Anti-Melanoma Effects of Miconazole: Investigating the Mitochondria Involvement
Abstract
:1. Introduction
2. Results
2.1. Effects at 24 h: Viability, Proliferation, Vascular Mimicry
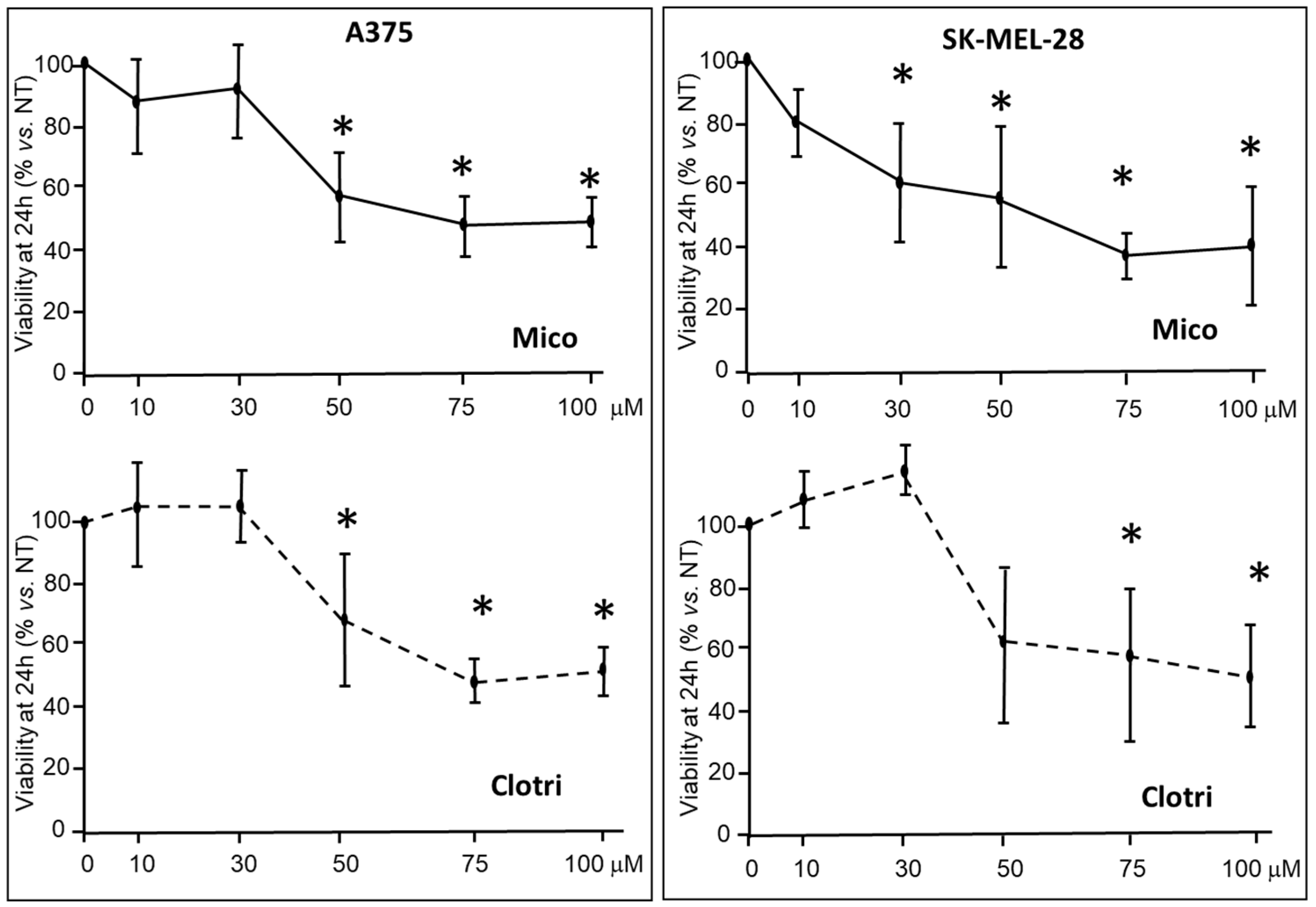
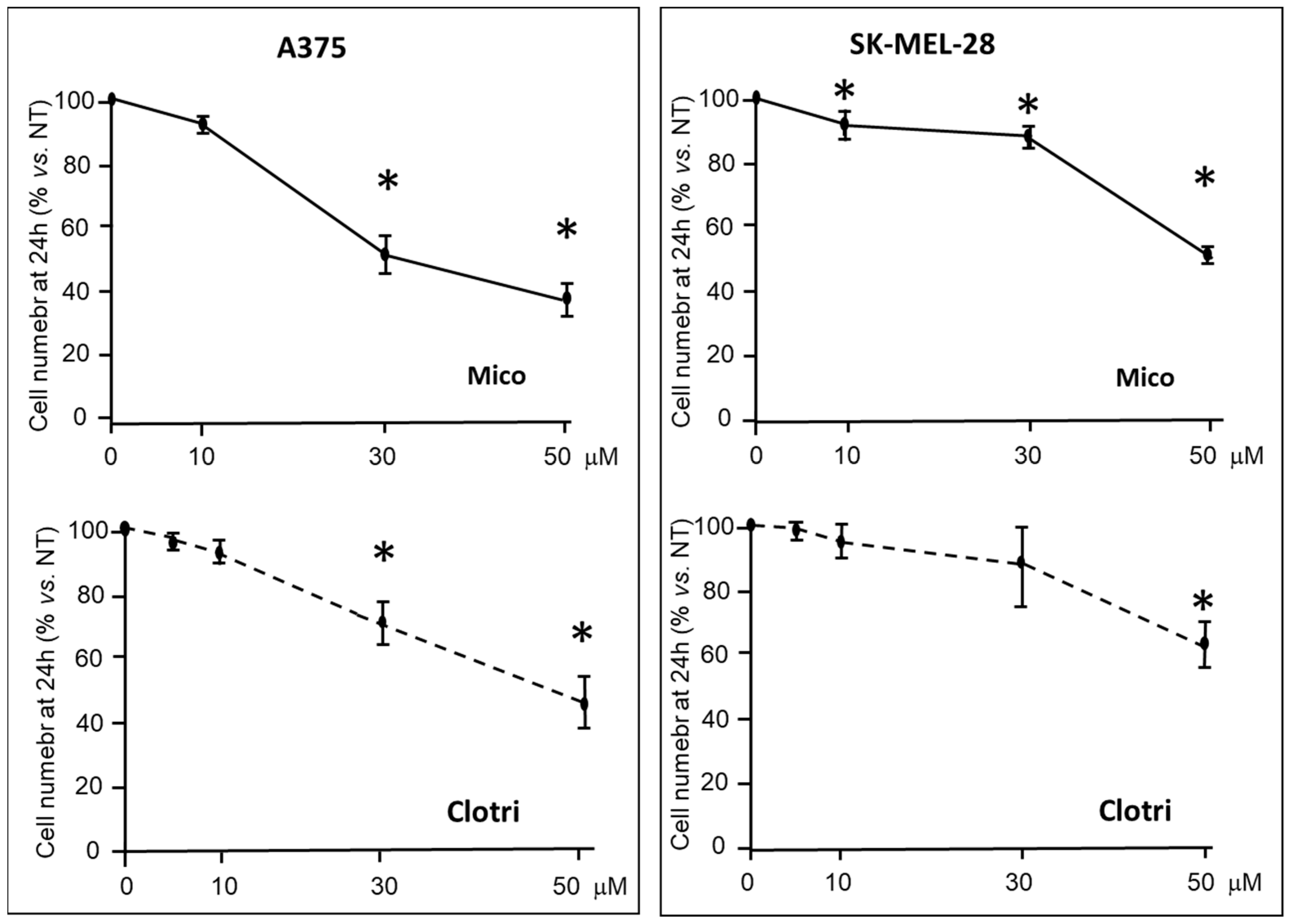
2.1.1. Viability and Proliferation
2.1.2. Vasculogenic Mimicry
2.2. Effects at 6 h: ATP, ROS, Mitochondria and Metabolomics
2.2.1. ATP and ROS Levels
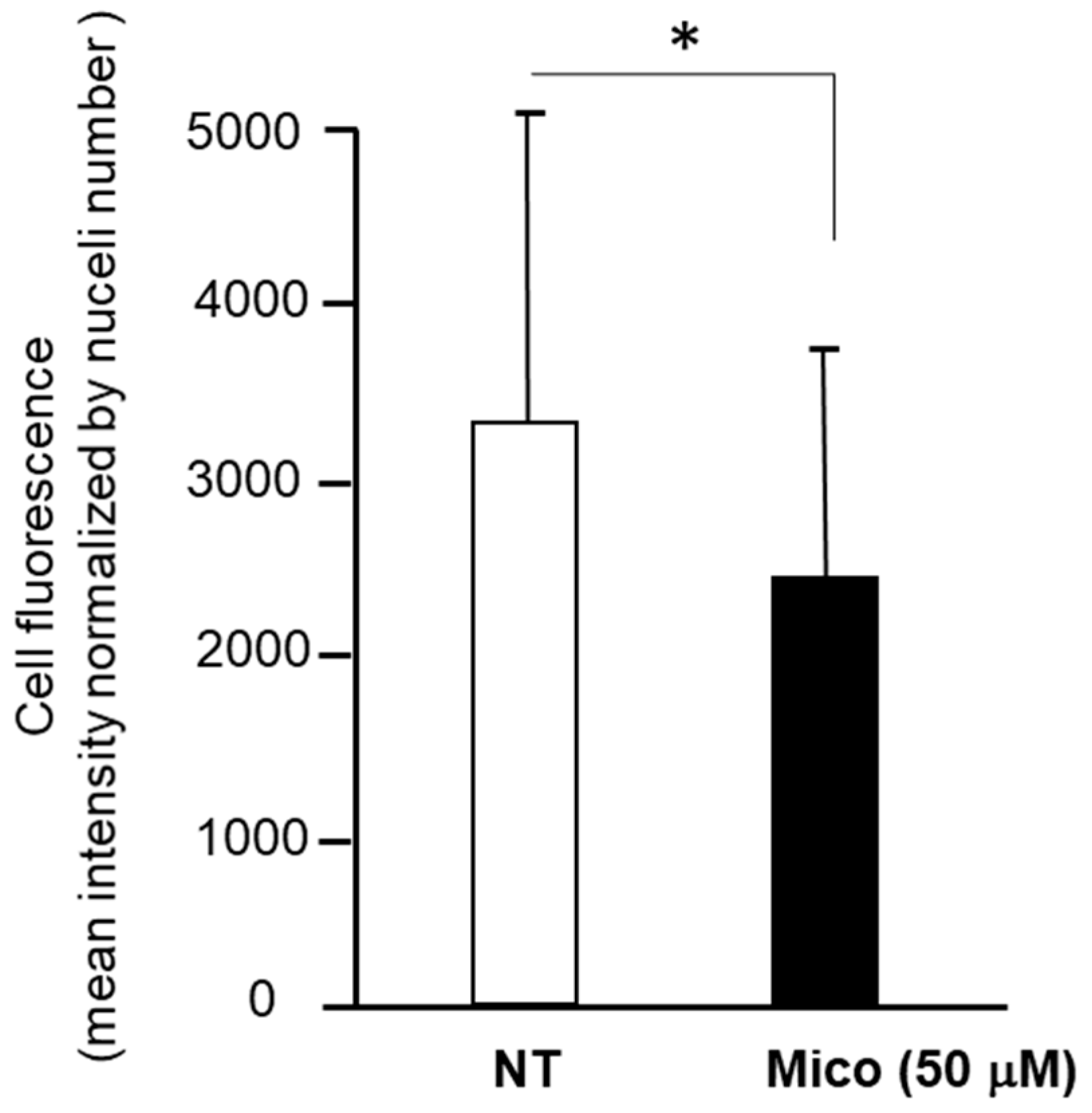
2.2.2. Mitochondria Investigation
2.2.3. Metabolomics Analysis
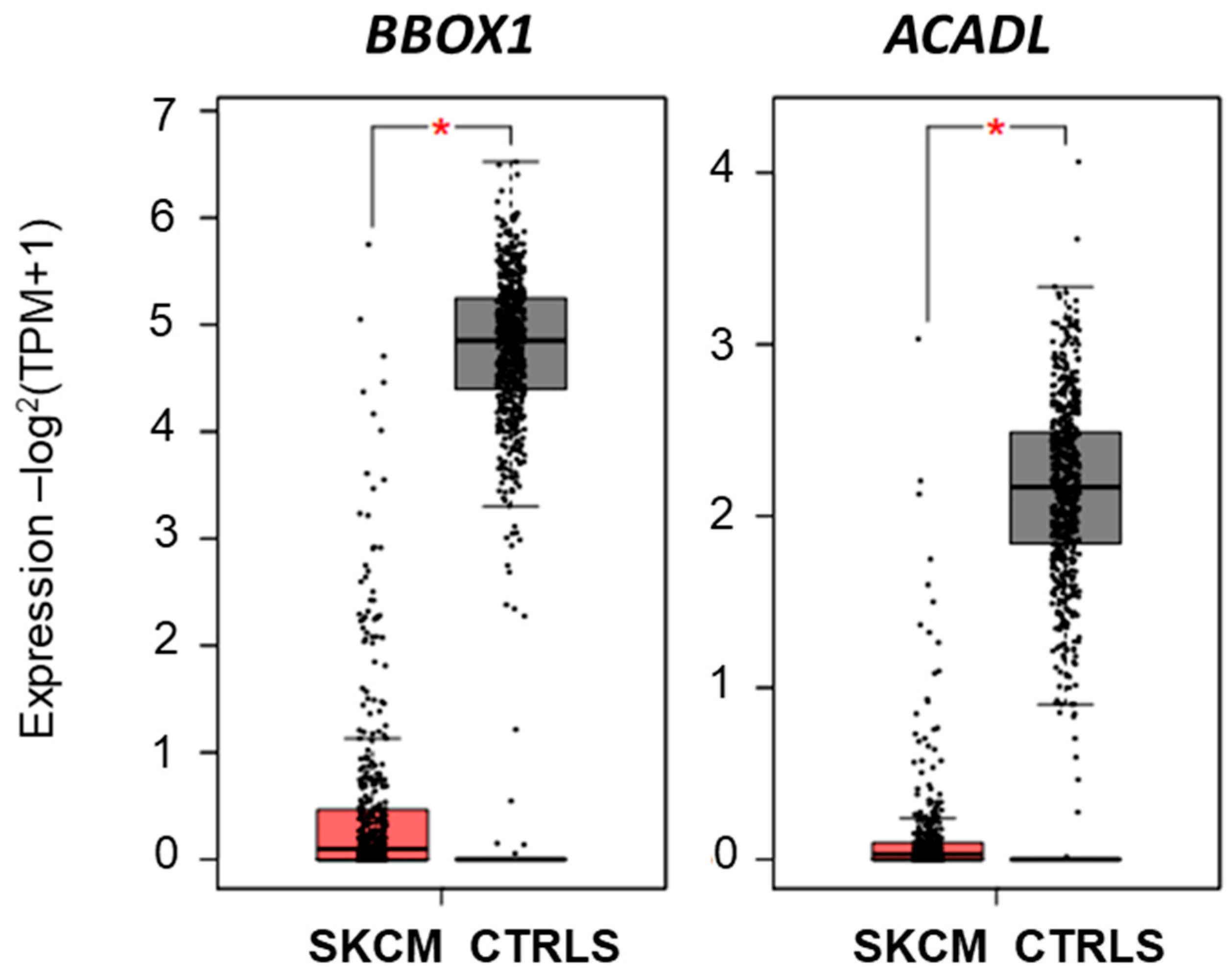
2.2.4. Investigating Carnitine’s Role
2.2.5. Investigating Pro-Apoptotic Caspases, Cell Death and Apoptosis
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Treatment for 24 h: Cell Proliferation, Cell Viability, Vascular Mimicry
4.2.1. Cell Proliferation
4.2.2. MTT Cell Viability Assay
4.2.3. Vasculogenic Mimicry Assay
4.3. Treatment for 6 h: ROS Release, ATP Release, Mitochondria Staining, Metabolomics Profiling
4.3.1. Evaluation of ROS and ATP Production
4.3.2. Mitochondria Investigation: Whole Fluorescence
4.3.3. Mitochondria Investigation: Confocal Cytofluorescence
4.3.4. Metabolomics Analysis
4.4. Analysis of Carnitine Role
4.5. Apoptosis Analyses
4.6. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ng, G.; Xu, W.; Atkinson, V. Treatment Approaches for Melanomas That Relapse After Adjuvant or Neoadjuvant Therapy. Curr. Oncol. Rep. 2022, 24, 1273–1280. [Google Scholar] [CrossRef] [PubMed]
- Kale, M.A.; Shamkuwar, P.B.; Mourya, V.K.; Deshpande, A.B.; Shelke, P.A. Drug Repositioning: A Unique Approach to Refurbish Drug Discovery. Curr. Drug Discov. Technol. 2022, 19, e140122192307. [Google Scholar] [CrossRef] [PubMed]
- Van Cutsem, J.M.; Thienpont, D. Miconazole, a broad-spectrum antimycotic agent with antibacterial activity. Chemotherapy 1972, 17, 392–404. [Google Scholar] [CrossRef] [PubMed]
- Botter, A.A. Topical treatment of nail and skin infections with miconazole, a new broad-spectrum antimycotic. Mycoses 1971, 14, 187–191. [Google Scholar] [CrossRef] [PubMed]
- Pierard, G.E.; Hermanns-Le, T.; Delvenne, P.; Pierard-Franchimont, C. Miconazole, a pharmacological barrier to skin fungal infections. Expert Opin. Pharmacother. 2012, 13, 1187–1194. [Google Scholar] [CrossRef]
- De Nollin, S.; Borgers, M. An ultrastructural and cytochemical study of Candida albicans after in vitro treatment with imidazoles. Mycoses 1976, 19, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Narashimamurthy, J.; Rao, A.R.; Sastry, G.N. Aromatase inhibitors: A new paradigm in breast cancer treatment. Curr. Med. Chem. Agents 2004, 4, 523–534. [Google Scholar] [CrossRef]
- Terrones-Gurrola, M.C.D.R.; Ponce-Peña, P.; Salas-Pacheco, J.M.; Camacho-Luis, A.; Pozos-Guillén, A.J.; Nieto-Delgado, G.; López-Guzmán, O.D.; Vértiz-Hernández, A.A. Arsenic: A Perspective on Its Effect on Pioglitazone Bioavailability. Int. J. Environ. Res. Public Health 2023, 20, 1901. [Google Scholar] [CrossRef]
- Kakimoto, A.; Ogura, H.; Suenaga, M.; Mishima, T.; Fujioka, S.; Ouma, S.; Matsunaga, Y.; Tsuboi, Y. Role of cytochrome P450 for vitamin D metabolisms in patients with neurodegenerative disorders. Clin. Park. Relat. Disord. 2022, 7, 100162. [Google Scholar] [CrossRef]
- Krkoška, M.; Svobodová, J.; Kabátková, M.; Zapletal, O.; Hyršlová Vaculová, A.; Nekvindová, J.; Vondráček, J. Deregulation of signaling pathways controlling cell survival and proliferation in cancer cells alters induction of cytochrome P450 family 1 enzymes. Toxicology 2021, 461, 152897. [Google Scholar] [CrossRef]
- Shetkar, S.S.; Ramakrishnan, S.; Seth, S.; Chandna, P.; Verma, S.K.; Bhargava, B.; Bahl, V.K. CYP 450 2C19 polymorphisms in Indian patients with coronary artery disease. Indian Heart J. 2014, 66, 16–24. [Google Scholar] [CrossRef]
- Nguyen, T.M.L.; McGowan, J.C.; Gardier, A.M. CYP 450 enzymes influence (R,S)-ketamine brain delivery and its antidepressant activity. Neuropharmacology 2022, 206, 108936. [Google Scholar] [CrossRef] [PubMed]
- Pikuleva, I.A.; Waterman, M.R. We have previously shown Cytochromes P450: Roles in Diseases. J. Biol. Chem. 2013, 288, 17091–17098. [Google Scholar] [CrossRef] [PubMed]
- Stavropoulou, E.; Pircalabioru, G.G.; Bezirtzoglou, E. The Role of Cytochromes P450 in Infection. Front. Immunol. 2018, 9, 89. [Google Scholar] [CrossRef] [PubMed]
- Villeneuve, J.P.; Pichette, V. Cytochrome P450 and Liver Diseases. Curr. Drug Metab. 2004, 5, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Tan, E.Y.; Bharwani, L.; Chia, Y.-H.; Soong, R.C.T.; Lee, S.S.Y.; Chen, J.J.C.; Chan, P.M.Y. Impact of cytochrome P450 2D6 polymorphisms on decision-making and clinical outcomes in adjuvant hormonal therapy for breast cancer. World J. Clin. Oncol. 2022, 13, 712–724. [Google Scholar] [CrossRef] [PubMed]
- Thorén, L.; Margolin, S.; Eliasson, E.; Bergh, J.; Lindh, J.D. Adherence to endocrine therapy in early breast cancer in relation to Cytochrome P450 2D6 genotype: A comparison between pharmacy dispensation data and medical records. Breast Cancer Res. Treat. 2023, 198, 499–508. [Google Scholar] [CrossRef] [PubMed]
- Korbecki, J.; Rębacz-Maron, E.; Kupnicka, P.; Chlubek, D.; Baranowska-Bosiacka, I. Synthesis and Significance of Arachidonic Acid, a Substrate for Cyclooxygenases, Lipoxygenases, and Cytochrome P450 Pathways in the Tumorigenesis of Glioblastoma Multiforme, Including a Pan-Cancer Comparative Analysis. Cancers 2023, 15, 946. [Google Scholar] [CrossRef]
- Uthansingh, K.; Parida, P.K.; Pati, G.K.; Sahu, M.K.; Padhy, R.N. Evaluating the Association of Genetic Polymorphism of Cytochrome p450 (CYP2C9*3) in Gastric Cancer Using Polymerase Chain Reaction-Restriction Fragment Length Polymorphism (PCR-RFLP). Cureus 2022, 14, e27220. [Google Scholar] [CrossRef]
- Sonesson, A.; Buur Rasmussen, B.B. In vitro studies investigating the interactions between degarelix, a decapeptide gonadotropin-releasing hormone blocker, and cytochrome P450. Basic Clin. Pharmacol. Toxicol. 2011, 109, 195–202. [Google Scholar] [CrossRef]
- Mo, H.Y.; Wei, Q.Y.; Zhong, Q.H.; Zhao, X.Y.; Guo, D.; Han, J.; Noracharttiyapot, W.; Visser, L.; Berg, A.V.D.; Xu, Y.-M.; et al. Cytochrome P450 27C1 Level Dictates Lung Cancer Tumorigenicity and Sensitivity towards Multiple Anticancer Agents and Its Potential Interplay with the IGF-1R/Akt/p53 Signaling Pathway. Int. J. Mol. Sci. 2022, 23, 7853. [Google Scholar] [CrossRef]
- Khamis, Z.I.; Pang, X.; Cui, Z.; Sang, Q.A.; Zhang, J. Cytochrome P450-2D6: A novel biomarker in liver cancer health disparity. PLoS ONE 2021, 16, e0257072. [Google Scholar] [CrossRef]
- Presa, D.; Khurram, S.A.; Zubir, A.Z.A.; Smarakan, S.; Cooper, P.A.; Morais, G.R.; Sadiq, M.; Sutherland, M.; Loadman, P.M.; McCaul, J.; et al. Cytochrome P450 isoforms 1A1, 1B1 AND 2W1 as targets for therapeutic intervention in head and neck cancer. Sci. Rep. 2021, 11, 18930. [Google Scholar] [CrossRef]
- Kong, C.; Yan, X.; Zhu, Y.; Zhu, H.; Luo, Y.; Liu, P.; Ferrandon, S.; Kalady, M.F.; Gao, R.; He, J.; et al. Fusobacterium Nucleatum Promotes the Development of Colorectal Cancer by Activating a Cytochrome P450/Epoxyoctadecenoic Acid Axis via TLR4/Keap1/NRF2 Signaling. Cancer Res. 2021, 81, 4485–4498. [Google Scholar] [CrossRef]
- Singh, R.D.; Avadhesh, A.; Sharma, G.; Dholariya, S.; Shah, R.B.; Goyal, B.; Gupta, S.C. Potential of Cytochrome P450, a Family of Xenobiotic Metabolizing Enzymes, in Cancer Therapy. Antioxid. Redox Signal. 2023, 38, 853–876. [Google Scholar] [CrossRef]
- Yada, E.; Kasajima, R.; Niida, A.; Imoto, S.; Miyano, S.; Miyagi, Y.; Sasada, T.; Wada, S. Possible Role of Cytochrome P450 1B1 in the Mechanism of Gemcitabine Resistance in Pancreatic Cancer. Biomedicines 2021, 9, 1396. [Google Scholar] [CrossRef]
- Flaherty, K.T.; Lathia, C.; Frye, R.F.; Schuchter, L.; Redlinger, M.; Rosen, M.; O’dwyer, P.J. Interaction of sorafenib and cytochrome P450 isoenzymes in patients with advanced melanoma: A phase I/II pharmacokinetic interaction study. Cancer Chemother. Pharmacol. 2011, 68, 1111–1118. [Google Scholar] [CrossRef]
- D’arcangelo, D.; Scatozza, F.; Giampietri, C.; Marchetti, P.; Facchiano, F.; Facchiano, A. Ion Channel Expression in Human Melanoma Samples: In Silico Identification and Experimental Validation of Molecular Targets. Cancers 2019, 11, 446. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, J.; Montero, M.; Garcia-Sancho, J. High affinity inhibition of Ca(2+)-dependent K+ channels by cytochrome P-450 inhibitors. J. Biol. Chem. 1992, 267, 11789–11793. [Google Scholar] [CrossRef] [PubMed]
- Hatton, C.J.; Peers, C. Effects of cytochrome P-450 inhibitors on ionic currents in isolated rat type I carotid body cells. Am. J. Physiol. 1996, 271, C85–C92. [Google Scholar] [CrossRef] [PubMed]
- Mandy, S.J.; Garrott, T.C. Miconazole treatment for severe dermatophytoses. JAMA 1974, 230, 72–75. [Google Scholar] [CrossRef] [PubMed]
- Pyo, S.M.; Hespeler, D.; Keck, C.M.; Müller, R.H. Dermal miconazole nitrate nanocrystals—Formulation development, increased antifungal efficacy & skin penetration. Int. J. Pharm. 2017, 531, 350–359. [Google Scholar] [CrossRef]
- Firooz, A.; Namdar, R.; Nafisi, S.; Maibach, H.I. Nano-Sized Technologies for Miconazole Skin Delivery. Curr. Pharm. Biotechnol. 2016, 17, 524–531. [Google Scholar] [CrossRef] [PubMed]
- Stepanenko, A.A.; Dmitrenko, V.V. Pitfalls of the MTT assay: Direct and off-target effects of inhibitors can result in over/underestimation of cell viability. Gene 2015, 574, 193–203. [Google Scholar] [CrossRef]
- Scatozza, F.; Moschella, F.; D’arcangelo, D.; Rossi, S.; Tabolacci, C.; Giampietri, C.; Proietti, E.; Facchiano, F.; Facchiano, A. Nicotinamide inhibits melanoma in vitro and in vivo. J. Exp. Clin. Cancer Res. 2020, 39, 211. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Kim, B.Y.; Son, Y.; Cho, H.R.; Lee, D.; Eo, S.K.; Kim, K. Miconazole Suppresses 27-Hydroxycholesterol-induced Inflammation by Regulating Activation of Monocytic Cells to a Proinflammatory Phenotype. Front. Pharmacol. 2021, 12, 691019. [Google Scholar] [CrossRef]
- Stryjska, K.; Radko, L.; Chęcińska, L.; Kusz, J.; Posyniak, A.; Ochocki, J. Synthesis, Spectroscopy, Light Stability, Single-Crystal Analysis, and In Vitro Cytotoxic Activity on HepG2 Liver Cancer of Two Novel Silver(I) Complexes of Miconazole. Int. J. Mol. Sci. 2020, 21, 3629. [Google Scholar] [CrossRef]
- Tsai, T.F.; Chen, P.C.; Lin, Y.C.; Chou, K.Y.; Chen, H.E.; Ho, C.Y.; Lin, J.F.; Hwang, T.I.S. Miconazole Contributes to NRF2 Activation by Noncanonical P62-KEAP1 Pathway in Bladder Cancer Cells. Drug Des. Dev. Ther. 2020, 2020, 1209–1218. [Google Scholar] [CrossRef]
- Wu, C.; Gao, M.; Shen, L.; Li, B.; Bai, X.; Gui, J.; Li, H.; Huo, Q.; Ma, T. Miconazole triggers various forms of cell death in human breast cancer MDA-MB-231 cells. Pharmazie 2019, 74, 290–294. [Google Scholar] [CrossRef]
- Jung, H.J.; Seo, I.; Jha, B.K.; Suh, S.I.; Baek, W.K. Miconazole induces autophagic death in glioblastoma cells via reactive oxygen species-mediated endoplasmic reticulum stress. Oncol. Lett. 2021, 21, 335. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.Y.; Chang, A.C.; Hsu, C.H.; Tsai, T.F.; Lin, Y.C.; Chou, K.Y.; Chen, H.; Lin, J.; Chen, P.; Hwang, T.I. Miconazole induces protective autophagy in bladder cancer cells. Environ. Toxicol. 2021, 36, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Precazzini, F.; Pancher, M.; Gatto, P.; Tushe, A.; Adami, V.; Anelli, V.; Mione, M.C. Automated in vivo screen in zebrafish identifies Clotrimazole as targeting a metabolic vulnerability in a melanoma model. Dev. Biol. 2020, 457, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Amado, J.; Pires, P.C.; Veiga, F.; Santos, A.C. Miconazole Nanoemulsions to Treat Melanoma: A Study of Formulation Development, Droplet Size and Solubility. Med. Sci. Forum. 2022, 14, 119. [Google Scholar] [CrossRef]
- Bertram, R.; Pedersen, M.G.; Luciani, D.S.; Sherman, A. A simplified model for mitochondrial ATP production. J. Theor. Biol. 2006, 243, 575–586. [Google Scholar] [CrossRef]
- Tirichen, H.; Yaigoub, H.; Xu, W.; Wu, C.; Li, R.; Li, Y. Mitochondrial Reactive Oxygen Species and Their Contribution in Chronic Kidney Disease Progression Through Oxidative Stress. Front. Physiol. 2021, 12, 627837. [Google Scholar] [CrossRef]
- Yuan, S.Y.; Shiau, M.Y.; Ou, Y.C.; Huang, Y.C.; Chen, C.C.; Cheng, C.L.; Chiu, K.Y.; Wang, S.S.; Tsai, K.J. Miconazole induces apoptosis via the death receptor 5-dependent and mitochondrial-mediated pathways in human bladder cancer cells. Oncol. Rep. 2017, 37, 3606–3616. [Google Scholar] [CrossRef]
- Vaz, F.M.; Wanders, R.J.A. Carnitine biosynthesis in mammals. Biochem. J. 2002, 361, 417–429. [Google Scholar] [CrossRef]
- Lee, Y.G.; Chou, H.C.; Chen, Y.T.; Tung, S.Y.; Ko, T.L.; Buyandelger, B.; Wen, L.L.; Juan, S.H. L-Carnitine reduces reactive oxygen species/endoplasmic reticulum stress and maintains mitochondrial function during autophagy-mediated cell apoptosis in perfluorooctanesulfonate-treated renal tubular cells. Sci. Rep. 2022, 12, 4673. [Google Scholar] [CrossRef]
- Mancilla, R.F.; Lindeboom, L.; Grevendonk, L.; Hoeks, J.; Koves, T.R.; Muoio, D.M.; Schrauwen, P.; Schrauwen-Hinderling, V.; Hesselink, M.K. Skeletal muscle mitochondrial inertia is associated with carnitine acetyltransferase activity and physical function in humans. JCI Insight 2023, 8, e163855. [Google Scholar] [CrossRef]
- Knottnerus, S.J.G.; Bleeker, J.C.; Wüst, R.C.I.; Ferdinandusse, S.; Ijlst, L.; Wijburg, F.A.; Wanders, R.J.A.; Visser, G.; Houtkooper, R.H. Disorders of mitochondrial long-chain fatty acid oxidation and the carnitine shuttle. Rev. Endocr. Metab. Disord. 2018, 19, 93–106. [Google Scholar] [CrossRef]
- Yao, M.; Zhou, P.; Qin, Y.Y.; Wang, L.; Yao, D.F. Mitochondrial carnitine palmitoyltransferase-II dysfunction: A possible novel mechanism for nonalcoholic fatty liver disease in hepatocarcinogenesis. World J. Gastroenterol. 2023, 29, 1765–1778. [Google Scholar] [CrossRef]
- Hassan, N.; Rashad, M.; Elleithy, E.; Sabry, Z.; Ali, G.; Elmosalamy, S. L-Carnitine alleviates hepatic and renal mitochondrial-dependent apoptotic progression induced by letrozole in female rats through modulation of Nrf-2, Cyt c and CASP-3 signaling. Drug Chem. Toxicol. 2023, 46, 357–368. [Google Scholar] [CrossRef]
- Virmani, M.A.; Cirulli, M. The Role of l-Carnitine in Mitochondria, Prevention of Metabolic Inflexibility and Disease Initiation. Int. J. Mol. Sci. 2022, 23, 2717. [Google Scholar] [CrossRef]
- Tonazzi, A.; Giangregorio, N.; Console, L.; Palmieri, F.; Indiveri, C. The Mitochondrial Carnitine Acyl-carnitine Carrier (SLC25A20): Molecular Mechanisms of Transport, Role in Redox Sensing and Interaction with Drugs. Biomolecules 2021, 11, 521. [Google Scholar] [CrossRef]
- McCann, M.R.; De la Rosa, M.V.G.; Rosania, G.R.; Stringer, K.A. L-Carnitine and Acylcarnitines: Mitochondrial Biomarkers for Precision Medicine. Metabolites 2021, 11, 51. [Google Scholar] [CrossRef] [PubMed]
- Awa, H.; Futamura, A.; Higashiguchi, T.; Ito, A.; Mori, N.; Murai, M.; Ohara, H.; Chihara, T.; Kaneko, T. Effects of Combined Treatment with Branched-Chain Amino Acids, Citric Acid, L-Carnitine, Coenzyme Q10, Zinc, and Various Vitamins in Tumor-Bearing Mice. Biol. Pharm. Bull. 2017, 40, 266–271. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Ma, X. Serum lactate dehydrogenase is a predictor of poor survival in malignant melanoma. Panminerva Medica 2017, 59, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Batra, S.; Zhang, J. Asparagine: A Metabolite to Be Targeted in Cancers. Metabolites 2021, 11, 402. [Google Scholar] [CrossRef]
- Sari, I.N.; Setiawan, T.; Kim, K.S.; Wijaya, Y.T.; Cho, K.W.; Kwon, H.Y. Metabolism and function of polyamines in cancer progression. Cancer Lett. 2021, 519, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Casero, R.A.; Stewart, T.M.; Pegg, A.E. Polyamine metabolism and cancer: Treatments, challenges and opportunities. Nat. Rev. Cancer 2018, 18, 681–695. [Google Scholar] [CrossRef]
- Ueland, P.M. Choline and betaine in health and disease. J. Inherit. Metab. Dis. 2011, 34, 3–15. [Google Scholar] [CrossRef]
- Korotkov, S.M. Mitochondrial Oxidative Stress Is the General Reason for Apoptosis Induced by Different-Valence Heavy Metals in Cells and Mitochondria. Int. J. Mol. Sci. 2023, 24, 14459. [Google Scholar] [CrossRef]
- Nandi, S.; Sikder, R.; Rapior, S.; Arnould, S.; Simal-Gandara, J.; Acharya, K. A review for cancer treatment with mushroom metabolites through targeting mitochondrial signaling pathway: In vitro and in vivo evaluations, clinical studies and future prospects for mycomedicine. Fitoterapia 2024, 172, 105681. [Google Scholar] [CrossRef]
- Gao, X.; Deng, B.; Ran, S.; Li, S. The effect of GSK-3β in arsenic-induced apoptosis of malignant tumor cells: A systematic review and me-ta-analysis. Toxicol. Mech. Methods 2022, 32, 477–487. [Google Scholar] [CrossRef]
- Boice, A.; Bouchier-Hayes, L. Targeting apoptotic caspases in cancer. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118688. [Google Scholar] [CrossRef] [PubMed]
- Weng, N.; Zhang, Z.; Tan, Y.; Zhang, X.; Wei, X.; Zhu, Q. Repurposing antifungal drugs for cancer therapy. J. Adv. Res. 2023, 48, 259–273. [Google Scholar] [CrossRef] [PubMed]
- Trisciuoglio, D.; Tupone, M.G.; Desideri, M.; Di Martile, M.; Gabellini, C.; Buglioni, S.; Pallocca, M.; Alessandrini, G.; D’aguanno, S.; Del Bufalo, D. BCL-XL overexpression promotes tumor progression-associated properties. Cell Death Dis. 2017, 8, 3216. [Google Scholar] [CrossRef] [PubMed]
- Chaudhry, A.; Shi, R.; Luciani, D.S. A pipeline for multidimensional confocal analysis of mitochondrial morphology, function, and dynamics in pancreatic β-cells. Am. J. Physiol. Metab. 2020, 318, E87–E101. [Google Scholar] [CrossRef] [PubMed]
- Fernadez Casafuz, A.B.; De Rossi, M.C.; Bruno, L. Mitochondrial cellular organization and shape fluctuations are differentially modulated by cytoskeletal networks. Sci. Rep. 2023, 13, 4065. [Google Scholar] [CrossRef] [PubMed]







| N | Metabolite or Metabolite Indicator | Ratio | p Value | Formula |
|---|---|---|---|---|
| 1 | Short-Chain Acyl-Coenzyme A Dehydrogenase (SCAD) Deficiency (NBS) | 12.46 | 3.3−13 | C4/C3 |
| 2 | Lactate Dehydrogenase Activity | 5.39 | 1.3−7 | Lac/H1 |
| 3 | Isobutyryl-Coenzyme A Dehydrogenase Deficiency (NBS) | 5.37 | 2.6−11 | C4/C2 |
| 4 | Monoglyceride 16:1 | 5.16 | 0.005 | |
| 5 | Asparagine Synthesis | 3.33 | 5.0−7 | Asn/Asp |
| 6 | Putrescine Synthesis | 2.87 | 0.007 | Putrescine/Orn |
| 7 | Lysophosphatidyl-choline 17:0 | 2.67 | 0.018 | |
| 8 | Polyamine Synthesis | 2.45 | 0.023 | (Putrescine + Spermidine + Spermine)/Orn |
| 9 | Sarcosine Synthesis from Choline | 2.37 | 0.002 | Sarcosine/Choline |
| 10 | Glutaminolysis Rate | 2.24 | 8.5−6 | (Ala + Asp + Glu + Lac + Suc)/Gln |
| 11 | Lysophosphatidyl-glycerol 16:1 | 2.07 | 4.6−4 | |
| 12 | Betaine Synthesis | 2.01 | 0.004 | Betaine/Choline |
| 13 | Asparagine | 1.96 | 0.012 | |
| 14 | Phosphatidyl-ethanolamine P-16:0/15:0 | 1.95 | 0.008 | |
| 15 | Butyrylcarnitine | 1.84 | 0.002 | |
| 16 | Lysophosphatidyl-choline 16:0 | 1.81 | 0.031 | |
| 17 | Sphingosine phosphate d18:1 | 1.73 | 4.0−4 | |
| 18 | 2-Methylbutyrylglycinuria (NBS) | 1.69 | 0.011 | C5/C3 |
| 19 | Taurine Synthesis | 1.67 | 0.025 | Taurine/Cys |
| 20 | Gamma-Aminobutyric Acid Synthesis | 1.47 | 0.005 | GABA/Glu |
| 21 | Methionine Oxidation | 0.85 | 0.017 | Met-SO/Met |
| 22 | Glycine Synthesis | 0.80 | 1.6−4 | Gly/Ser |
| 23 | Dihydrolipoamide Dehydrogenase Deficiency (NBS) | 0.71 | 0.008 | Pro/Phe |
| 24 | 5-Aminovaleric acid | 0.70 | 0.017 | |
| 25 | beta-Alanine | 0.69 | 0.025 | |
| 26 | Hydroxyglutaric acid | 0.68 | 0.003 | |
| 27 | Serine | 0.67 | 0.010 | |
| 28 | Alanine | 0.63 | 0.031 | |
| 29 | Taurine | 0.62 | 0.006 | |
| 30 | Carnitine | 0.60 | 4.4−4 | |
| 31 | Triacylglyceride 18:1_33:3 | 0.60 | 0.009 | |
| 32 | Threonine | 0.59 | 0.007 | |
| 33 | Glutamine | 0.59 | 0.002 | |
| 34 | Aspartic acid | 0.59 | 0.028 | |
| 35 | Sum of Amino Acids | 0.59 | 0.004 | |
| 36 | alpha-Aminoadipic acid | 0.59 | 0.018 | |
| 37 | trans-4-Hydroxyproline | 0.58 | 0.010 | |
| 38 | Ratio of Acetylcarnitine to Carnitine | 0.57 | 0.008 | C2/C0 |
| 39 | Sum of Solely Glucogenic Amino Acids | 0.57 | 0.002 | Ala + Arg + Asn + Asp + Cys + Gln + Glu + Gly + His + Met + Pro + Ser + Thr + Val |
| 40 | beta-Aminobutyric acid | 0.56 | 0.012 | |
| 41 | Sum of Non-Essential Amino Acids | 0.56 | 8.9 × 10−4 | Ala + Arg + Asn + Asp + Cys + Gln + Glu + Gly + Pro + Ser + Tyr |
| 42 | Cysteine Synthesis | 0.55 | 6.0 × 10−4 | Cys/(Ser + Met) |
| 43 | Glutamic acid | 0.55 | 1.7 × 10−4 | |
| 44 | Glicine | 0.54 | 0.002 | |
| 45 | Taurolithocholic acid | 0.51 | 0.013 | |
| 46 | Carnosine Synthesis | 0.51 | 0.029 | Carnosine/His |
| 47 | Glutathione Constituents | 0.50 | 1.2 × 10−4 | Glu + Gly + Cys |
| 48 | Proline | 0.48 | 8.2 × 10−4 | |
| 49 | Sum of Conjugated Primary Bile Acids | 0.47 | 0.024 | GCA + GCDCA + TCA + TCDCA |
| 50 | Taurochenodeoxy-cholic acid | 0.46 | 0.019 | |
| 51 | Beta-Oxidation | 0.44 | 4.8 × 10−4 | (C2 + C3)/C0 |
| 52 | Sum of Taurine-Conjugated Bile Acids | 0.43 | 0.014 | TCA + TCDCA + TDCA + TLCA |
| 53 | Malonic Aciduria (NBS) | 0.43 | 1.6 × 10−5 | C3/C2 |
| 54 | Sum of Sulfur-Containing Amino Acids | 0.43 | 0.002 | Met + Cys |
| 55 | Short/Branched-Chain Acyl-Coenzyme A Dehydrogenase Deficiency (NBS) | 0.42 | 5.4 × 10−5 | C5/C0 |
| 56 | Taurodeoxycholic acid | 0.41 | 0.016 | |
| 57 | Phosphatidyl-choline O-38:3 | 0.39 | 0.023 | |
| 58 | Phosphatidyl-choline O-36:5 | 0.38 | 0.024 | |
| 59 | Ornithine | 0.38 | 0.031 | |
| 60 | Phosphatidyl-choline O-36:3 | 0.38 | 0.020 | |
| 61 | Phosphatidyl-choline O-38:6 | 0.38 | 0.018 | |
| 62 | Cysteine | 0.37 | 7.8 × 10−4 | |
| 63 | Choline | 0.37 | 6.3 × 10−4 | |
| 64 | Phosphatidyl-choline 36:3 | 0.35 | 0.02 | |
| 65 | Phosphatidyl-choline 38:4 | 0.35 | 0.022 | |
| 66 | Phosphatidyl-choline 40:6 | 0.35 | 0.027 | |
| 67 | Acetylcarnitine | 0.34 | 3.9 × 10−5 | |
| 68 | Aconitic acid | 0.33 | 5.0 × 10−4 | |
| 69 | Hexose | 0.32 | 0.013 | |
| 70 | Phosphatidyl-choline 38:5 | 0.32 | 0.011 | |
| 71 | Phosphatidyl-choline O-36:4 | 0.32 | 0.008 | |
| 72 | Taurocholic acid | 0.32 | 0.005 | |
| 73 | Phosphatidyl-choline 36:4 | 0.31 | 0.009 | |
| 74 | Phosphatidyl-choline 36:5 | 0.31 | 0.008 | |
| 75 | Phosphatidyl-choline O-38:4 | 0.31 | 0.007 | |
| 76 | Phosphatidyl-choline 38:6 | 0.29 | 0.007 | |
| 77 | Succinic acid | 0.26 | 2.5 × 10−4 | |
| 78 | Valerylcarnitine | 0.25 | 7.8 × 10−7 | C5 |
| 79 | Methylmalonic Acidemia (NBS) | 0.25 | 1.8 × 10−5 | C3/C0 |
| 80 | Sphinganine d14:0 | 0.21 | 0.004 | |
| 81 | Propionylcarnitine | 0.15 | 8.6 × 10−7 | C3 |
| 82 | Triacylglyceride 18:1_33:0 | 0.07 | 0.004 | |
| 83 | Phosphatidyl-inositol (18:1_20:2) | 0.04 | 0.011 |
| Enzymes Related to Carnitine Metabolism | Expression in Melanoma vs. Ctrls log2(TPM-1) | Patients’ Survival Hazard Ratio (p < 0.05) |
|---|---|---|
| Related to carnitine synthesis | ||
| TMLD | n.s. | n.s. |
| TMABADH | n.s | n.s. |
| SHMT1 | n.s | n.s. |
| SHMT2 | 6.8 vs. 5.5 * | HR 1.4 (p = 0.02) |
| BBOX1 | 0.1 vs. 4.9 * | n.s. |
| ALDH9A1 | n.s. | n.s. |
| TMLHE | n.s. | n.s. |
| ALDH9A1 | n.s. | n.s. |
| Carnitine Carriers | ||
| SLC22A5 | 2.1 vs. 3.5 * | n.s. |
| SLC25A20 | n.s. | n.s. |
| SLC22A4 | n.s. | HR 0.66 (p = 0.002) |
| SLC22A16 | n.s. | n.s. |
| SLC25A29 | 3.9 vs. 5.4 * | n.s. |
| SLC16A9 | n.s. | n.s. |
| Related to Carnitine Palmitoyltransferases | ||
| CPT2 | n.s. | n.s. |
| CPT1A | n.s. | n.s. |
| CPT1B | 3 vs. 4.2 * | HR 0.74 (p = 0.02) |
| CPT1C | n.s. | n.s. |
| ACACB | 2.1 vs. 3.9 * | n.s. |
| CHKB-CPT1B | n.s. | HR 0.75 (p = 0.03) |
| Carnitine O-Acetyltransferase | ||
| CRAT | n.s. | HR 1.3 (p = 0.04) |
| Carnitine O-Octanoyltransferase | ||
| CROT | 2.9 vs. 3.9 * | n.s. |
| Related to acethylcanitine metabolism | ||
| ACADM | 5 vs. 4 * | n.s. |
| ACAD8 | 3.6 vs. 5 * | n.s. |
| ACADS | n.s. | n.s. |
| ACADVL | 7.5 vs. 9 * | n.s. |
| ACADL | 0.1 vs. 2.1 * | n.s. |
| Related to carnitine deficiency | ||
| HADHA | n.s. | HR = 1.4 (p = 0.009) |
| IFT81 | 3.6 vs. 2.7 * | n.s. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scatozza, F.; Giardina, M.M.; Valente, C.; Vigiano Benedetti, V.; Facchiano, A. Anti-Melanoma Effects of Miconazole: Investigating the Mitochondria Involvement. Int. J. Mol. Sci. 2024, 25, 3589. https://doi.org/10.3390/ijms25073589
Scatozza F, Giardina MM, Valente C, Vigiano Benedetti V, Facchiano A. Anti-Melanoma Effects of Miconazole: Investigating the Mitochondria Involvement. International Journal of Molecular Sciences. 2024; 25(7):3589. https://doi.org/10.3390/ijms25073589
Chicago/Turabian StyleScatozza, Francesca, Maria Miriam Giardina, Carola Valente, Virginia Vigiano Benedetti, and Antonio Facchiano. 2024. "Anti-Melanoma Effects of Miconazole: Investigating the Mitochondria Involvement" International Journal of Molecular Sciences 25, no. 7: 3589. https://doi.org/10.3390/ijms25073589





