Identification and Expression Patterns of WOX Transcription Factors under Abiotic Stresses in Pinus massoniana
Abstract
:1. Introduction
2. Results
2.1. Identification and Cloning of WOX Genes in P. massoniana
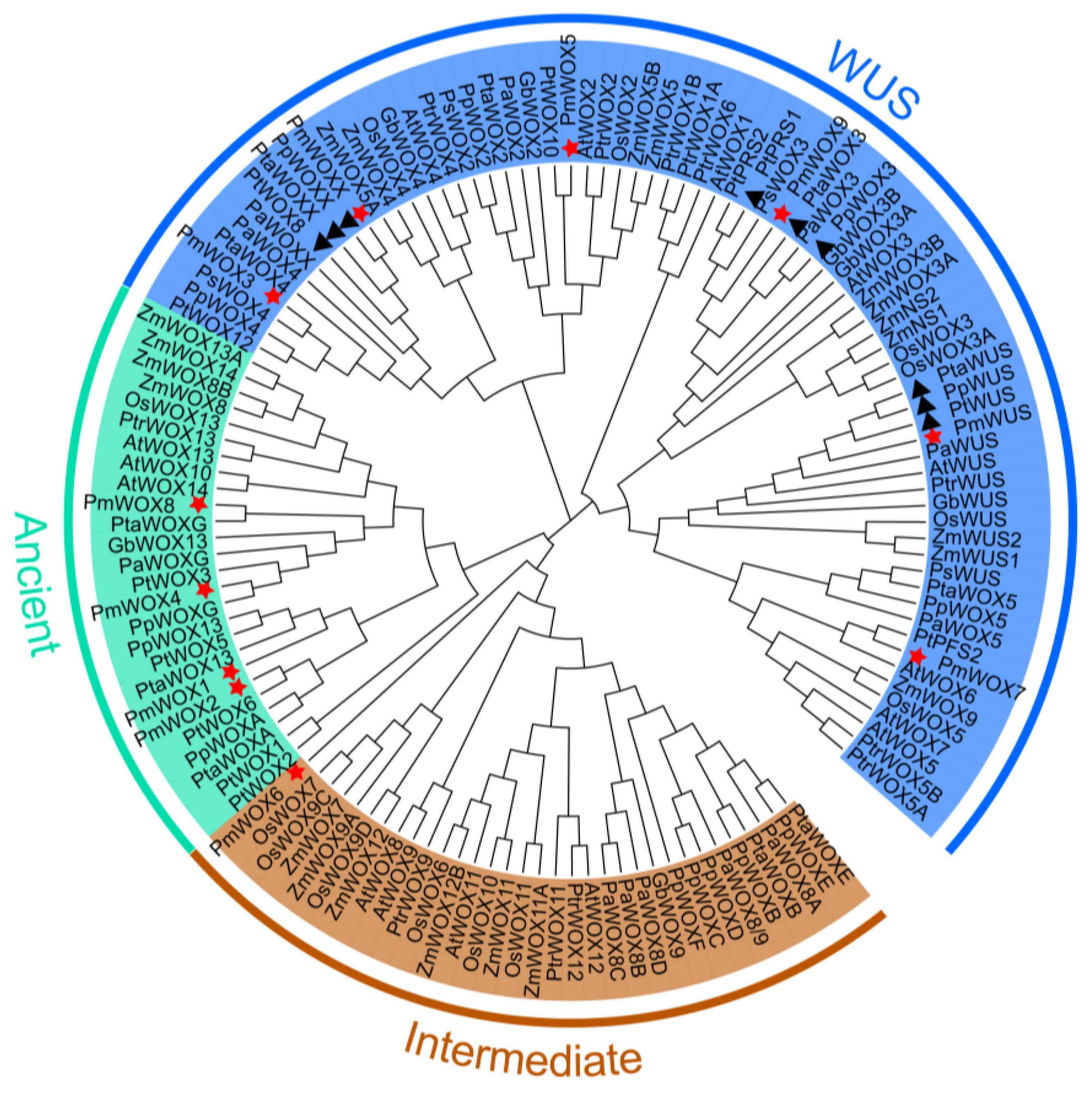
2.2. Phylogenetic Analysis of PmWOX
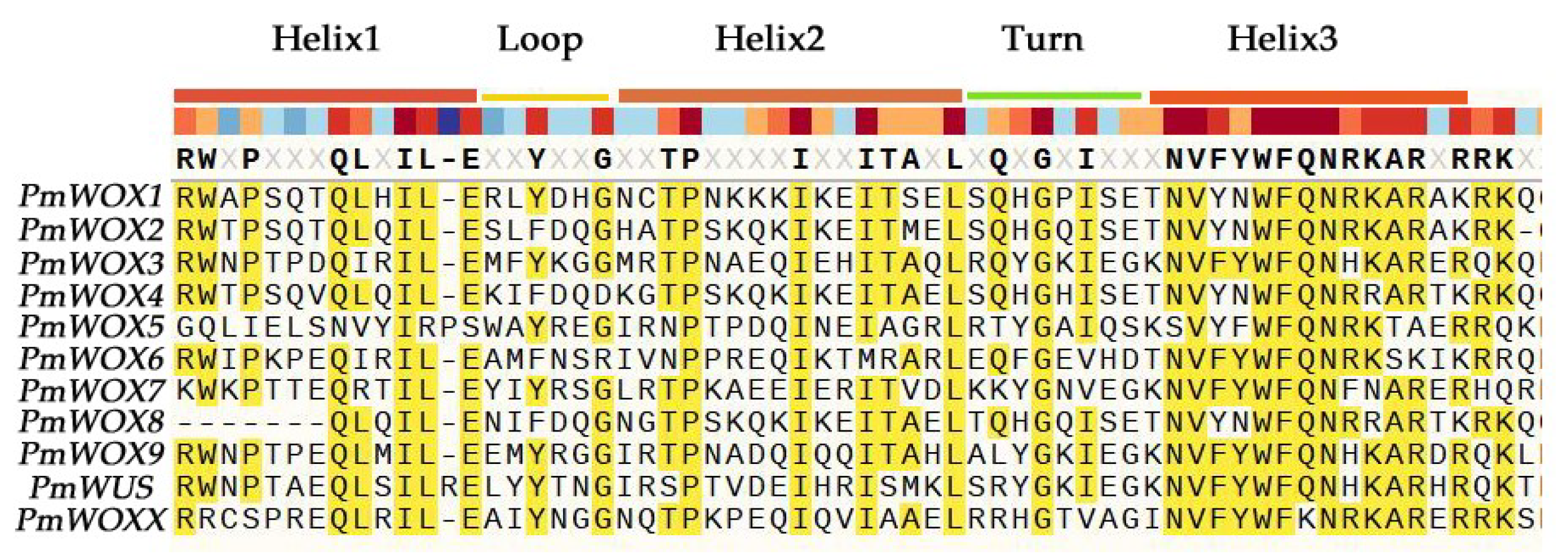
2.3. Protein Domain Analysis of PmWOX Proteins
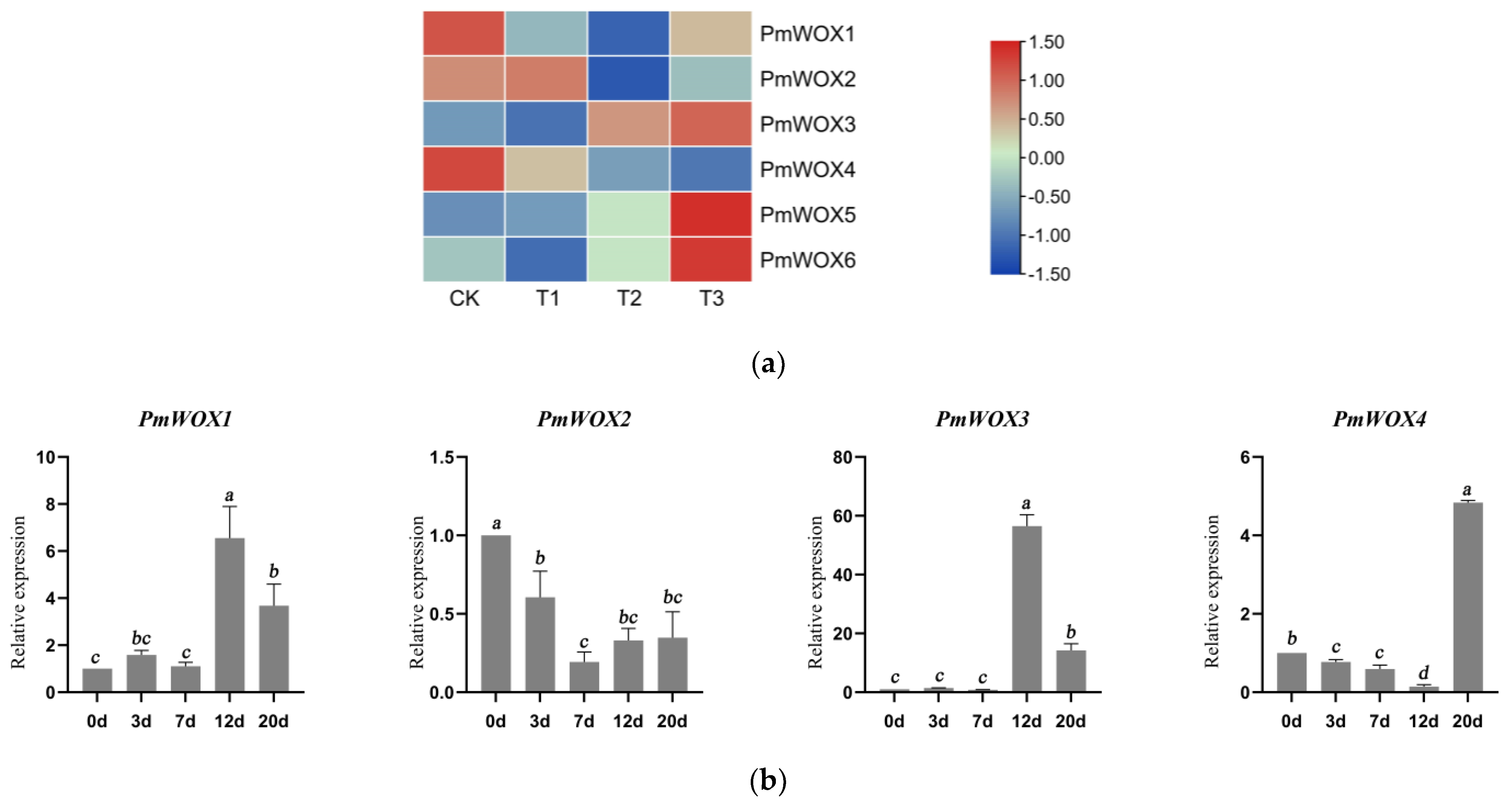
2.4. Analysis of the Transcriptional Profiles of PmWOX Genes under Drought Stress
2.5. Expression Patterns of PmWOX Genes in Different Tissues
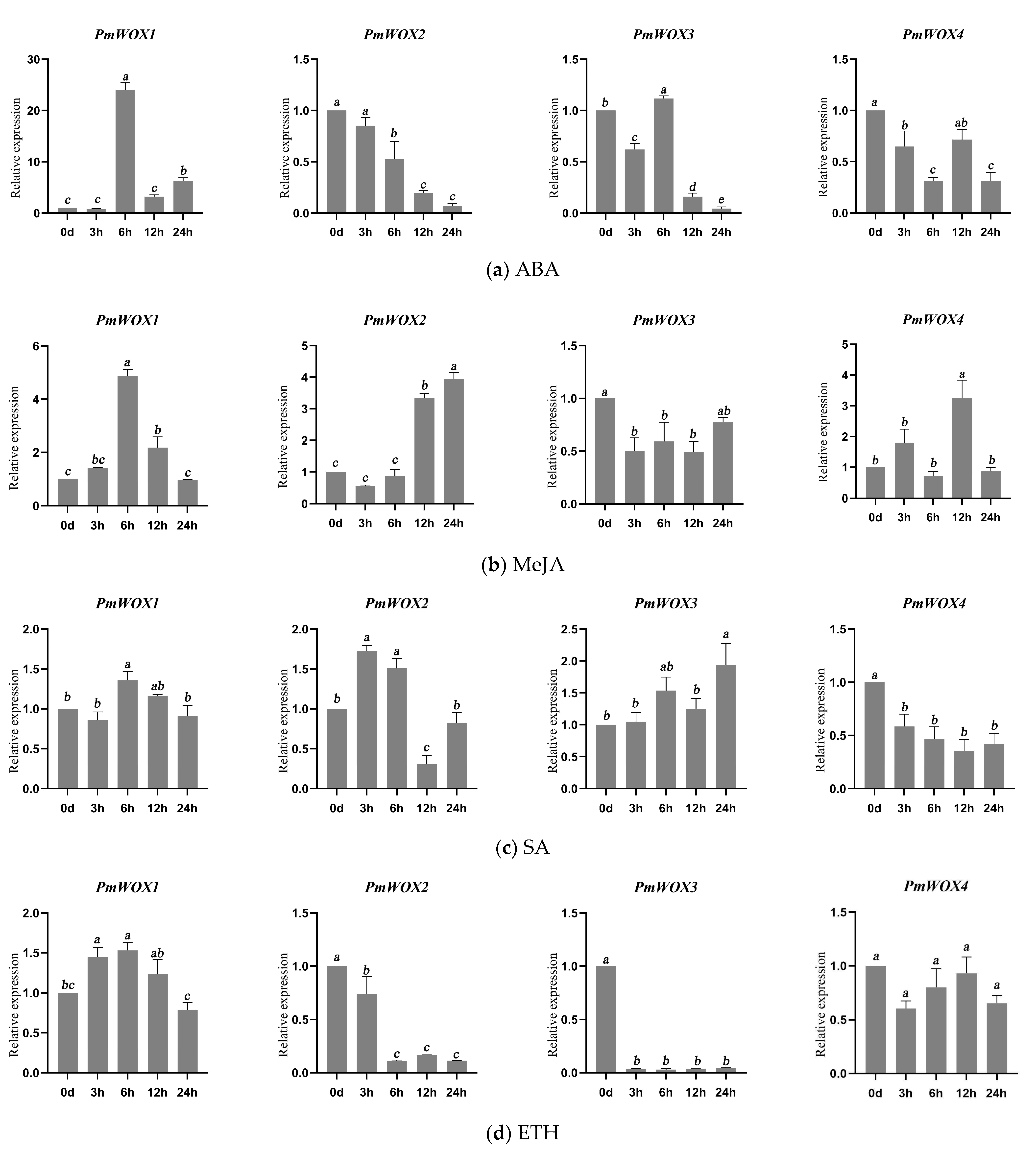
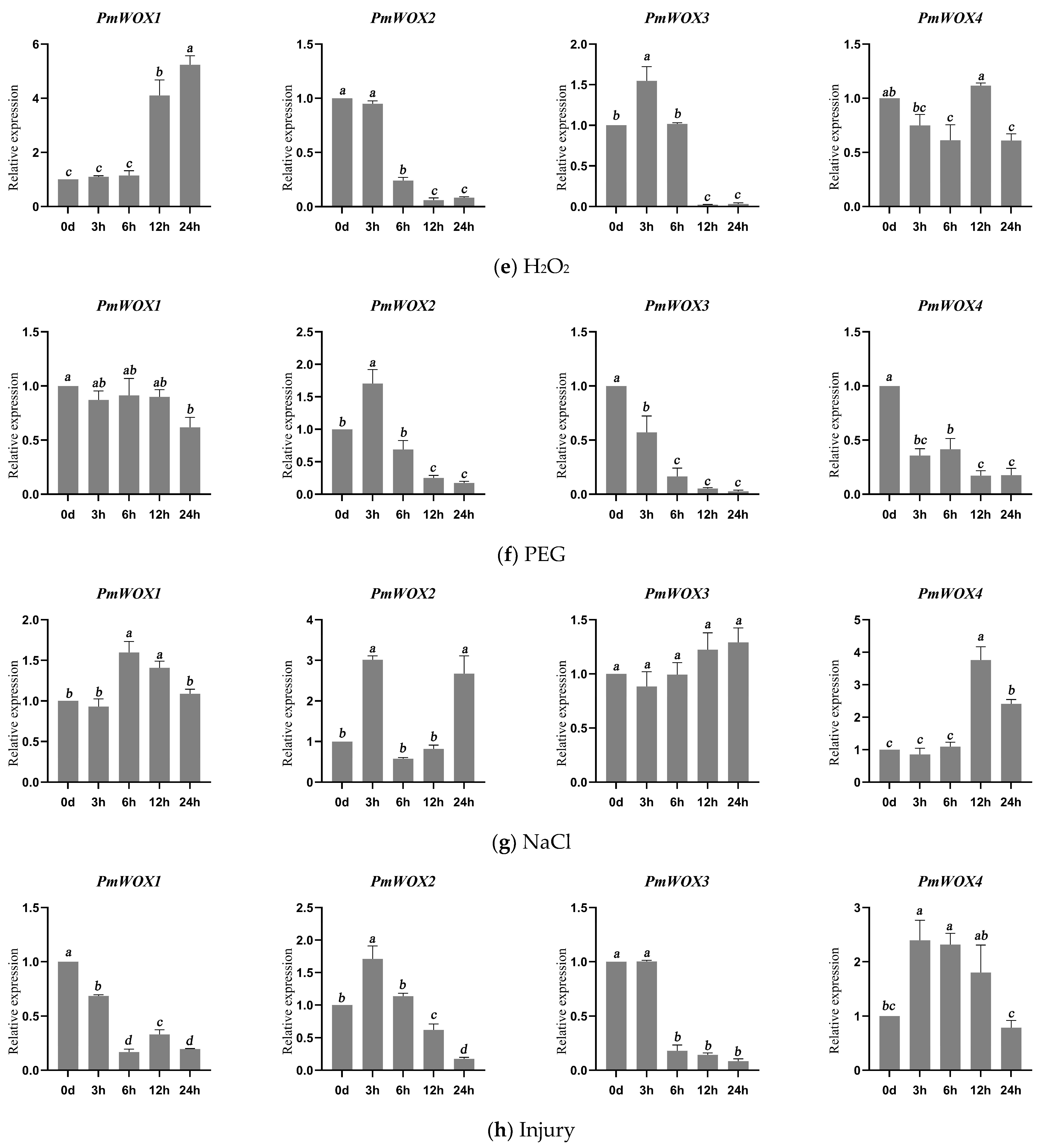
2.6. Expression Patterns of PmWOX Genes under Different Treatments
2.7. Transcriptional Activation Activity
3. Discussion
4. Materials and Methods
4.1. Identification and Cloning of WOX Genes in P. massoniana
4.2. Bioinformatics and Phylogenetic Analysis of PmWOX Proteins
4.3. RNA-Seq Data Analysis under Drought Stress
4.4. Plant Materials and Abiotic Stress Treatments
4.5. RNA Extraction and qRT-PCR Analysis
4.6. Transcriptional-Activation Activity Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bueno, N.; Cuesta, C.; Centeno, M.L.; Ordás, R.J.; Alvarez, J.M. In vitro plant regeneration in conifers: The role of WOX and KNOX gene families. Genes 2021, 12, 438. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.S.; Yang, Z.Q.; Huang, Y.L. Analysis and evaluation of resin productivity and resin component among different half sibling families of Pinus massoniana. J. Beijing For. Univ. 2019, 41, 53–61. [Google Scholar]
- van der Graaff, E.; Laux, T.; Rensing, S.A. The WUS homeobox-containing (WOX) protein family. Genome Biol. 2009, 10, 248. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, K.; Brocchieri, L.; Bürglin, T.R. A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol. Biol. Evol. 2009, 26, 2775–2794. [Google Scholar] [CrossRef] [PubMed]
- Lkeda, M.; Mitsuda, N.; Ohme-Takagi, M. Arabidopsis WUSCHEL is abifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell 2009, 21, 3493–3505. [Google Scholar]
- Nardmann, J.; Reisewitz, P.; Werr, W. Discrete shoot and root stem cell-promoting WUS/WOX5 functions are an evolutionary innovation of angiosperms. Mol. Biol. Evol. 2009, 26, 1745–1755. [Google Scholar] [CrossRef]
- Haecker, A.; Gross-Hardt, R.; Geiges, B.; Sarkar, A.; Breuninger, H.; Herrmann, M.; Laux, T. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 2004, 131, 657–668. [Google Scholar] [CrossRef]
- Alvarez, J.M.; Bueno, N.; Cañas, R.A.; Avila, C.; Cánovas, F.M.; Ordás, R.J. Analysis of the WUSCHELrelated HOMEOBOX gene family in Pinus pinaster: New insights into the gene family evolution. Plant Physiol. Biochem. 2018, 123, 304–318. [Google Scholar] [CrossRef]
- Cheng, S.F.; Huang, Y.L.; Zhu, N.; Zhao, Y. The rice WUSCHEL-related homeobox genes are involved in reproductive organ development, hormone signaling and abiotic stress response. Gene 2014, 549, 266–274. [Google Scholar] [CrossRef]
- Nardmann, J.; Zimmermann, R.; Durantini, D.; Kranz, E.; Werr, W. WOX Gene Phylogeny in Poaceae: A comparative approach addressing leaf and embryo development. Mol. Biol. Evol. 2007, 24, 2474–2484. [Google Scholar] [CrossRef]
- Zhang, X.; Zong, J.; Liu, J.; Yin, J.; Zhang, D. Genome-wide analysis of WOX gene family in rice, sorghum, maize, Arabidopsis and poplar. J. Integr. Plant Biol. 2010, 52, 1016–1026. [Google Scholar] [CrossRef]
- Niu, S.; Li, J.; Bo, W.; Yang, W.; Zuccolo, A.; Giacomello, S.; Chen, X.; Han, F.; Yang, J.; Song, Y.; et al. The Chinese pine genome and methylome unveil key features of conifer evolution. Cell 2022, 185, 204–217. [Google Scholar] [CrossRef]
- Hedman, H.; Zhu, T.; von Arnold, S.; Sohlberg, J.J. Analysis of the WUSCHEL-related homeobox gene family in the conifer Picea abies reveals extensive conservation as well as dynamic patterns. BMC Plant Biol. 2013, 13, 89. [Google Scholar] [CrossRef]
- Nardmann, J.; Werr, W. Symplesiomorphies in the WUSCHEL clade suggest that the last common ancestor of seed plants contained at least four independent stem cell niches. New Phytol. 2013, 199, 1081–1092. [Google Scholar] [CrossRef]
- Li, Z.; Liu, D.; Xia, Y.; Li, Z.; Jing, D.; Du, J.J.; Niu, N.; Ma, S.C.; Wang, J.W.; Song, Y.L.; et al. Identification of the WUSCHEL-related homeobox (WOX) gene family, and interaction and functional analysis of TaWOX9 and TaWUS in wheat. Int. J. Mol. Sci. 2020, 21, 1581. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Gong, Q.; Qin, W.Q.; Yang, Z.R.; Cheng, Y.; Lu, L.L.; Ge, X.Y.; Zhang, C.J.; Wu, Z.X.; Li, F.G. Genome-wide analysis of WOX genes in upland cotton and their expression pattern under different stresses. BMC Plant Biol. 2017, 17, 113. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Shi, H.; Lee, B.H.; Damsz, B.; Cheng, S.; Stirm, V.; Zhu, J.K.; Hasegawa, P.M.; Bressan, R.A. An Arabidopsis homeodomain transcription factor gene, HOS9, mediates cold tolerance through a CBF-independent pathway. Proc. Natl. Acad. Sci. USA 2004, 26, 9873–9878. [Google Scholar] [CrossRef]
- Chen, G.; Feng, H.; Hu, Q.; Qu, H.; Chen, A.; Yu, L.; Xu, G. Improving rice tolerance to potassium deficiency by enhancing OsHAK16p: WOX11-controlledroot development. Plant Biotechnol. J. 2015, 6, 833–848. [Google Scholar] [CrossRef] [PubMed]
- Minh-Thu, P.T.; Kim, J.S.; Chae, S.; Jun, K.M.; Lee, G.; Kim, D.; Cheong, J.; Song, S.I.; Nahm, B.H.; Kim, Y. A WUSCHEL homeobox transcription factor, OsWOX13, enhances drought tolerance and triggers early flowering in rice. Mol. Cells 2018, 41, 781–798. [Google Scholar] [PubMed]
- Wang, L.Q.; Wen, S.S.; Wang, R.; Wang, C.; Gao, B.; Lu, M.Z. PagWOX11/12a activates PagCYP736A12 gene that facilitates salt tolerance in poplar. Plant Biotechnol. J. 2021, 11, 2249–2260. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Wen, S.S.; Sun, T.T.; Wang, R.; Zuo, W.T.; Yang, T.; Wang, C.; Hu, J.J.; Lu, M.Z.; Wang, L.Q. PagWOX11/12a positively regulates the PagSAUR36 gene that enhances adventitious root development in poplar. J. Exp. Bot. 2022, 22, 7298–7311. [Google Scholar] [CrossRef]
- Wang, L.Q.; Li, Z.; Wen, S.S.; Wang, J.N.; Zhao, S.T.; Lu, M.Z. WUSCHEL-related homeobox gene PagWOX11/12a responds to drought stress by enhancing root elongation and biomass growth in poplar. J. Exp. Bot. 2020, 71, 1503–1513. [Google Scholar] [CrossRef]
- Liu, R.; Wang, R.; Lu, M.Z.; Wang, L.Q. WUSCHEL-related homeobox gene PagWOX11/12a is involved in drought tolerance through modulating reactive oxygen species scavenging in poplar. Plant Signal. Behav. 2021, 16, 1866312. [Google Scholar] [CrossRef]
- Chang, Y.; Song, X.; Li, M.; Zhang, Q.; Zhang, P.; Lei, X.; Pei, D. Characterization of walnut JrWOX11 and its overexpression provide insights into adventitious root formation and development and abiotic stress tolerance. Front. Plant Sci. 2022, 13, 951737. [Google Scholar] [CrossRef]
- Tang, Y.H.; Li, H.; Guan, Y.X.; Li, S.; Xun, C.F.; Dong, Y.Y.; Huo, R.; Guo, Y.X.; Bao, X.X.; Pei, E.Q.; et al. Genome-wide identification of the physic nut WUSCHEL-related homeobox gene family and functional analysis of the abiotic stress responsive gene JcWOX5. Front. Genet. 2020, 11, 670. [Google Scholar] [CrossRef] [PubMed]
- Sajjad, M.; Wei, X.; Liu, L.; Li, F.; Ge, X. Transcriptome analysis revealed GhWOX4 intercedes myriad regulatory pathways to modulate drought tolerance and vascular growth in cotton. Int. J. Mol. Sci. 2021, 2, 898. [Google Scholar] [CrossRef]
- Ditengou, F.A.; Teale, W.D.; Kochersperger, P.; Flittner, K.A.; Kneuper, I.; van der Graaff, E.; Nziengui, H.; Pinosa, F.; Li, X.; Nitschke, R.; et al. Mechanical induction of lateral root initiation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2008, 105, 18818–18823. [Google Scholar] [CrossRef]
- Palovaara, J.; Hakman, I. Conifer WOX-related homeodomain transcription factors, developmental consideration and expression dynamic of WOX2 during Picea abies somatic embryogenesis. Plant Mol. Biol. 2008, 66, 533–549. [Google Scholar] [CrossRef]
- Matsumoto, N.; Okada, K. A homeobox gene, PRESSED FLOWER, regulates lateral axis-dependent development of Arabidopsis flowers. Genes Dev. 2001, 15, 3355–3364. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, Y.; Kondo, Y.; Fukuda, H. TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell 2010, 22, 2618–2629. [Google Scholar] [CrossRef] [PubMed]
- Mayer, K.F.; Schoof, H.; Haecker, A.; Lenhard, M.; Jürgens, G.; Laux, T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 1998, 95, 805–815. [Google Scholar] [CrossRef]
- Park, S.O.; Zheng, Z.; Oppenheimer, D.G.; Hauser, B.A. The PRETTY FEW SEEDS2 gene encodes an Arabidopsis homeodomain protein that regulates ovule development. Development 2005, 132, 841–849. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Hu, Y.; Dai, M.; Huang, L.; Zhou, D.X. The WUSCHEL-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. Plant Cell 2009, 21, 736–748. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, G.; Zhao, J.; Chu, H.; Lin, W.; Zhang, D.; Wang, Z.; Liang, W. DWARFT ILLER1, a WUSCHEL-related homeobox transcription factor, is required for tiller growth in rice. PLoS Genet. 2013, 10, 1340. [Google Scholar]
- Liu, B.; Wang, L.; Zhang, J.; Li, J.; Zheng, H.; Chen, J.; Lu, M. WUSCHEL-related homeobox genes in Populus tomentosa; diversified expression patterns and a functional similarity in adventitious root formation. BMC Genom. 2014, 15, 296. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Feng, Y.; Jiang, L.; Zhang, G.; Wu, T.; Zhang, X.; Xu, X.; Wang, Y.; Han, Z. Genome-wide identification of WOX family members in nine Rosaceae species and a functional analysis of MdWOX13-1 in drought resistance. Plant Sci. 2023, 328, 111564. [Google Scholar] [CrossRef] [PubMed]
- Hao, Q.; Zhang, L.; Yang, Y.; Shan, Z.; Zhou, X.A. Genome-wide analysis of the WOX gene family and function exploration of GmWOX18 in soybean. Plants 2019, 7, 215. [Google Scholar] [CrossRef] [PubMed]
- Dai, M.; Hu, Y.; Zhao, Y.; Liu, H.; Zhou, D. A WUSCHEL-like homeobox gene represses a YABBY gene expression required for rice leaf development. Plant Physiol. 2007, 144, 380–390. [Google Scholar] [CrossRef]
- Ueda, M.; Zhang, Z.; Laux, T. Transcriptional activation of Arabidopsis axis patterning genes WOX8/9 links zygote polarity to embryo development. Dev. Cell 2011, 20, 264–270. [Google Scholar] [CrossRef]
- Nadarajah, K.K. ROS homeostasis in abiotic stress tolerance in plants. Int. J. Mol. Sci. 2020, 21, 5208. [Google Scholar] [CrossRef]
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2017, 90, 856–867. [Google Scholar] [CrossRef]
- Udvardi, M.K.; Kakar, K.; Wandrey, M.; Montanri, O.; Murray, J.; Andraiankaja, A.; Zhang, J.Y.; Benedito, V.; Hofer, J.M.; Cheng, F.; et al. Legume transcription factors: Global regulators of plant development and response to the environment. Plant Physiol. 2007, 144, 538–549. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Sun, X.; Zou, B.; Zhu, P.; Ji, K. Transcriptional analysis of Masson pine (Pinus massoniana) under high CO2 stress. Genes 2019, 10, 804. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, D.; Chen, P.; Zhang, C.; Yao, S.; Hao, Q.; Agassin, R.H.; Ji, K. The transcriptomic analysis of the response of pinus massoniana to drought stress and a functional study on the ERF1 transcription factor. Int. J. Mol. Sci. 2023, 24, 11103. [Google Scholar] [CrossRef] [PubMed]
- Zhu, P.; Ma, Y.; Zhu, L.; Chen, Y.; Li, R.; Ji, K. Selection of suitable reference genes in Pinus massoniana Lamb. under different abiotic stresses for qPCR normalization. Forests 2019, 10, 632. [Google Scholar] [CrossRef]
- Wang, D.; Yao, S.; Agassin, R.H.; Zhang, M.; Lou, X.; Huang, Z.; Zhang, J.; Ji, K. Transcriptome-Wide Identification of CCCH-Type Zinc Finger Proteins Family in Pinus massoniana and RR-TZF Proteins in Stress Response. Genes 2022, 13, 1639. [Google Scholar] [CrossRef]




| Protein ID | Prediction of Subcellular Localization | Amino Acid Number (aa) | Protein Molecular Weight (KD) | Isoelectric Point | Instability Index | Grand Average of Hydropathicity |
|---|---|---|---|---|---|---|
| PmWOX1 | nucleus | 354 | 40.03 | 6.21 | 47.92 | −0.857 |
| PmWOX2 | nucleus | 366 | 40.68 | 5.92 | 43.82 | −0.783 |
| PmWOX3 | nucleus | 482 | 53.15 | 7.11 | 57.57 | −0.654 |
| PmWOX4 | nucleus | 294 | 33.23 | 6.61 | 52.45 | −0.657 |
| PmWOX5 | nucleus | 171 | 19.64 | 8.80 | 38.32 | −0.753 |
| PmWOX6 | nucleus | 117 | 13.64 | 10.26 | 58.31 | −0.997 |
| PmWOX7 | nucleus | 80 | 9.71 | 9.35 | 52.38 | −1.290 |
| PmWOX8 | nucleus | 110 | 12.75 | 6.47 | 55.07 | −1.286 |
| PmWOX9 | nucleus | 189 | 21.95 | 8.19 | 63.33 | −0.695 |
| PmWUS | nucleus | 215 | 24.88 | 8.05 | 63.64 | −0.926 |
| PmWOXX | nucleus | 168 | 18.83 | 5.45 | 65.06 | −0.656 |
| Motif | Sequence | Number of Amino Acids in the Motif |
|---|---|---|
| 1 | ILEAJYDQGIRTPSKEQIKEITAELSQYGKIEGTNVFYWFQNRKARERRK | 50 |
| 2 | QVMTEEQLETLRRQICVYSTICSQLVEMHRAMSQQ | 35 |
| 3 | FRPSARTRWNPTPEQL | 16 |
| 4 | DGKQWZVPVGVVDVRRMFGENAVLLDSRGHMVPTNDMGMSFHPLQGSEGY | 50 |
| 5 | GESEVDTDLESPKEKKVKMDH | 21 |
| 6 | TLZLFPVHPE | 10 |
| 7 | CCWGGC | 10 |
| 8 | RMLYDL | 6 |
| 9 | CEEAFCC | 7 |
| 10 | CVEEYDC | 7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, D.; Qiu, Z.; Xu, T.; Yao, S.; Zhang, M.; Cheng, X.; Zhao, Y.; Ji, K. Identification and Expression Patterns of WOX Transcription Factors under Abiotic Stresses in Pinus massoniana. Int. J. Mol. Sci. 2024, 25, 1627. https://doi.org/10.3390/ijms25031627
Wang D, Qiu Z, Xu T, Yao S, Zhang M, Cheng X, Zhao Y, Ji K. Identification and Expression Patterns of WOX Transcription Factors under Abiotic Stresses in Pinus massoniana. International Journal of Molecular Sciences. 2024; 25(3):1627. https://doi.org/10.3390/ijms25031627
Chicago/Turabian StyleWang, Dengbao, Zimo Qiu, Tao Xu, Sheng Yao, Mengyang Zhang, Xiang Cheng, Yulu Zhao, and Kongshu Ji. 2024. "Identification and Expression Patterns of WOX Transcription Factors under Abiotic Stresses in Pinus massoniana" International Journal of Molecular Sciences 25, no. 3: 1627. https://doi.org/10.3390/ijms25031627






