The Predictive Value of Gut Microbiota Composition for Sustained Immunogenicity following Two Doses of CoronaVac
Abstract
:1. Introduction
2. Results
2.1. Demographics and Baseline Characteristics
2.2. Baseline Gut Microbiota Composition Was Associated with Immune Response to CoronaVac at Day 180
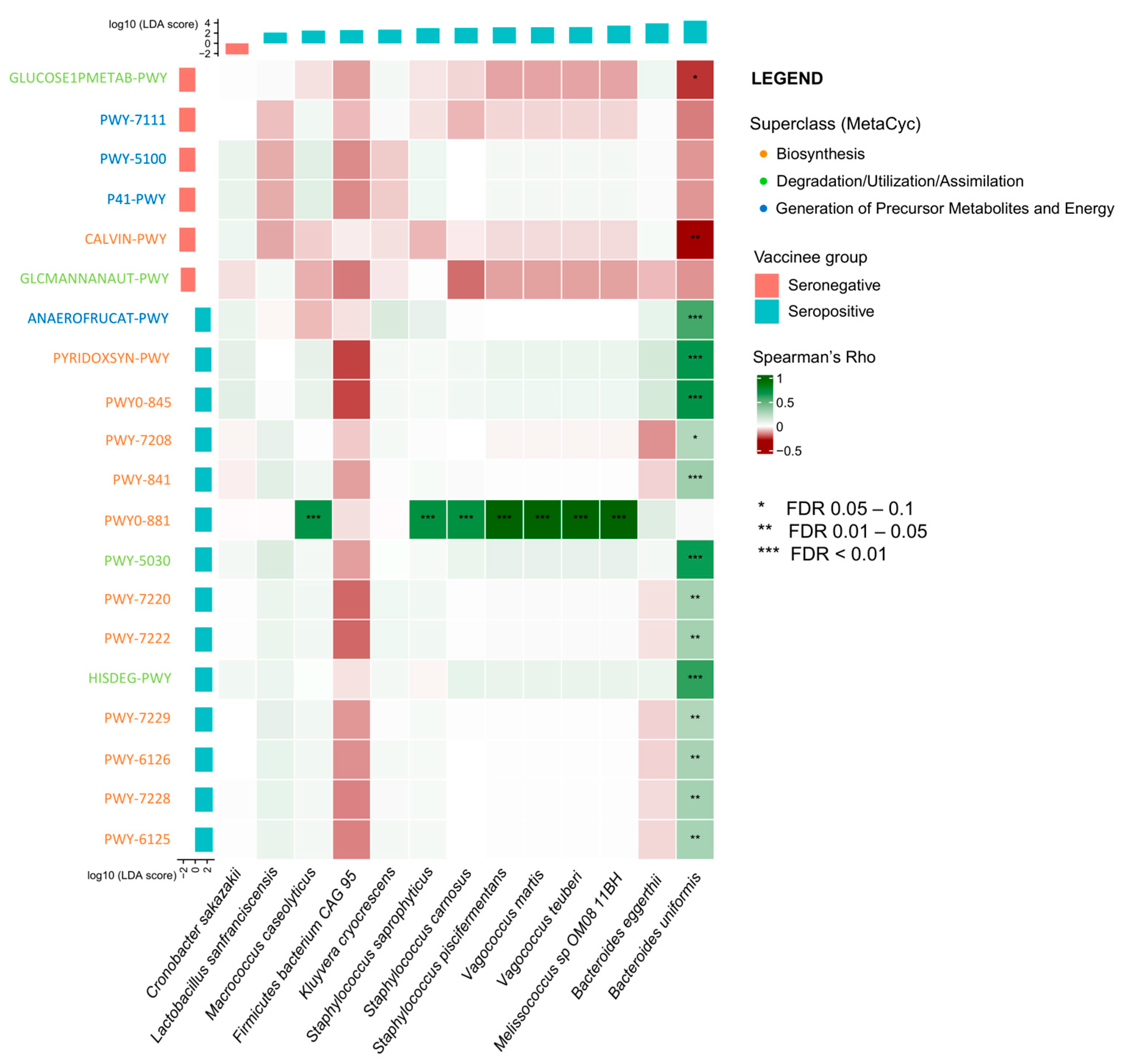
2.3. Association between Baseline Gut Microbiota and Metabolic Pathways and Vaccine Immunogenicity at Day 180
2.4. Correlation between Gut Microbiota and Metabolic Pathways on Vaccine Immunogenicity
2.5. Association between Baseline Gut Microbiota Composition and CoronaVac-Related Adverse Events
2.5.1. After 1st Dose Vaccine
2.5.2. After 2nd Dose Vaccine
3. Discussion
4. Methods
4.1. Study Design and Participants
4.2. Collection of Demographics, Blood and Stool Samples
4.3. Primary and Secondary Outcomes of Interest
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. WHO Coronavirus (COVID-19) Dashboard; WHO: Geneva, Switzerland, 2022; Volume 2022. [Google Scholar]
- Haas, E.J.; Angulo, F.J.; McLaughlin, J.M.; Anis, E.; Singer, S.R.; Khan, F.; Brooks, N.; Smaja, M.; Mircus, G.; Pan, K.; et al. Impact and effectiveness of mRNA BNT162b2 vaccine against SARS-CoV-2 infections and COVID-19 cases, hospitalisations, and deaths following a nationwide vaccination campaign in Israel: An observational study using national surveillance data. Lancet 2021, 397, 1819–1829. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, M.H.G.; de Souza, T.F.G.; de Carvalho Araujo, F.M.; de Andrade, L.O.M. Dynamics of antibody response to CoronaVac vaccine. J. Med. Virol. 2022, 94, 2139–2148. [Google Scholar] [CrossRef] [PubMed]
- Cowling, B.J.; Wong, I.O.L.; Shiu, E.Y.C.; Lai, A.Y.T.; Cheng, S.M.S.; Chaothai, S.; Kwan, K.K.H.; Martin-Sanchez, M.; Poon, L.L.M.; Ip, D.K.M.; et al. Strength and durability of antibody responses to BNT162b2 and CoronaVac. Vaccine 2022, 40, 4312–4317. [Google Scholar] [CrossRef] [PubMed]
- Lynn, D.J.; Benson, S.C.; Lynn, M.A.; Pulendran, B. Modulation of immune responses to vaccination by the microbiota: Implications and potential mechanisms. Nat. Rev. Immunol. 2022, 22, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Georg, P.; Sander, L.E. Innate sensors that regulate vaccine responses. Curr. Opin. Immunol. 2019, 59, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.Z.; Ravindran, R.; Chassaing, B.; Carvalho, F.A.; Maddur, M.S.; Bower, M.; Hakimpour, P.; Gill, K.P.; Nakaya, H.I.; Yarovinsky, F.; et al. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity 2014, 41, 478–492. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Kim, Y.G.; Seo, S.U.; Kim, D.J.; Kamada, N.; Prescott, D.; Chamaillard, M.; Philpott, D.J.; Rosenstiel, P.; Inohara, N.; et al. Nod2-mediated recognition of the microbiota is critical for mucosal adjuvant activity of cholera toxin. Nat. Med. 2016, 22, 524–530. [Google Scholar] [CrossRef]
- Kim, M.; Qie, Y.; Park, J.; Kim, C.H. Gut Microbial Metabolites Fuel Host Antibody Responses. Cell Host Microbe 2016, 20, 202–214. [Google Scholar] [CrossRef]
- Hagan, T.; Cortese, M.; Rouphael, N.; Boudreau, C.; Linde, C.; Maddur, M.S.; Das, J.; Wang, H.; Guthmiller, J.; Zheng, N.Y.; et al. Antibiotics-Driven Gut Microbiome Perturbation Alters Immunity to Vaccines in Humans. Cell 2019, 178, 1313–1328.e13. [Google Scholar] [CrossRef]
- Ng, H.Y.; Leung, W.K.; Cheung, K.S. Association between Gut Microbiota and SARS-CoV-2 Infection and Vaccine Immunogenicity. Microorganisms 2023, 11, 452. [Google Scholar] [CrossRef]
- Cheung, K.S.; Lam, L.K.; Zhang, R.; Ooi, P.H.; Tan, J.T.; To, W.P.; Hui, C.H.; Chan, K.H.; Seto, W.K.; Hung, I.F.N.; et al. Association between Recent Usage of Antibiotics and Immunogenicity within Six Months after COVID-19 Vaccination. Vaccines 2022, 10, 1122. [Google Scholar] [CrossRef]
- Cheung, K.S.; Yan, V.K.C.; Lam, L.K.; Ye, X.; Hung, I.F.N.; Chan, E.W.; Leung, W.K. Antibiotic Use Prior to COVID-19 Vaccine Is Associated with Higher Risk of COVID-19 and Adverse Outcomes: A Propensity-Scored Matched Territory-Wide Cohort. Vaccines 2023, 11, 1341. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.C.; Peng, Y.; Zhang, L.; Mok, C.K.; Zhao, S.; Li, A.; Ching, J.Y.; Liu, Y.; Yan, S.; Chan, D.L.S.; et al. Gut microbiota composition is associated with SARS-CoV-2 vaccine immunogenicity and adverse events. Gut 2022, 71, 1106–1116. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Tang, L.; He, W.; Jiang, X.; Hu, C.; Li, Y.; Zhang, Y.; Pang, K.; Lei, Y.; Li, S.; et al. Correlation of gut microbiota and metabolic functions with the antibody response to the BBIBP-CorV vaccine. Cell Rep. Med. 2022, 3, 100752. [Google Scholar] [CrossRef] [PubMed]
- Gauffin Cano, P.; Santacruz, A.; Moya, A.; Sanz, Y. Bacteroides uniformis CECT 7771 ameliorates metabolic and immunological dysfunction in mice with high-fat-diet induced obesity. PLoS ONE 2012, 7, e41079. [Google Scholar] [CrossRef]
- Lopez-Almela, I.; Romani-Perez, M.; Bullich-Vilarrubias, C.; Benitez-Paez, A.; Gomez Del Pulgar, E.M.; Frances, R.; Liebisch, G.; Sanz, Y. Bacteroides uniformis combined with fiber amplifies metabolic and immune benefits in obese mice. Gut Microbes 2021, 13, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wolin, M.J. Influence of heme and vitamin B12 on growth and fermentations of Bacteroides species. J. Bacteriol. 1981, 145, 466–471. [Google Scholar] [CrossRef]
- Dziarski, R.; Park, S.Y.; Kashyap, D.R.; Dowd, S.E.; Gupta, D. Pglyrp-Regulated Gut Microflora Prevotella falsenii, Parabacteroides distasonis and Bacteroides eggerthii Enhance and Alistipes finegoldii Attenuates Colitis in Mice. PLoS ONE 2016, 11, e0146162. [Google Scholar] [CrossRef]
- Li, M.; Liu, B.; Li, R.; Yang, P.; Leng, P.; Huang, Y. Exploration of the link between gut microbiota and purinergic signalling. Purinergic Signal. 2022, 19, 315–327. [Google Scholar] [CrossRef]
- Schenk, U.; Westendorf, A.M.; Radaelli, E.; Casati, A.; Ferro, M.; Fumagalli, M.; Verderio, C.; Buer, J.; Scanziani, E.; Grassi, F. Purinergic control of T cell activation by ATP released through pannexin-1 hemichannels. Sci. Signal 2008, 1, ra6. [Google Scholar] [CrossRef]
- Trautmann, A. Extracellular ATP in the immune system: More than just a “danger signal”. Sci. Signal 2009, 2, pe6. [Google Scholar] [CrossRef]
- Caspi, R.; Billington, R.; Fulcher, C.A.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Midford, P.E.; Ong, Q.; Ong, W.K.; et al. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 2018, 46, D633–D639. [Google Scholar] [CrossRef]
- Ganor, Y.; Levite, M. The neurotransmitter glutamate and human T cells: Glutamate receptors and glutamate-induced direct and potent effects on normal human T cells, cancerous human leukemia and lymphoma T cells, and autoimmune human T cells. J. Neural Transm. 2014, 121, 983–1006. [Google Scholar] [CrossRef] [PubMed]
- Kumrungsee, T.; Zhang, P.; Chartkul, M.; Yanaka, N.; Kato, N. Potential Role of Vitamin B6 in Ameliorating the Severity of COVID-19 and Its Complications. Front. Nutr. 2020, 7, 562051. [Google Scholar] [CrossRef] [PubMed]
- Kunisawa, J.; Kiyono, H. Vitamin-mediated regulation of intestinal immunity. Front. Immunol. 2013, 4, 189. [Google Scholar] [CrossRef] [PubMed]
- Kurashige, S.; Akuzawa, Y.; Fujii, N.; Kishi, S.; Takeshita, M.; Miyamoto, Y. Effect of vitamin B complex on the immunodeficiency produced by surgery of gastric cancer patients. Jpn. J. Exp. Med. 1988, 58, 197–202. [Google Scholar] [PubMed]
- Chilton, P.M.; Hadel, D.M.; To, T.T.; Mitchell, T.C.; Darveau, R.P. Adjuvant activity of naturally occurring monophosphoryl lipopolysaccharide preparations from mucosa-associated bacteria. Infect. Immun. 2013, 81, 3317–3325. [Google Scholar] [CrossRef] [PubMed]
- Hickey, C.A.; Kuhn, K.A.; Donermeyer, D.L.; Porter, N.T.; Jin, C.; Cameron, E.A.; Jung, H.; Kaiko, G.E.; Wegorzewska, M.; Malvin, N.P.; et al. Colitogenic Bacteroides thetaiotaomicron Antigens Access Host Immune Cells in a Sulfatase-Dependent Manner via Outer Membrane Vesicles. Cell Host Microbe 2015, 17, 672–680. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, M.T.; Cresci, G.A.M. The Immunomodulatory Functions of Butyrate. J. Inflamm. Res. 2021, 14, 6025–6041. [Google Scholar] [CrossRef]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermudez-Humaran, L.G.; Gratadoux, J.J.; Blugeon, S.; Bridonneau, C.; Furet, J.P.; Corthier, G.; et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef]
- Shaw, K.A.; Bertha, M.; Hofmekler, T.; Chopra, P.; Vatanen, T.; Srivatsa, A.; Prince, J.; Kumar, A.; Sauer, C.; Zwick, M.E.; et al. Dysbiosis, inflammation, and response to treatment: A longitudinal study of pediatric subjects with newly diagnosed inflammatory bowel disease. Genome Med. 2016, 8, 75. [Google Scholar] [CrossRef]
- Nie, K.; Ma, K.; Luo, W.; Shen, Z.; Yang, Z.; Xiao, M.; Tong, T.; Yang, Y.; Wang, X. Roseburia intestinalis: A Beneficial Gut Organism From the Discoveries in Genus and Species. Front. Cell Infect. Microbiol. 2021, 11, 757718. [Google Scholar] [CrossRef]
- Luo, C.; Wei, X.; Song, J.; Xu, X.; Huang, H.; Fan, S.; Zhang, D.; Han, L.; Lin, J. Interactions between Gut Microbiota and Polyphenols: New Insights into the Treatment of Fatigue. Molecules 2022, 27, 7377. [Google Scholar] [CrossRef]
- Kittel, M.; Muth, M.C.; Zahn, I.; Roth, H.J.; Thiaucourt, M.; Gerhards, C.; Haselmann, V.; Neumaier, M.; Findeisen, P. Clinical evaluation of commercial automated SARS-CoV-2 immunoassays. Int. J. Infect. Dis. 2021, 103, 590–596. [Google Scholar] [CrossRef]
- Muena, N.A.; Garcia-Salum, T.; Pardo-Roa, C.; Avendano, M.J.; Serrano, E.F.; Levican, J.; Almonacid, L.I.; Valenzuela, G.; Poblete, E.; Strohmeier, S.; et al. Induction of SARS-CoV-2 neutralizing antibodies by CoronaVac and BNT162b2 vaccines in naive and previously infected individuals. EBioMedicine 2022, 78, 103972. [Google Scholar] [CrossRef]
- Earle, K.A.; Ambrosino, D.M.; Fiore-Gartland, A.; Goldblatt, D.; Gilbert, P.B.; Siber, G.R.; Dull, P.; Plotkin, S.A. Evidence for antibody as a protective correlate for COVID-19 vaccines. Vaccine 2021, 39, 4423–4428. [Google Scholar] [CrossRef] [PubMed]
- Khoury, D.S.; Cromer, D.; Reynaldi, A.; Schlub, T.E.; Wheatley, A.K.; Juno, J.A.; Subbarao, K.; Kent, S.J.; Triccas, J.A.; Davenport, M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021, 27, 1205–1211. [Google Scholar] [CrossRef] [PubMed]
- Bergwerk, M.; Gonen, T.; Lustig, Y.; Amit, S.; Lipsitch, M.; Cohen, C.; Mandelboim, M.; Levin, E.G.; Rubin, C.; Indenbaum, V.; et al. COVID-19 Breakthrough Infections in Vaccinated Health Care Workers. N. Engl. J. Med. 2021, 385, 1474–1484. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.H.; Leung, K.Y.; Zhang, R.R.; Liu, D.; Fan, Y.; Chen, H.; Yuen, K.Y.; Hung, I.F. Performance of a Surrogate SARS-CoV-2-Neutralizing Antibody Assay in Natural Infection and Vaccination Samples. Diagnostics 2021, 11, 1757. [Google Scholar] [CrossRef] [PubMed]
- Norquist, J.M.; Khawaja, S.S.; Kurian, C.; Mast, T.C.; Liaw, K.L.; Robertson, M.N.; Evans, B.; Gutsch, D.; Saddier, P. Adaptation of a previously validated vaccination report card for use in adult vaccine clinical trials to align with the 2007 FDA Toxicity Grading Scale Guidance. Hum. Vaccines Immunother. 2012, 8, 1208–1212. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Truong, D.T.; Franzosa, E.A.; Tickle, T.L.; Scholz, M.; Weingart, G.; Pasolli, E.; Tett, A.; Huttenhower, C.; Segata, N. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods. 2015, 12, 902–903. [Google Scholar] [CrossRef] [PubMed]
- Beghini, F.; McIver, L.J.; Blanco-Míguez, A.; Dubois, L.; Asnicar, F.; Maharjan, S.; Mailyan, A.; Manghi, P.; Scholz, M.; Thomas, A.M.; et al. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. eLife 2021, 10, e65088. [Google Scholar] [CrossRef]


| Characteristic | Overall, n = 119 1 | Seronegative, n = 111 1 | Seropositive, n = 8 1 | p-Value 2 |
|---|---|---|---|---|
| Age (years) ≥ 55 (n, %) | 51 (42.9%) | 46 (41.4%) | 5 (62.5%) | 0.286 |
| Male sex (n, %) | 39 (32.8%) | 39 (35.1%) | 0 (0.0%) | 0.052 |
| Antibiotics * (n, %) | 5 (4.2%) | 4 (3.6%) | 1 (12.5%) | 0.298 |
| Proton pump inhibitors * (n, %) | 14 (11.8%) | 13 (11.7%) | 1 (12.5%) | >0.999 |
| Prebiotics/Probiotics (n, %) | 3 (2.5%) | 3 (2.7%) | 0 (0.0%) | >0.999 |
| Univariate Logistic Regression | Univariate Linear Regression | |||||
|---|---|---|---|---|---|---|
| Characteristic | OR 1 | 95% CI 1 | p-Value | Beta | 95% CI 1 | p-Value |
| Clinical factors | ||||||
| Age ≥ 55 years old | 2.36 | 0.55, 11.94 | 0.257 | 1.68 | −0.33, 3.70 | 0.101 |
| Sex | NA * | NA * | 0.992 | −2.03 | −4.14, 0.09 | 0.060 |
| Antibiotics | 3.82 | 0.18, 30.70 | 0.258 | 1.21 | −3.81, 6.23 | 0.635 |
| Proton pump inhibitors | 1.08 | 0.06, 6.78 | 0.947 | −0.83 | −3.96, 2.29 | 0.598 |
| High relative abundance of baseline gut microbiota species | ||||||
| Bacteroides uniformis | 3.27 | 0.73, 14.72 | 0.110 | 0.69 | −1.63, 3.01 | 0.556 |
| Bacteroides eggerthii | 5.73 | 1.32, 29.55 | 0.022 | 2.52 | 0.24, 4.79 | 0.031 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ng, H.-Y.; Liao, Y.; Zhang, R.; Chan, K.-H.; To, W.-P.; Hui, C.-H.; Seto, W.-K.; Leung, W.K.; Hung, I.F.N.; Lam, T.T.Y.; et al. The Predictive Value of Gut Microbiota Composition for Sustained Immunogenicity following Two Doses of CoronaVac. Int. J. Mol. Sci. 2024, 25, 2583. https://doi.org/10.3390/ijms25052583
Ng H-Y, Liao Y, Zhang R, Chan K-H, To W-P, Hui C-H, Seto W-K, Leung WK, Hung IFN, Lam TTY, et al. The Predictive Value of Gut Microbiota Composition for Sustained Immunogenicity following Two Doses of CoronaVac. International Journal of Molecular Sciences. 2024; 25(5):2583. https://doi.org/10.3390/ijms25052583
Chicago/Turabian StyleNg, Ho-Yu, Yunshi Liao, Ruiqi Zhang, Kwok-Hung Chan, Wai-Pan To, Chun-Him Hui, Wai-Kay Seto, Wai K. Leung, Ivan F. N. Hung, Tommy T. Y. Lam, and et al. 2024. "The Predictive Value of Gut Microbiota Composition for Sustained Immunogenicity following Two Doses of CoronaVac" International Journal of Molecular Sciences 25, no. 5: 2583. https://doi.org/10.3390/ijms25052583





