; He, X. YTHDF1 promotes hepatocellular carcinoma progression via activating PI3K/AKT/mTOR signaling pathway and inducing epithelial-mesenchymal transition. Exp. Hematol. Oncol. 2021, 10, 35. [Google Scholar] [CrossRef]
- Liu, X.; Qin, J.; Gao, T.; Li, C.; He, B.; Pan, B.; Xu, X.; Chen, X.; Zeng, K.; Xu, M.; et al. YTHDF1 Facilitates the Progression of Hepatocellular Carcinoma by Promoting FZD5 mRNA Translation in an m6A-Dependent Manner. Mol. Ther. Nucleic Acids 2020, 22, 750–765. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Liao, D.; Zhang, M.; Zeng, C.; Li, X.; Zhang, R.; Ma, H.; Kang, T. YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett. 2019, 442, 252–261. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Zhang, H.; Liu, J.; Zhao, Z.; Wang, J.; Lu, Z.; Hu, B.; Zhou, J.; Zhao, Z.; Feng, M.; et al. YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol. Cancer 2019, 18, 163. [Google Scholar] [CrossRef]
- Yang, Y.; Yan, Y.; Yin, J.; Tang, N.; Wang, K.; Huang, L.; Hu, J.; Feng, Z.; Gao, Q.; Huang, A. O-GlcNAcylation of YTHDF2 promotes HBV-related hepatocellular carcinoma progression in an N(6)-methyladenosine-dependent manner. Signal Transduct. Target. Ther. 2023, 8, 63. [Google Scholar] [CrossRef]
- Zhang, C.; Huang, S.; Zhuang, H.; Ruan, S.; Zhou, Z.; Huang, K.; Ji, F.; Ma, Z.; Hou, B.; He, X. YTHDF2 promotes the liver cancer stem cell phenotype and cancer metastasis by regulating OCT4 expression via m6A RNA methylation. Oncogene 2020, 39, 4507–4518. [Google Scholar] [CrossRef]
- Sheth, U.; Parker, R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 2003, 300, 805–808. [Google Scholar] [CrossRef]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef]





| Parameter | BMP9-High (n = 26) | BMP9-Low (n = 25) | p Value * |
|---|---|---|---|
| Age (years, mean, SEM) | 53.38, 1.981 | 54.32, 1.892 | 0.7345 |
| Sex (M/F) | 23/3 | 22/3 | 0.7013 |
| AFP (ng/mL, median) | 262.5 | 8.06 | 0 |
| PIVKA-II (ng/mL, mean, SEM) | 9230, 3763 | 4743, 2173 | 0.3118 |
| BCLC stage (0–B, C–D) | 16, 10 | 21, 4 | 0.1381 |
| Liver cirrhosis (F1–F2/F3–F4) | 2/24 | 3/22 | 0.9632 |
| Microscopic PV invasion (yes/no) | 12/14 | 12/13 | 0.8949 |
| Tumor size (cm3, mean, SEM) | 239, 79.62 | 106.5, 34.08 | 0.1377 |
| Recurrence (yes/no) | 15/11 | 10/15 | 0.2064 |
| Recurrence pattern (single/multiple/extrahepatic) | 2/9/4 | 1/5/4 | 0.7803 |
| Metastasis (yes/no) | 13/13 | 8/17 | 0.1917 |
| Metastasis pattern (intrahepatic/extrahepatic) | 10/3 | 4/4 | 0.3458 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, H.; Zhang, M.; Li, J.; Liu, M.; Cao, D.; Li, Y.-Y.; Yamashita, T.; Nio, K.; Tang, H. BMP9-ID1 Pathway Attenuates N6-Methyladenosine Levels of CyclinD1 to Promote Cell Proliferation in Hepatocellular Carcinoma. Int. J. Mol. Sci. 2024, 25, 981. https://doi.org/10.3390/ijms25020981
Chen H, Zhang M, Li J, Liu M, Cao D, Li Y-Y, Yamashita T, Nio K, Tang H. BMP9-ID1 Pathway Attenuates N6-Methyladenosine Levels of CyclinD1 to Promote Cell Proliferation in Hepatocellular Carcinoma. International Journal of Molecular Sciences. 2024; 25(2):981. https://doi.org/10.3390/ijms25020981
Chicago/Turabian StyleChen, Han, Mingming Zhang, Jianhao Li, Miao Liu, Dan Cao, Ying-Yi Li, Taro Yamashita, Kouki Nio, and Hong Tang. 2024. "BMP9-ID1 Pathway Attenuates N6-Methyladenosine Levels of CyclinD1 to Promote Cell Proliferation in Hepatocellular Carcinoma" International Journal of Molecular Sciences 25, no. 2: 981. https://doi.org/10.3390/ijms25020981






