Evolution of the WRKY Family in Angiosperms and Functional Diversity under Environmental Stress
Abstract
:1. Introduction
2. Results
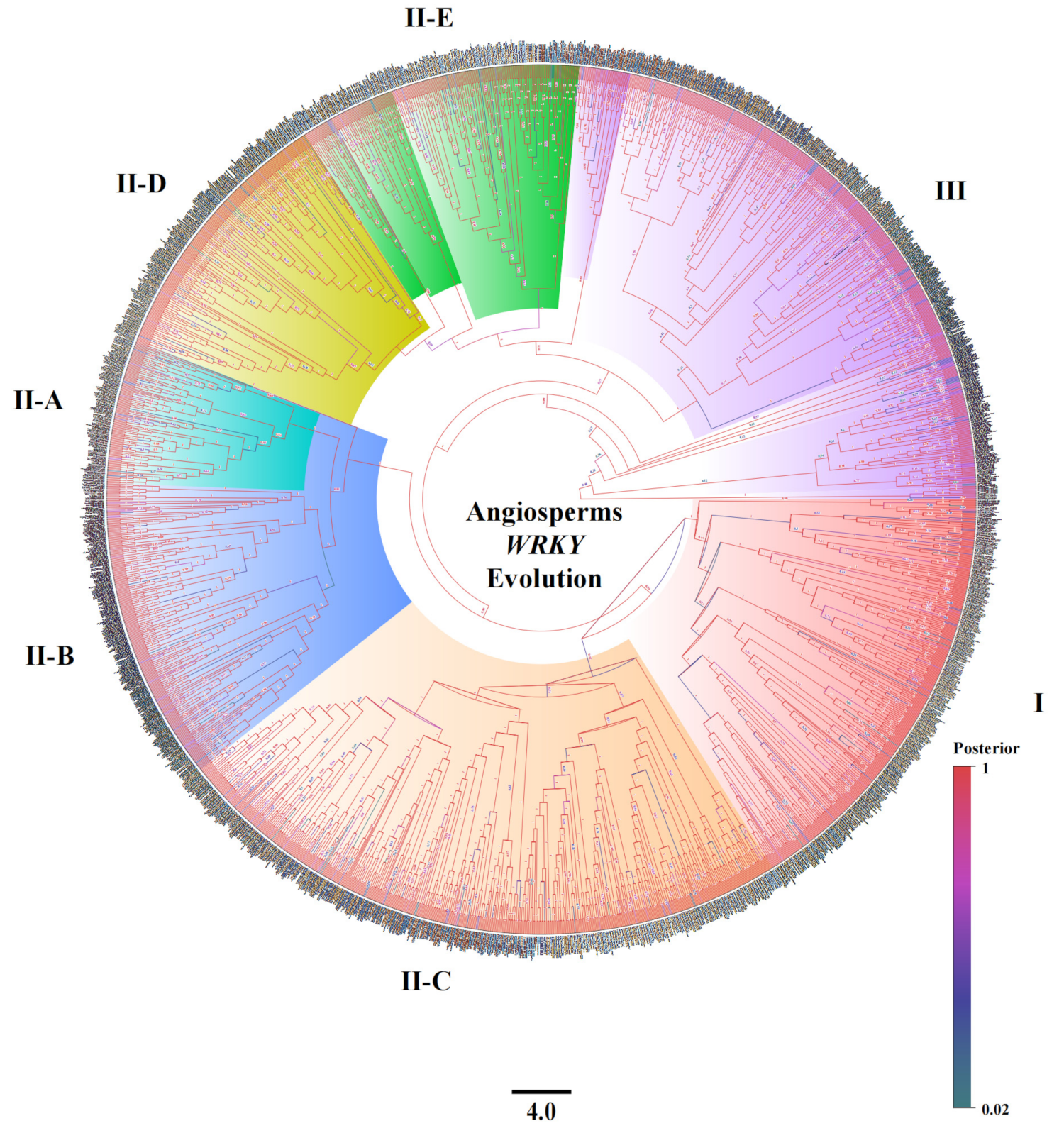
2.1. Grouping Characteristics of the WRKY Family
2.2. The Structural Characteristics and Grouping of the WRKY Family
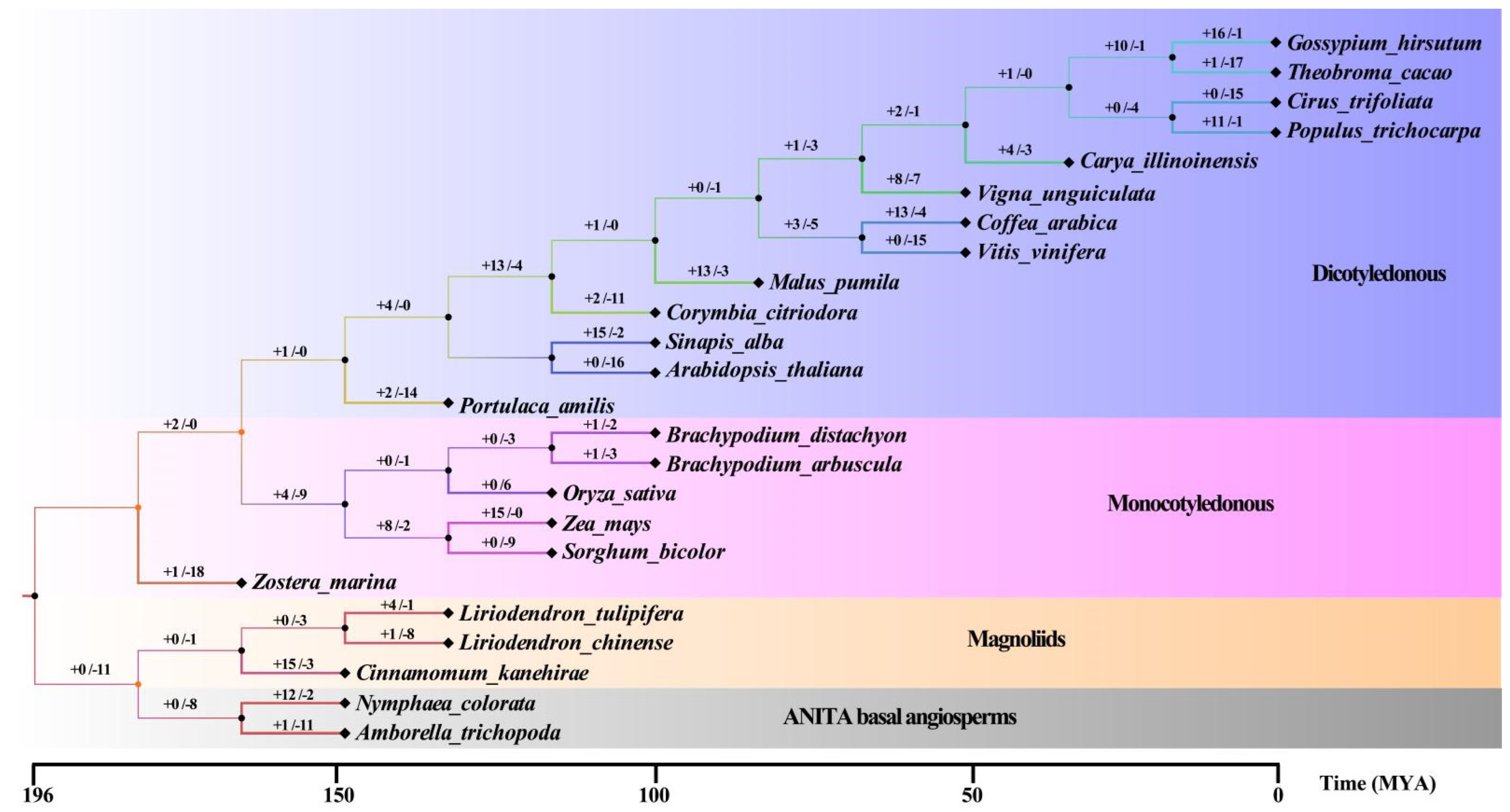
2.3. Origin and Evolution of the WRKY Family
2.4. The Environmental Selection Pressure of the WRKY Family
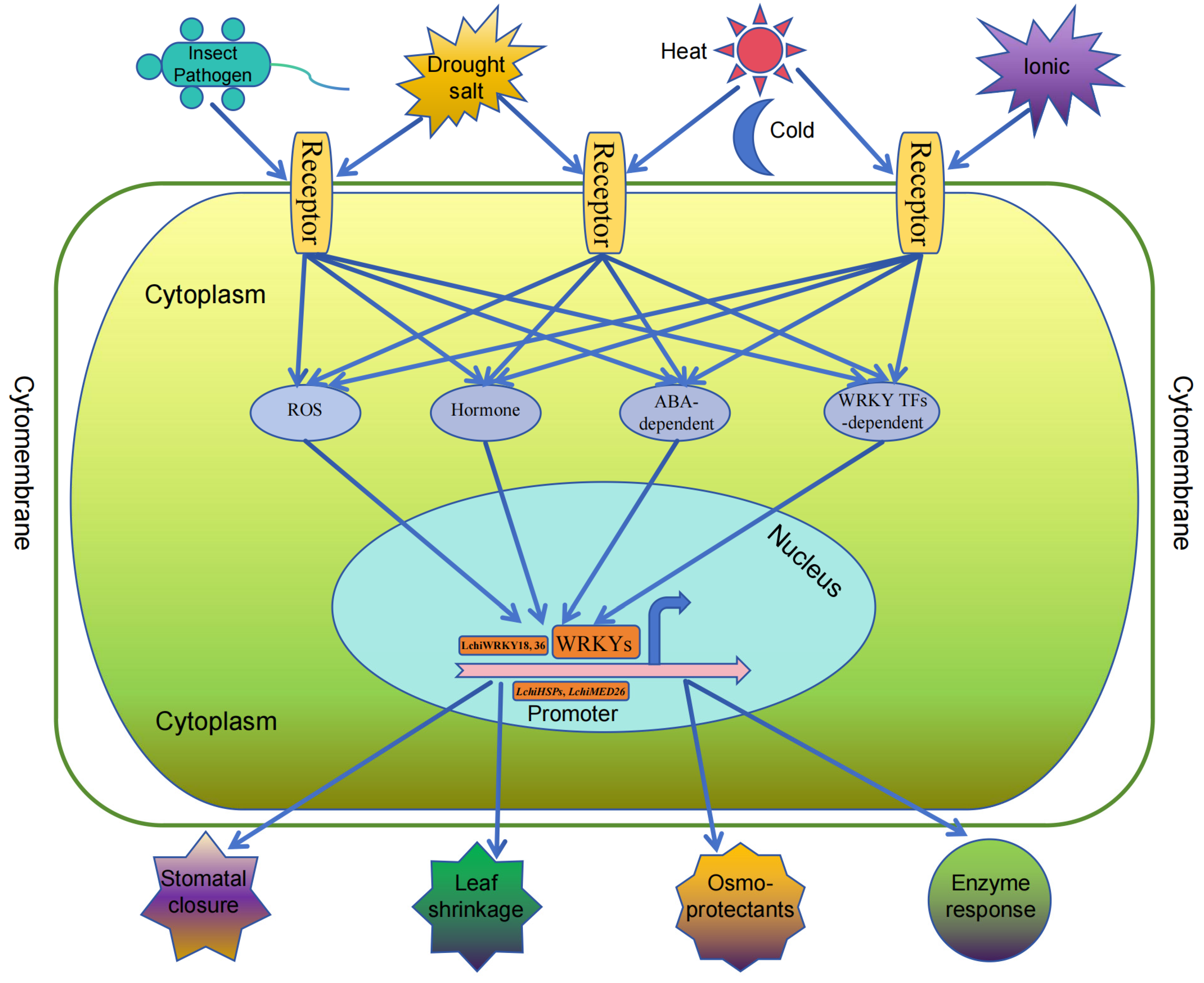
2.5. Development Function of WRKY Transcription Factor
2.6. Functions of the WRKY Family in Biotic Stress
2.7. Functions of the WRKY Family in Abiotic Stress
3. Discussion
3.1. The Evolution of the WRKY Family in Higher Plants Is Relatively Conservative
3.2. The Environment Selection Preserves the Existing WRKY Number of Family Members and Different Expansion–Contraction Ratios
3.3. The Conservatism of Domain and 3D Structure of the WRKY Family
3.4. Functional Diversity of the WRKY Family in Angiosperms
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Javed, T.; Gao, S. WRKY transcription factors in plant defense. Trends Genet. 2023, 39, 787–801. [Google Scholar] [CrossRef] [PubMed]
- Goyal, P.; Devi, R.; Verma, B.; Hussain, S.; Arora, P.; Tabassum, R.; Gupta, S. WRKY transcription factors: Evolution, regulation, and functional diversity in plants. Protoplasma 2023, 260, 331–348. [Google Scholar] [CrossRef] [PubMed]
- Rai, G.K.; Mishra, S.; Chouhan, R.; Mushtaq, M.; Chowdhary, A.A.; Rai, P.K.; Kumar, R.R.; Kumar, P.; Perez-Alfocea, F.; Colla, G.; et al. Plant salinity stress, sensing, and its mitigation through WRKY. Front. Plant Sci. 2023, 14, 1238507. [Google Scholar] [CrossRef] [PubMed]
- Khoso, M.A.; Hussain, A.; Ritonga, F.N.; Ali, Q.; Channa, M.M.; Alshegaihi, R.M.; Meng, Q.; Ali, M.; Zaman, W.; Brohi, R.D.; et al. WRKY transcription factors (TFs): Molecular switches to regulate drought, temperature, and salinity stresses in plants. Front. Plant Sci. 2022, 13, 1039329. [Google Scholar] [CrossRef] [PubMed]
- Hsin, K.T.; Hsieh, M.-C.; Lee, Y.-H.; Lin, K.-C.; Cheng, Y.-S. Insight into the Phylogeny and Binding Ability of WRKY Transcription Factors. Int. J. Mol. Sci. 2022, 23, 2895. [Google Scholar] [CrossRef] [PubMed]
- Wani, S.H.; Anand, S.; Singh, B.; Bohra, A.; Joshi, R. WRKY transcription factors and plant defense responses: Latest discoveries and future prospects. Plant Cell Rep. 2021, 40, 1071–1085. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Luan, Y.; Meng, J.; Sun, J.; Tao, J.; Zhao, D. WRKY Transcription Factor Response to High-Temperature Stress. Plants 2021, 10, 2211. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Wang, X.; Mostafa, S.; Noor, I.; Lin, X.; Ren, S.; Cui, J.; Jin, B. WRKY Transcription Factors in Jasminum sambac: An Insight into the Regulation of Aroma Synthesis. Biomolecules 2023, 13, 1679. [Google Scholar] [CrossRef]
- Guo, X.; Ullah, A.; Siuta, D.; Kukfisz, B.; Iqbal, S. Role of WRKY Transcription Factors in Regulation of Abiotic Stress Responses in Cotton. Life 2022, 12, 1410. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, H.; Chen, L.; Lin, J.; Wang, Z.; Pan, J.; Yang, F.; Ni, X.; Wang, Y.; Wang, Y.; et al. Multifaceted roles of WRKY transcription factors in abiotic stress and flavonoid biosynthesis. Front. Plant Sci. 2023, 14, 1303667. [Google Scholar] [CrossRef]
- Wang, H.; Cheng, X.; Yin, D.; Chen, D.; Luo, C.; Liu, H.; Huang, C. Advances in the Research on Plant WRKY Transcription Factors Responsive to External Stresses. Curr. Issues Mol. Biol. 2023, 45, 2861–2880. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, T.B.; Ribas, A.F.; de Souza, S.G.H.; Budzinski, I.G.F.; Domingues, D.S. Physiological Responses to Drought, Salinity, and Heat Stress in Plants: A Review. Stresses 2022, 2, 113–135. [Google Scholar] [CrossRef]
- Chen, J.; Hao, Z.; Guang, X.; Zhao, C.; Wang, P.; Xue, L.; Zhu, Q.; Yang, L.; Sheng, Y.; Zhou, Y.; et al. Liriodendron genome sheds light on angiosperm phylogeny and species-pair differentiation. Nat. Plants 2019, 5, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Chaw, S.M.; Liu, Y.-C.; Wu, Y.-W.; Wang, H.-Y.; Lin, C.-Y.I.; Wu, C.-S.; Ke, H.-M.; Chang, L.-Y.; Hsu, C.-Y.; Yang, H.-T.; et al. Stout camphor tree genome fills gaps in understanding of flowering plant genome evolution. Nat. Plants 2019, 5, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Fu, H.; Mi, F.; Yang, Y.; Wang, Y.; Li, Z.; He, Y.; Yue, Z. Genomic characterization of WRKY transcription factors related to secoiridoid biosynthesis in Gentiana macrophylla. BMC Plant Biol. 2024, 24, 66. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Hong, S.; Wang, J.; Shang, L.; Zhang, G.; Zhao, Y.; Ma, Q.; Ma, D. Identification and expression analysis of the bZIP and WRKY gene families during anthocyanins biosynthesis in Lagerstroemia indica L. Hortic. Environ. Biotechnol. 2024, 65, 169–180. [Google Scholar] [CrossRef]
- Yang, M.; Wang, Y.; Chen, C.; Xin, X.; Dai, S.; Meng, C.; Ma, N. Transcription factor WRKY75 maintains auxin homeostasis to promote tomato defense against Pseudomonas syringae. Plant Physiol. 2024, kiae025. [Google Scholar] [CrossRef]
- Li, D.; Li, X.; Wang, Z.; Wang, H.; Gao, J.; Liu, X.; Zhang, Z. Transcription factors RhbZIP17 and RhWRKY30 enhance resistance to Botrytis cinerea by increasing lignin content in rose petals. J. Exp. Bot. 2024, 75, 1633–1646. [Google Scholar] [CrossRef]
- Tang, R.; Zhu, Y.; Yang, S.; Wang, F.; Chen, G.; Chen, J.; Zhao, K.; Liu, Z.; Peng, D. Genome-Wide Identification and Analysis of WRKY Gene Family in Melastoma dodecandrum. Int. J. Mol. Sci. 2023, 24, 14904. [Google Scholar] [CrossRef]
- Meng, L.; Yang, H.; Yang, J.; Ye, T.; Xiang, L.; Chan, Z.; Wang, Y. Tulip transcription factor TgWRKY75 activates salicylic acid and abscisic acid biosynthesis to synergistically promote petal senescence. J. Exp. Bot. 2024, erae021. [Google Scholar] [CrossRef]
- Jia, M.; Ni, Y.; Zhao, H.; Liu, X.; Yan, W.; Zhao, X.; Wang, J.; He, B.; Liu, H. Full-length transcriptome and RNA-Seq analyses reveal the resistance mechanism of sesame in response to Corynespora cassiicola. BMC Plant Biol. 2024, 24, 64. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kim, J.; Choi, D.S.; Kim, M.-S.; Deslandes, L.; Jayaraman, J.; Sohn, K.H. Molecular basis for the interference of the Arabidopsis WRKY54-mediated immune response by two sequence-unrelated bacterial effectors. Plant J. Cell Mol. Biol. 2024. [Google Scholar] [CrossRef]
- Deokar, A.A.; Sagi, M.; Tar’an, B. Genetic Analysis of Partially Resistant and Susceptible Chickpea Cultivars in Response to Ascochyta rabiei Infection. Int. J. Mol. Sci. 2024, 25, 1360. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Wang, J.; Wang, Z.; Zhu, T.; Zheng, Y.; Hawar, A.; Chang, Y.; Wang, X.; Li, D.; Wang, G.; et al. Capture of regulatory factors via CRISPR-dCas9 for mechanistic analysis of fine-tuned SERRATE expression in Arabidopsis. Nat. Plants 2024, 10, 86–99. [Google Scholar] [CrossRef] [PubMed]
- Czarnocka, W.; Karpiński, S. Friend or foe? Reactive oxygen species production, scavenging, and signaling in plant response to environmental stresses. Free Radic. Biol. Med. 2018, 122, 4–20. [Google Scholar] [CrossRef] [PubMed]
- Ahanger, M.A.; Akram, N.A.; Ashraf, M.; Alyemeni, M.N.; Wijaya, L.; Ahmad, P. Plant responses to environmental stresses—From gene to biotechnology. AoB Plants 2017, 9, plx025. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; He, Z.; Chen, N.; Tang, Z.; Wang, Q.; Cai, Y. The Roles of Environmental Factors in Regulation of Oxidative Stress in Plant. BioMed Res. Int. 2019, 2019, 9732325. [Google Scholar] [CrossRef]
- Doebley, J.; Lukens, L. Transcriptional regulators and the evolution of plant form. Plant Cell 1998, 10, 1075–1082. [Google Scholar] [CrossRef]
- Luo, D.; Xian, C.; Zhang, W.; Qin, Y.; Li, Q.; Usman, M.; Sun, S.; Xing, Y.; Dong, D. Physiological and Transcriptomic Analyses Reveal Commonalities and Specificities in Wheat in Response to Aluminum and Manganese. Curr. Issues Mol. Biol. 2024, 46, 367–397. [Google Scholar] [CrossRef]
- Li, X.Z.; Zhang, X.-T.; Bie, X.-M.; Zhang, J.; Jiang, D.-J.; Tang, H.; Wang, F. Transcriptome analysis of axillary buds in low phosphorus stress and functional analysis of TaWRKY74s in wheat. BMC Plant Biol. 2024, 24, 1. [Google Scholar] [CrossRef]
- Ma, P.; Guo, G.; Xu, X.; Luo, T.; Sun, Y.; Tang, X.; Heng, W.; Jia, B.; Liu, L. Transcriptome Analysis Reveals Key Genes Involved in the Response of Pyrus betuleafolia to Drought and High-Temperature Stress. Plants 2024, 13, 309. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Guo, Q.; Ji, W.; Wang, H.; Tao, J.; Xu, P.; Chen, X.; Ali, W.; Wu, X.; Shen, X.; et al. Transcriptome Expression Profiling Reveals the Molecular Response to Salt Stress in Gossypium anomalum Seedlings. Plants 2024, 13, 312. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhang, X.; Zhang, T.; Bai, Y.; Chen, C.; Guo, D.; Guo, C.; Shu, Y. Genome-wide analysis of the WRKY genes and their important roles during cold stress in white clover. PeerJ 2023, 11, e15610. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, W.; Manzoor, M.A.; Sabir, I.A.; Zhang, P.; Cao, Y.; Song, C. Differential involvement of WRKY genes in abiotic stress tolerance of Dendrobium huoshanense. Ind. Crops Prod. 2023, 204, 117295. [Google Scholar] [CrossRef]
- Wu, W.; Zhu, S.; Xu, L.; Zhu, L.; Wang, D.; Liu, Y.; Liu, S.; Hao, Z.; Lu, Y.; Yang, L.; et al. Genome-wide identification of the Liriodendron chinense WRKY gene family and its diverse roles in response to multiple abiotic stress. BMC Plant Biol. 2022, 22, 25. [Google Scholar] [CrossRef] [PubMed]
- Soltis, D.E.; Bell, C.D.; Kim, S.; Soltis, P.S. Origin and early evolution of angiosperms. Ann. N. Y. Acad. Sci. 2008, 1133, 3–25. [Google Scholar] [CrossRef] [PubMed]
- Wikstrom, N.; Savolainen, V.; Chase, M.W. Evolution of the angiosperms: Calibrating the family tree. Proc. Biol. Sci. 2001, 268, 2211–2220. [Google Scholar] [CrossRef]
- Chen, M.; Li, M.; Zhao, L.; Song, H. Deciphering evolutionary dynamics of WRKY genes in Arachis species. BMC Genom. 2023, 24, 48. [Google Scholar] [CrossRef]
- Tang, W.; Wang, F.; Chu, H.; You, M.; Lv, Q.; Ji, W.; Deng, X.; Zhou, B.; Peng, D. WRKY transcription factors regulate phosphate uptake in plants. Environ. Exp. Bot. 2023, 208, 105241. [Google Scholar] [CrossRef]
- An, X.; Liu, Q.; Jiang, H.; Dong, G.; Tian, D.; Luo, X.; Chen, C.; Li, W.; Liu, T.; Zou, L.; et al. Bioinformatics Analysis of WRKY Family Genes in Flax (Linum usitatissimum). Life 2023, 13, 1258. [Google Scholar] [CrossRef]
- Li, W.; Pang, S.; Lu, Z.; Jin, B. Function and Mechanism of WRKY Transcription Factors in Abiotic Stress Responses of Plants. Plants 2020, 9, 1515. [Google Scholar] [CrossRef] [PubMed]
- Mohanta, T.K.; Park, Y.; Bae, H. Novel Genomic and Evolutionary Insight of WRKY Transcription Factors in Plant Lineage. Sci. Rep. 2016, 6, 37309. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Huang, T.; Song, W.; An, Z.; Lai, Z.; Liu, S. Identification of WRKY gene family members in amaranth based on a transcriptome database and functional analysis of AtrWRKY42-2 in betalain metabolism. Front. Plant Sci. 2023, 14, 1300522. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Asaf, S.; Jan, R.; Bilal, S.; Lubna; Khan, A.L.; Kim, K.-M.; Al-Harrasi, A. Genome-wide annotation and expression analysis of WRKY and bHLH transcriptional factor families reveal their involvement under cadmium stress in tomato (Solanum lycopersicum L.). Front. Plant Sci. 2023, 14, 1100895. [Google Scholar] [CrossRef] [PubMed]
- Ge, M.; Tang, Y.; Guan, Y.; Lv, M.; Zhou, C.; Ma, H.; Lv, J. TaWRKY31, a novel WRKY transcription factor in wheat, participates in regulation of plant drought stress tolerance. BMC Plant Biol. 2024, 24, 27. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Hou, Y.; Sun, Y.; Chen, X.; Wang, H.; Zhu, B.; Du, X. ZmB12D, a target of transcription factor ZmWRKY70, enhances the tolerance of Arabidopsis to submergence. Plant Physiol. Biochem. 2024, 206, 108322. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Dong, B.; Wang, N.; Zheng, Z.; Yang, L.; Zhong, S.; Fang, Q.; Xiao, Z.; Zhao, H. A WRKY Transcription Factor PmWRKY57 from Prunus mume Improves Cold Tolerance in Arabidopsis thaliana. Mol. Biotechnol. 2023, 65, 1359–1368. [Google Scholar] [CrossRef]
- Wang, L.; Guo, D.; Zhao, G.; Wang, J.; Zhang, S.; Wang, C.; Guo, X. Group IIc WRKY transcription factors regulate cotton resistance to Fusarium oxysporum by promoting GhMKK2-mediated flavonoid biosynthesis. New Phytol. 2022, 236, 249–265. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015, 16, 157. [Google Scholar] [CrossRef]
- Kumar, S.; Suleski, M.; Craig, J.M.; Kasprowicz, A.E.; Sanderford, M.; Li, M.; Stecher, G.; Hedges, S.B. TimeTree 5: An Expanded Resource for Species Divergence Times. Mol. Biol. Evol. 2022, 39, msac174. [Google Scholar] [CrossRef]
- Barido-Sottani, J.; Morlon, H. The ClaDS rate-heterogeneous birth-death prior for full phylogenetic inference in BEAST2. Syst. Biol. 2023, 72, 1180–1187. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Z.; Vang, S.; Yu, J.; Wong, G.K.-S.; Wang, J. Correlation between Ka/Ks and Ks is related to substitution model and evolutionary lineage. J. Mol. Evol. 2009, 68, 414–423. [Google Scholar] [CrossRef]
- Peterson, G.I.; Masel, J. Quantitative prediction of molecular clock and ka/ks at short timescales. Mol. Biol. Evol. 2009, 26, 2595–2603. [Google Scholar] [CrossRef]
- Hurst, L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 2002, 18, 486. [Google Scholar] [CrossRef] [PubMed]

| Species | Group I | Group II-A | Group II-B | Group II-C | Group II-D | Group II-E | Group III |
|---|---|---|---|---|---|---|---|
| Amborella trichopoda | 7 | 2 | 5 | 6 | 3 | 3 | 8 |
| Nymphaea colorata | 18 | 6 | 9 | 10 | 7 | 10 | 5 |
| Cinnamomum kanehirae | 15 | 6 | 8 | 12 | 9 | 12 | 11 |
| Liriodendron chinense | 7 | 3 | 6 | 8 | 4 | 7 | 9 |
| Liriodendron tulipifera | 9 | 6 | 7 | 13 | 6 | 7 | 10 |
| Brachypodium arbuscula | 13 | 4 | 8 | 23 | 7 | 10 | 26 |
| Brachypodium distachyon | 17 | 5 | 5 | 16 | 10 | 10 | 26 |
| Oryza sativa | 7 | 4 | 8 | 19 | 6 | 11 | 51 |
| Sorghum bicolor | 9 | 5 | 8 | 20 | 7 | 11 | 36 |
| Zea mays | 15 | 7 | 12 | 30 | 12 | 17 | 44 |
| Zostera marina | 7 | 2 | 8 | 10 | 6 | 6 | 7 |
| Arabidopsis thaliana | 12 | 3 | 8 | 18 | 7 | 7 | 28 |
| Carya illinoinensis | 17 | 6 | 13 | 24 | 9 | 10 | 12 |
| Cirus trifoliata | 10 | 3 | 8 | 13 | 5 | 7 | 9 |
| Coffea arabica | 23 | 5 | 15 | 28 | 13 | 11 | 32 |
| Corymbia citriodora | 16 | 6 | 11 | 18 | 5 | 8 | 15 |
| Gossypium hirsutum | 36 | 16 | 31 | 69 | 28 | 26 | 32 |
| Malus pumila | 23 | 6 | 15 | 25 | 14 | 13 | 28 |
| Populus trichocarpa | 23 | 5 | 9 | 24 | 13 | 12 | 14 |
| Portulaca amilis | 12 | 5 | 5 | 14 | 10 | 8 | 21 |
| Sinapis alba | 41 | 8 | 26 | 64 | 24 | 19 | 51 |
| Theobroma cacao | 10 | 3 | 8 | 14 | 6 | 6 | 18 |
| Vigna unguiculata | 15 | 6 | 15 | 22 | 7 | 11 | 22 |
| Vitis vinifera | 14 | 3 | 8 | 15 | 9 | 6 | 12 |
| Species | Family | Taxonomy | WGD | Total Number |
|---|---|---|---|---|
| Amborella trichopoda | Amborellaceae | ANITA Basal angiosperms | 1 | 34 |
| Nymphaea colorata | Nymphaeaceae | ANITA Basal angiosperms | 1 | 65 |
| Liriodendron chinense | Magnoliaceae | Magnoliids | 1 | 44 |
| Liriodendron tulipifera | Magnoliaceae | Magnoliids | 1 | 58 |
| Cinnamomum kanehirae | Lauraceae | Magnoliids | 2 | 73 |
| Zostera marina | Zosteraceae | Monocotyledonous | 1 | 46 |
| Brachypodium distachyon | Poaceae | Monocotyledonous | 1 | 89 |
| Brachypodium arbuscula | Poaceae | Monocotyledonous | 1 | 91 |
| Sorghum bicolor | Poaceae | Monocotyledonous | 1 | 96 |
| Oryza sativa | Poaceae | Monocotyledonous | 1 | 106 |
| Zea mays | Poaceae | Monocotyledonous | 1 | 137 |
| Cirus trifoliata | Rutaceae | Dicotyledonous | 1 | 55 |
| Theobroma cacao | Malvaceae | Dicotyledonous | 1 | 65 |
| Vitis vinifera | Vitaceae | Dicotyledonous | 2 | 67 |
| Portulaca amilis | Portulacaceae | Dicotyledonous | 1 | 75 |
| Corymbia citriodora | Myrtaceae | Dicotyledonous | 1 | 79 |
| Arabidopsis thaliana | Brassicaceae | Dicotyledonous | 1 | 83 |
| Carya illinoinensis | Juglandaceae | Dicotyledonous | 2 | 91 |
| Vigna unguiculata | Fabaceae | Dicotyledonous | 1 | 98 |
| Populus trichocarpa | Salicaceae | Dicotyledonous | 1 | 100 |
| Malus pumila | Rosaceae | Dicotyledonous | 1 | 124 |
| Coffea arabica | Rubiaceae | Dicotyledonous | 1 | 127 |
| Sinapis alba | Brassicaceae | Dicotyledonous | 2 | 233 |
| Gossypium hirsutum | Malvaceae | Dicotyledonous | 1 | 238 |
| Species | Taxonomy | Gene Pairs | Ka/Ks < 1 | Ka/Ks > 1 | Selection Pressure | Singe Copy |
|---|---|---|---|---|---|---|
| Amborella trichopoda | ANITA Basal angiosperms | 0 | 0 | 0 | \ | 6 |
| Nymphaea colorata | ANITA Basal angiosperms | 18 | 12 | 6 | Negative | 10 |
| Cinnamomum kanehirae | Magnoliids | 0 | 0 | 0 | \ | 9 |
| Liriodendron chinense | Magnoliids | 5 | 3 | 2 | Negative | 7 |
| Liriodendron tulipifera | Magnoliids | 6 | 2 | 4 | Positive | 7 |
| Brachypodium arbuscula | Monocotyledonous | 105 | 57 | 48 | Negative | 14 |
| Brachypodium distachyon | Monocotyledonous | 0 | 0 | 0 | \ | 15 |
| Oryza sativa | Monocotyledonous | 229 | 140 | 89 | Negative | 13 |
| Sorghum bicolor | Monocotyledonous | 157 | 67 | 90 | Positive | 11 |
| Zea mays | Monocotyledonous | 366 | 201 | 165 | Negative | 19 |
| Zostera marina | Monocotyledonous | 7 | 5 | 2 | Negative | 7 |
| Arabidopsis thaliana | Dicotyledonous | 18 | 5 | 13 | Positive | 11 |
| Carya illinoinensis | Dicotyledonous | 0 | 0 | 0 | \ | 15 |
| Cirus trifoliata | Dicotyledonous | 7 | 3 | 4 | Positive | 8 |
| Coffea arabica | Dicotyledonous | 0 | 0 | 0 | \ | 21 |
| Corymbia citriodora | Dicotyledonous | 0 | 0 | 0 | \ | 12 |
| Gossypium hirsutum | Dicotyledonous | 115 | 71 | 44 | Negative | 32 |
| Malus pumila | Dicotyledonous | 27 | 21 | 6 | Negative | 17 |
| Populus trichocarpa | Dicotyledonous | 18 | 8 | 10 | Positive | 18 |
| Portulaca amilis | Dicotyledonous | 0 | 0 | 0 | \ | 13 |
| Sinapis alba | Dicotyledonous | 55 | 30 | 25 | Negative | 33 |
| Theobroma cacao | Dicotyledonous | 7 | 4 | 3 | Negative | 9 |
| Vigna unguiculata | Dicotyledonous | 15 | 9 | 6 | Negative | 13 |
| Vitis vinifera | Dicotyledonous | 9 | 8 | 1 | Negative | 11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, W.; Yang, J.; Yu, N.; Li, R.; Yuan, Z.; Shi, J.; Chen, J. Evolution of the WRKY Family in Angiosperms and Functional Diversity under Environmental Stress. Int. J. Mol. Sci. 2024, 25, 3551. https://doi.org/10.3390/ijms25063551
Wu W, Yang J, Yu N, Li R, Yuan Z, Shi J, Chen J. Evolution of the WRKY Family in Angiosperms and Functional Diversity under Environmental Stress. International Journal of Molecular Sciences. 2024; 25(6):3551. https://doi.org/10.3390/ijms25063551
Chicago/Turabian StyleWu, Weihuang, Jinchang Yang, Niu Yu, Rongsheng Li, Zaixiang Yuan, Jisen Shi, and Jinhui Chen. 2024. "Evolution of the WRKY Family in Angiosperms and Functional Diversity under Environmental Stress" International Journal of Molecular Sciences 25, no. 6: 3551. https://doi.org/10.3390/ijms25063551






