Implementing a New Algorithm for Reinterpretation of Ambiguous Variants in Genetic Dilated Cardiomyopathy
Abstract
:1. Introduction
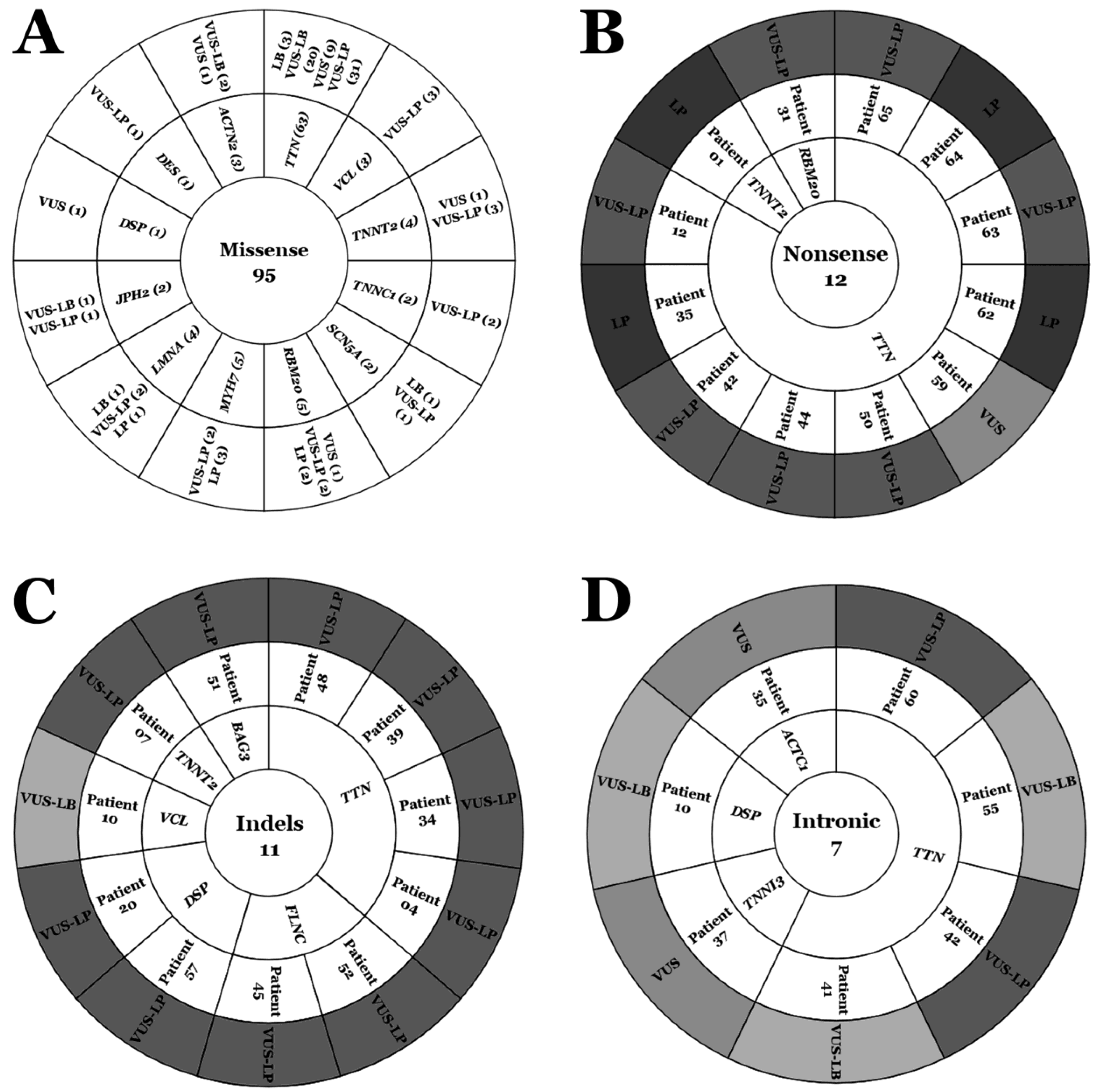
2. Results
3. Discussion
4. Materials and Methods
4.1. Study Cohort
4.2. Genetic Analysis
4.3. Data Sources
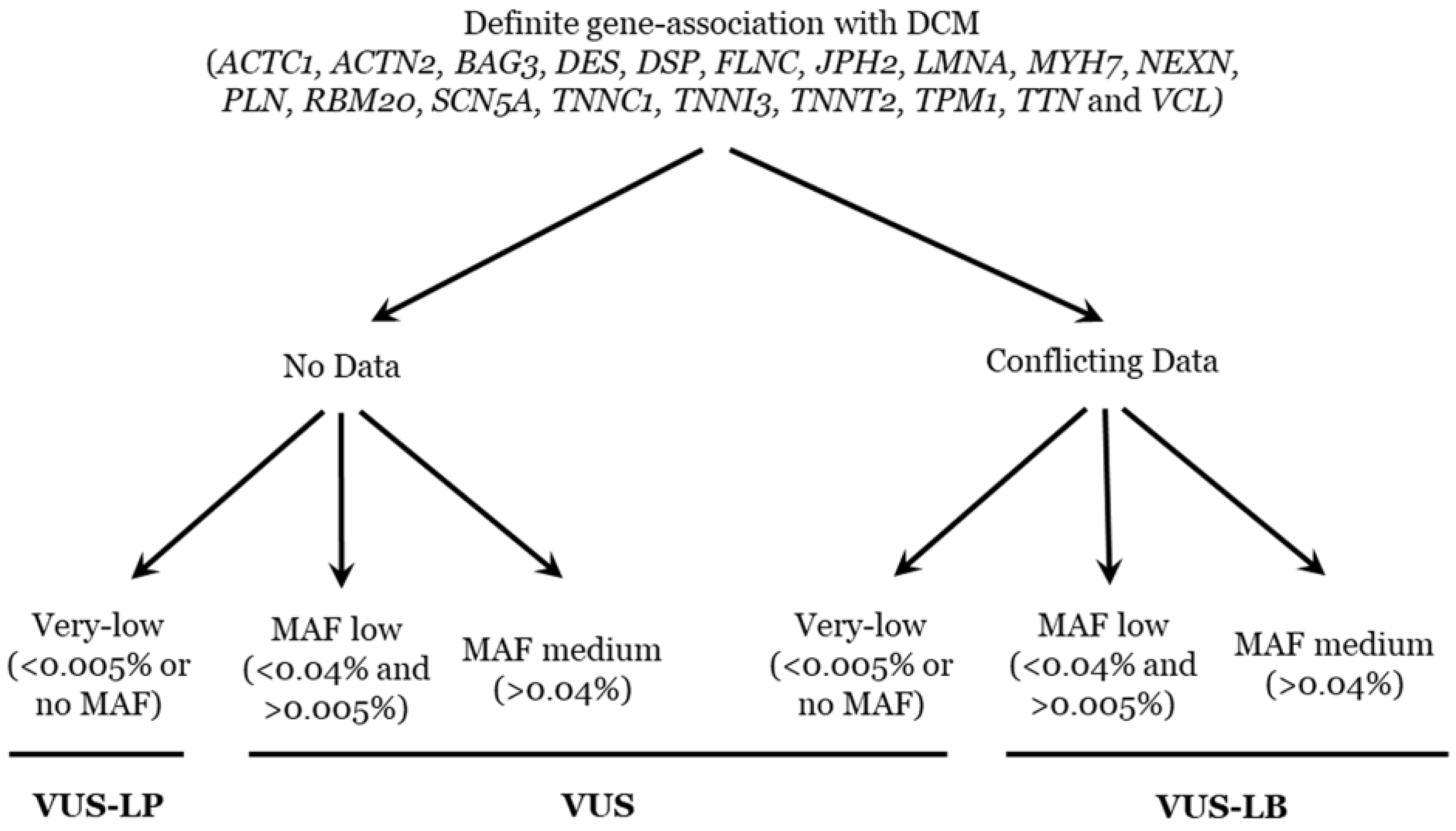
4.4. Classification/Interpretation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Arbelo, E.; Protonotarios, A.; Gimeno, J.R.; Arbustini, E.; Barriales-Villa, R.; Basso, C.; Bezzina, C.R.; Biagini, E.; A Blom, N.; A de Boer, R.; et al. 2023 ESC Guidelines for the management of cardiomyopathies. Eur. Heart J. 2023, 44, 3503–3626. [Google Scholar] [CrossRef] [PubMed]
- Wilde, A.A.M.; Semsarian, C.; Marquez, M.F.; Shamloo, A.S.; Ackerman, M.J.; Ashley, E.A.; Sternick, E.B.; Barajas-Martinez, H.; Behr, E.R.; Bezzina, C.R.; et al. European Heart Rhythm Association (EHRA)/Heart Rhythm Society (HRS)/Asia Pacific Heart Rhythm Society (APHRS)/Latin American Heart Rhythm Society (LAHRS) Expert Consensus Statement on the State of Genetic Testing for Cardiac Diseases. Europace 2022, 24, 1307–1367. [Google Scholar] [CrossRef]
- Muller, R.D.; McDonald, T.; Pope, K.; Cragun, D. Evaluation of Clinical Practices Related to Variants of Uncertain Significance Results in Inherited Cardiac Arrhythmia and Inherited Cardiomyopathy Genes. Circ. Genom. Precis. Med. 2020, 13, e002789. [Google Scholar] [CrossRef] [PubMed]
- Jordan, E.; Peterson, L.; Ai, T.; Asatryan, B.; Bronicki, L.; Brown, E.; Celeghin, R.; Edwards, M.; Fan, J.; Ingles, J.; et al. An Evidence-Based Assessment of Genes in Dilated Cardiomyopathy. Circulation 2021, 144, 7–19. [Google Scholar] [CrossRef]
- Landstrom, A.P.; Chahal, A.A.; Ackerman, M.J.; Cresci, S.; Milewicz, D.M.; Morris, A.A.; Sarquella-Brugada, G.; Semsarian, C.; Shah, S.H.; Sturm, A.C.; et al. Interpreting Incidentally Identified Variants in Genes Associated with Heritable Cardiovascular Disease: A Scientific Statement from the American Heart Association. Circ. Genom. Precis. Med. 2023, 16, e000092. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Asatryan, B.; Shah, R.A.; Sharaf Dabbagh, G.; Landstrom, A.P.; Darbar, D.; Khanji, M.Y.; Lopes, L.R.; van Duijvenboden, S.; Muser, D.; Lee, A.M.; et al. Predicted Deleterious Variants in Cardiomyopathy Genes Prognosticate Mortality and Composite Outcomes in UK Biobank. JACC Heart Fail. 2023. ahead of print. [Google Scholar] [CrossRef]
- Hofmeyer, M.; Haas, G.J.; Jordan, E.; Cao, J.; Kransdorf, E.; Ewald, G.A.; Morris, A.A.; Owens, A.; Lowes, B.; Stoller, D.; et al. Rare Variant Genetics and Dilated Cardiomyopathy Severity: The DCM Precision Medicine Study. Circulation 2023, 148, 872–881. [Google Scholar] [CrossRef]
- Stroeks, S.; Verdonschot, J.A.J. The next step toward personalized recommendations for genetic cardiomyopathies. Eur. J. Hum. Genet. 2023, 31, 1201–1203. [Google Scholar] [CrossRef]
- Campuzano, O.; Sarquella-Brugada, G.; Fernandez-Falgueras, A.; Coll, M.; Iglesias, A.; Ferrer-Costa, C.; Cesar, S.; Arbelo, E.; García-Álvarez, A.; Jordà, P.; et al. Reanalysis and reclassification of rare genetic variants associated with inherited arrhythmogenic syndromes. EBioMedicine 2020, 54, 102732. [Google Scholar] [CrossRef]
- Towbin, J.A.; McKenna, W.J.; Abrams, D.J.; Ackerman, M.J.; Calkins, H.; Darrieux, F.C.C.; Daubert, J.P.; de Chillou, C.; DePasquale, E.C.; Desai, M.Y.; et al. 2019 HRS expert consensus statement on evaluation, risk stratification, and management of arrhythmogenic cardiomyopathy. Heart Rhythm 2019, 16, e301–e372. [Google Scholar] [CrossRef] [PubMed]
- Hershberger, R.E.; Givertz, M.M.; Ho, C.Y.; Judge, D.P.; Kantor, P.F.; McBride, K.L.; Morales, A.; Taylor, M.R.G.; Vatta, M.; Ware, S.M.; et al. Genetic evaluation of cardiomyopathy: A clinical practice resource of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. 2018, 20, 899–909. [Google Scholar] [CrossRef] [PubMed]
- Musunuru, K.; Hershberger, R.E.; Day, S.M.; Klinedinst, N.J.; Landstrom, A.P.; Parikh, V.N.; Prakash, S.; Semsarian, C.; Sturm, A.C.; on behalf of the American Heart Association Council on Genomic and Precision Medicine; et al. Genetic Testing for Inherited Cardiovascular Diseases: A Scientific Statement from the American Heart Association. Circ. Genom. Precis. Med. 2020, 13, e000067. [Google Scholar] [CrossRef] [PubMed]
- Stiles, M.K.; Wilde, A.A.; Abrams, D.J.; Ackerman, M.J.; Albert, C.M.; Behr, E.R.; Chugh, S.S.; Cornel, M.C.; Gardner, K.; Ingles, J.; et al. 2020 APHRS/HRS expert consensus statement on the investigation of decedents with sudden unexplained death and patients with sudden cardiac arrest, and of their families. Heart Rhythm 2021, 18, e1–e50. [Google Scholar] [CrossRef] [PubMed]
- Sarquella-Brugada, G.; Fernandez-Falgueras, A.; Cesar, S.; Arbelo, E.; Coll, M.; Perez-Serra, A.; Puigmulé, M.; Iglesias, A.; Alcalde, M.; Vallverdú-Prats, M.; et al. Clinical impact of rare variants associated with inherited channelopathies: A 5-year update. Hum. Genet. 2022, 141, 1579–1589. [Google Scholar] [CrossRef] [PubMed]
- Bennett, J.S.; Bernhardt, M.; McBride, K.L.; Reshmi, S.C.; Zmuda, E.; Kertesz, N.J.; Garg, V.; Fitzgerald-Butt, S.; Kamp, A.N. Reclassification of Variants of Uncertain Significance in Children with Inherited Arrhythmia Syndromes is Predicted by Clinical Factors. Pediatr. Cardiol. 2019, 40, 1679–1687. [Google Scholar] [CrossRef] [PubMed]
- Vallverdú-Prats, M.; Alcalde, M.; Sarquella-Brugada, G.; Cesar, S.; Arbelo, E.; Fernandez-Falgueras, A.; Coll, M.; Pérez-Serra, A.; Puigmulé, M.; Iglesias, A.; et al. Rare Variants Associated with Arrhythmogenic Cardiomyopathy: Reclassification Five Years Later. J. Pers. Med. 2021, 11, 162. [Google Scholar] [CrossRef] [PubMed]
- Quiat, D.; Witkowski, L.; Zouk, H.; Daly, K.P.; Roberts, A.E. Retrospective Analysis of Clinical Genetic Testing in Pediatric Primary Dilated Cardiomyopathy: Testing Outcomes and the Effects of Variant Reclassification. J. Am. Heart Assoc. 2020, 9, e016195. [Google Scholar] [CrossRef]
- Stroeks, S.; Hellebrekers, D.; Claes, G.R.F.; Tayal, U.; Krapels, I.P.C.; Vanhoutte, E.K.; van den Wijngaard, A.; Henkens, M.T.H.M.; Ware, J.S.; Heymans, S.R.B.; et al. Clinical impact of re-evaluating genes and variants implicated in dilated cardiomyopathy. Genet. Med. 2021, 23, 2186–2193. [Google Scholar] [CrossRef]
- Martínez-Barrios, E.; Sarquella-Brugada, G.; Pérez-Serra, A.; Fernández-Falgueras, A.; Cesar, S.; Coll, M.; Puigmulé, M.; Iglesias, A.; Alcalde, M.; Vallverdú-Prats, M.; et al. Discerning the Ambiguous Role of Missense TTN Variants in Inherited Arrhythmogenic Syndromes. J. Pers. Med. 2022, 12, 241. [Google Scholar] [CrossRef]
- Martinez-Barrios, E.; Sarquella-Brugada, G.; Perez-Serra, A.; Fernandez-Falgueras, A.; Cesar, S.; Alcalde, M.; Coll, M.; Puigmulé, M.; Iglesias, A.; Ferrer-Costa, C.; et al. Reevaluation of ambiguous genetic variants in sudden unexplained deaths of a young cohort. Int. J. Leg. Med. 2023, 137, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Ware, S.M.; Bhatnagar, S.; Dexheimer, P.J.; Wilkinson, J.D.; Sridhar, A.; Fan, X.; Shen, Y.; Tariq, M.; Schubert, J.A.; Colan, S.D.; et al. The genetic architecture of pediatric cardiomyopathy. Am. J. Hum. Genet. 2022, 109, 282–298. [Google Scholar] [CrossRef]
- Stroeks, S.L.V.M.; Hellebrekers, D.; Claes, G.R.F.; Krapels, I.P.C.; Henkens, M.H.T.M.; Sikking, M.; Vanhoutte, E.K.; Enden, A.H.-V.D.; Brunner, H.G.; Wijngaard, A.v.D.; et al. Diagnostic and prognostic relevance of using large gene panels in the genetic testing of patients with dilated cardiomyopathy. Eur. J. Hum. Genet. 2023, 31, 776–783. [Google Scholar] [CrossRef] [PubMed]
- McAfee, Q.; Chen, C.Y.; Yang, Y.; Caporizzo, M.A.; Morley, M.; Babu, A.; Jeong, S.; Brandimarto, J.; BediJr, K.C.; Flam, E.; et al. Truncated titin proteins in dilated cardiomyopathy. Sci. Transl. Med. 2021, 13, eabd7287. [Google Scholar] [CrossRef]
- Begay, R.L.; Graw, S.; Sinagra, G.; Merlo, M.; Slavov, D.; Gowan, K.; Jones, K.L.; Barbati, G.; Spezzacatene, A.; Brun, F.; et al. Role of Titin Missense Variants in Dilated Cardiomyopathy. J. Am. Heart Assoc. 2015, 4, e002645. [Google Scholar] [CrossRef] [PubMed]
- Rich, K.A.; Moscarello, T.; Siskind, C.; Brock, G.; Tan, C.A.; Vatta, M.; Winder, T.L.; Elsheikh, B.; Vicini, L.; Tucker, B.; et al. Novel heterozygous truncating titin variants affecting the A-band are associated with cardiomyopathy and myopathy/muscular dystrophy. Mol. Genet. Genom. Med. 2020, 8, e1460. [Google Scholar] [CrossRef] [PubMed]
- Akinrinade, O.; Heliö, T.; Deprez, R.H.L.; Jongbloed, J.D.H.; Boven, L.G.; Berg, M.P.v.D.; Pinto, Y.M.; Alastalo, T.-P.; Myllykangas, S.; van Spaendonck-Zwarts, K.; et al. Relevance of Titin Missense and Non-Frameshifting Insertions/Deletions Variants in Dilated Cardiomyopathy. Sci. Rep. 2019, 9, 4093. [Google Scholar] [CrossRef] [PubMed]
- Domínguez, F.; Lalaguna, L.; Martínez-Martín, I.; Piqueras-Flores, J.; Rasmussen, T.B.; Zorio, E.; Giovinazzo, G.; Prados, B.; Ochoa, J.P.; Bornstein, B.; et al. Titin Missense Variants as a Cause of Familial Dilated Cardiomyopathy. Circulation 2023, 147, 1711–1713. [Google Scholar] [CrossRef] [PubMed]
- Rosamilia, M.B.; Markunas, A.M.; Kishnani, P.S.; Landstrom, A.P. Underrepresentation of Diverse Ancestries Drives Uncertainty in Genetic Variants Found in Cardiomyopathy-Associated Genes. JACC 2024, 3, 100767. [Google Scholar] [CrossRef]
- Priori, S.G.; Blomstrom-Lundqvist, C. 2015 European Society of Cardiology Guidelines for the management of patients with ventricular arrhythmias and the prevention of sudden cardiac death summarized by co-chairs. Eur. Heart J. 2015, 36, 2757–2759. [Google Scholar] [CrossRef]
- Al-Khatib, S.M.; Stevenson, W.G.; Ackerman, M.J.; Bryant, W.J.; Callans, D.J.; Curtis, A.B.; Deal, B.J.; Dickfeld, T.; Field, M.E.; Fonarow, G.C.; et al. 2017 AHA/ACC/HRS guideline for management of patients with ventricular arrhythmias and the prevention of sudden cardiac death: Executive summary: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines and the Heart Rhythm Society. Heart Rhythm 2018, 15, e190–e252. [Google Scholar] [PubMed]
- Kobayashi, Y.; Yang, S.; Nykamp, K.; Garcia, J.; Lincoln, S.E.; Topper, S.E. Pathogenic variant burden in the ExAC database: An empirical approach to evaluating population data for clinical variant interpretation. Genome Med. 2017, 9, 13. [Google Scholar] [CrossRef] [PubMed]
- Jordan, D.M.; Kiezun, A.; Baxter, S.M.; Agarwala, V.; Green, R.C.; Murray, M.F.; Pugh, T.; Lebo, M.S.; Rehm, H.L.; Funke, B.H.; et al. Development and validation of a computational method for assessment of missense variants in hypertrophic cardiomyopathy. Am. J. Hum. Genet. 2011, 88, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Kelly, M.A.; Caleshu, C.; Morales, A.; Buchan, J.; Wolf, Z.; Harrison, S.M.; Cook, S.; Dillon, M.W.; Garcia, J.; Haverfield, E.; et al. Adaptation and validation of the ACMG/AMP variant classification framework for MYH7-associated inherited cardiomyopathies: Recommendations by ClinGen’s Inherited Cardiomyopathy Expert Panel. Genet. Med. 2018, 20, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Bains, S.; Dotzler, S.M.; Krijger, C.; Giudicessi, J.R.; Ye, D.; Bikker, H.; Rohatgi, R.K.; Tester, D.J.; Bos, J.M.; Wilde, A.A.; et al. A phenotype-enhanced variant classification framework to decrease the burden of missense variants of uncertain significance in type 1 long QT syndrome. Heart Rhythm 2022, 19, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Giudicessi, J.R.; Lieve, K.V.V.; Rohatgi, R.K.; Koca, F.; Tester, D.J.; van der Werf, C.; Martijn Bos, J.; Wilde, A.A.M.; Ackerman, M.J. Assessment and Validation of a Phenotype-Enhanced Variant Classification Framework to Promote or Demote RYR2 Missense Variants of Uncertain Significance. Circ. Genom. Precis. Med. 2019, 12, e002510. [Google Scholar] [CrossRef] [PubMed]
- Arbustini, E.; Behr, E.R.; Carrier, L.; van Duijn, C.; Evans, P.; Favalli, V.; van der Harst, P.; Haugaa, K.H.; Jondeau, G.; Kääb, S.; et al. Interpretation and actionability of genetic variants in cardiomyopathies: A position statement from the European Society of Cardiology Council on cardiovascular genomics. Eur. Heart J. 2022, 43, 1901–1916. [Google Scholar] [CrossRef]
- Arbustini, E.; Urtis, M.; Elliott, P. Interpretation of genetic variants depends on a clinically guided integration of phenotype and molecular data. Eur. Heart J. 2022, 43, 2638–2639. [Google Scholar] [CrossRef]

| Patient | Gene | Nucleotide | Protein | dbSNP | ClinVar | GnomAD (MAF%) | Classification (Year) | Classification 2023 |
|---|---|---|---|---|---|---|---|---|
| 1 | TNNT2 | c.860G>A | p.Trp287Ter | rs727504247 | LP | NA | VUS (2016) | LP |
| 1 | TTN | c.47951G>A | p.Arg15984His | rs201774108 | VUSc | 0.0001 | VUS (2016) | VUS |
| 2 | TTN | c.73195G>A | p.Val24399Ile | rs1257567608 | NA | NA | VUS (2016) | VUS (VUS-LP) |
| 2 | TTN | c.57388C>T | p.Arg19130Cys | rs72646861 | LB | 0.827 | VUS (2016) | LB |
| 2 | TTN | c.53117C>T | p.Pro17706Leu | rs72646845 | LB | 0.369 | VUS (2016) | LB |
| 3 | ACTN2 | c.2051A>T | p.Asn684Ile | rs576783493 | VUSc | 0.0001 | VUS (2016) | VUS |
| 3 | TTN | c.93125G>A | p.Gly31042Asp | rs373754986 | VUS | 0.007 | VUS (2016) | VUS (VUS-LP) |
| 3 | TTN | c.76456G>C | p.Asp25486His | rs780958039 | VUS | 0.0008 | VUS (2016) | VUS (VUS-LP) |
| 4 | ACTN2 | c.1426G>A | p.Ala476Thr | rs142943120 | VUSc | 0.027 | VUS (2016) | VUS (VUS-LB) |
| 4 | TTN | c.57978del | p.Val19327PhefsTer10 | NA | NA | NA | VUS (2016) | VUS (VUS-LP) |
| 4 | TTN | c.76559G>A | p.Ser25520Asn | rs200450022 | VUSc | 0.061 | VUS (2016) | VUS (VUS-LB) |
| 4 | TTN | c.72764G>C | p.Gly24225Ala | rs114071241 | VUS | 0.001 | VUS (2016) | VUS (VUS-LP) |
| 4 | TTN | c.17066G>C | p.Gly5689Ala | rs200118743 | VUSc | 0.063 | VUS (2016) | VUS (VUS-LB) |
| 4 | TTN | c.4675G>A | p.Val1559Ile | rs538451328 | NA | 0.0003 | VUS (2016) | VUS (VUS-LP) |
| 5 | TTN | c.98971G>C | p.Glu32991Gln | rs199632397 | VUSc | 0.042 | VUS (2016) | VUS (VUS-LB) |
| 5 | TTN | c.27659G>A | p.Arg9220Gln | rs727504757 | VUS | 0.003 | VUS (2016) | VUS (VUS-LP) |
| 6 | TTN | c.45392G>A | p.Arg15131His | rs72646808 | VUSc | 0.185 | VUS (2016) | LB |
| 7 | TNNT2 | c.629_631del | p.Lys210del | rs45578238 | VUS | NA | VUS (2016) | VUS (VUS-LP) |
| 8 | MYH7 | c.1106G>A | p.Arg369Gln | rs397516089 | LP | 0.00006 | VUS (2016) | LP |
| 9 | TTN | c.73967A>G | p.Asn24656Ser | rs368443217 | VUSc | 0.008 | VUS (2016) | VUS (VUS-LB) |
| 9 | TTN | c.65012T>A | p.Met21671Lys | rs750298083 | VUSc | 0.004 | VUS (2016) | VUS |
| 9 | TTN | c.51830G>A | p.Arg17277His | rs201457934 | VUSc | 0.01 | VUS (2016) | VUS (VUS-LB) |
| 9 | TTN | c.9247G>A | p.Glu3083Lys | NA | NA | NA | VUS (2016) | VUS (VUS-LP) |
| 9 | MYH7 | c.2711G>A | p.Arg904His | rs397516165 | LP | 0.0001 | VUS (2016) | LP |
| 10 | DSP | c.1266+6G>T | NA | rs73375345 | LB | 0.037 | VUS (2016) | VUS (VUS-LB) |
| 10 | TTN | c.20920A>G | p.Ser6974Gly | rs72648980 | VUSc | 0.062 | VUS (2016) | VUS (VUS-LB) |
| 10 | VCL | c.2862_2864del | p.Leu955del | rs397517237 | VUSc | 0.021 | VUS (2016) | VUS (VUS-LB) |
| 11 | SCN5A | c.6010T>C | p.Phe2004Leu | rs41311117 | VUSc | 0.198 | VUS (2016) | LB |
| 12 | TTN | c.72970C>T | p.Gln24324Ter | NA | NA | NA | VUS (2016) | VUS (VUS-LP) |
| 13 | TTN | c.49249G>A | p.Asp16417Asn | rs1244503464 | NA | 0.0004 | VUS (2016) | VUS (VUS-LP) |
| 14 | TTN | c.12814G>T | p.Asp4272Tyr | rs72648940 | VUSc | 0.0005 | VUS (2016) | VUS |
| 15 | TTN | c.77076A>C | p.Glu25692Asp | rs370547473 | NA | 0.004 | VUS (2016) | VUS (VUS-LP) |
| 16 | TTN | c.32932C>A | p.Pro10978Thr | rs1393076582 | VUSc | NA | VUS (2016) | VUS |
| 17 | DES | c.179C>T | p.Ser60Leu | rs868853251 | NA | 0.0001 | VUS (2016) | VUS (VUS-LP) |
| 17 | TTN | c.46877G>A | p.Gly15626Asp | rs201802447 | VUS | 0.007 | VUS (2016) | VUS |
| 17 | TTN | c.1066G>C | p.Glu356Gln | rs144531477 | VUSc | 0.015 | VUS (2016) | VUS (VUS-LB) |
| 18 | TTN | c.95137A>G | p.Ile31713Val | rs758945559 | NA | 0.0008 | VUS (2016) | VUS (VUS-LP) |
| 18 | TTN | c.30484C>A | p.Pro10162Thr | rs532102837 | VUSc | 0.058 | VUS (2016) | VUS (VUS-LB) |
| 18 | DSP | c.6799A>T | p.Thr2267Ser | rs181378432 | LB | 0.009 | VUS (2016) | VUS |
| 18 | LMNA | c.1621C>T | p.Arg541Cys | rs56984562 | LP | 0.0001 | VUS (2016) | LP |
| 19 | RBM20 | c.1900C>T | p.Arg634Trp | rs796734066 | LP | NA | VUS (2016) | LP |
| 20 | DSP | del ex. 21_24 | NA | NA | NA | NA | VUS (2016) | VUS (VUS-LP) |
| 21 | MYH7 | c.1371A>G | p.Ile457Met | NA | NA | NA | VUS (2016) | VUS (VUS-LP) |
| 22 | TTN | c.94724T>C | p.Met31575Thr | rs397517786 | VUSc | 0.007 | VUS (2016) | VUS (VUS-LB) |
| 22 | TTN | c.62687G>T | p.Gly20896Val | rs549938348 | VUS | 0.0004 | VUS (2016) | VUS (VUS-LP) |
| 23 | RBM20 | c.1906C>T | p.Arg636Cys | rs267607002 | VUSc | 0.0001 | VUS (2016) | VUS |
| 24 | JPH2 | c.1736C>T | p.Pro579Leu | rs953353202 | VUS | 0.001 | VUS (2016) | VUS (VUS-LP) |
| 24 | RBM20 | c.1980C>A | p.Ser660Arg | NA | NA | 0.00007 | VUS (2016) | VUS (VUS-LP) |
| 25 | RBM20 | c.2200C>G | p.Arg734Gly | NA | NA | NA | VUS (2016) | VUS (VUS-LP) |
| 26 | TTN | c.57212G>A | p.Arg19071Gln | rs373282633 | VUS | 0.003 | VUS (2016) | VUS |
| 26 | TTN | c.48221T>A | p.Leu16074Gln | rs140714512 | VUSc | 0.051 | VUS (2016) | VUS (VUS-LB) |
| 26 | TTN | c.29645A>C | p.Lys9882Thr | rs760742068 | VUS | 0.001 | VUS (2016) | VUS (VUS-LP) |
| 27 | LMNA | c.1930C>T | p.Arg644Cys | rs1420000963 | VUSc | 0.201 | VUS (2016) | LB |
| 27 | TTN | c.77188C>T | p.Arg25730Trp | rs779581886 | VUS | 0.002 | VUS (2016) | VUS (VUS-LP) |
| 27 | TTN | c.60581T>C | p.Leu20194Pro | rs1359881893 | VUS | 0.0008 | VUS (2016) | VUS (VUS-LP) |
| 28 | RBM20 | c.1913C>T | p.Pro638Leu | rs267607003 | LP | 0.0003 | VUS (2016) | LP |
| 28 | TTN | c.74942C>T | p.Ala24981Val | rs749950083 | NA | NA | VUS (2016) | VUS (VUS-LP) |
| 28 | TTN | c.73501C>G | p.Pro24501Ala | rs770542451 | NA | 0.0004 | VUS (2016) | VUS (VUS-LP) |
| 28 | TTN | c.1333G>A | p.Ala445Thr | rs142414432 | VUSc | 0.021 | VUS (2016) | VUS (VUS-LB) |
| 29 | LMNA | c.1949A>G | p.Asn650Ser | rs775728847 | VUS | 0.0003 | VUS (2016) | VUS (VUS-LP) |
| 29 | TTN | c.38807A>G | p.Asn12936Ser | rs1184631064 | VUS | 0.0008 | VUS (2016) | VUS (VUS-LP) |
| 30 | ACTN2 | c.1040C>T | p.Thr347Met | rs727504590 | VUSc | 0.009 | VUS (2016) | VUS (VUS-LB) |
| 31 | RBM20 | c.3684A>G | p.Ter1228TrpextTer33 | rs1845123103 | NA | 0.0001 | VUS (2016) | VUS (VUS-LP) |
| 31 | TTN | c.40001G>A | p.Gly13334Glu | rs561284948 | VUS | 0.001 | VUS (2016) | VUS (VUS-LP) |
| 31 | TTN | c.20863C>T | p.Pro6955Ser | rs1438804317 | NA | NA | VUS (2016) | VUS (VUS-LP) |
| 32 | MYH7 | c.602T>C | p.Ile201Thr | rs397516258 | LP | 0.0001 | VUS (2016) | LP |
| 32 | TTN | c.71188G>A | p.Gly23730Arg | rs72648205 | VUSc | 0.034 | VUS (2016) | VUS (VUS-LB) |
| 33 | TTN | c.82400G>A | p.Arg27467His | rs199895320 | VUS | 0.002 | VUS (2016) | VUS (VUS-LP) |
| 33 | TTN | c.75575A>T | p.Asn25192IIe | rs200714263 | VUS | 0.002 | VUS (2016) | VUS (VUS-LP) |
| 34 | TTN | c.9139_9150del | p.Ser3047_Thr3050del | NA | NA | NA | VUS (2016) | VUS (VUS-LP) |
| 35 | ACTC1 | c.455-7C>T | NA | rs768363857 | LB | 0.003 | VUS (2017) | VUS |
| 35 | TTN | c.53791C>T | p.Arg17931Ter | rs869312112 | LP | NA | VUS (2017) | LP |
| 35 | TTN | c.33568T>G | p.Cys11190Gly | NA | NA | NA | VUS (2017) | VUS (VUS-LP) |
| 36 | TTN | c.58846G>A | p.Gly19616Ser | rs1262240030 | NA | NA | VUS (2017) | VUS (VUS-LP) |
| 36 | TTN | c.58590G>C | p.Gln19530His | NA | NA | NA | VUS (2017) | VUS (VUS-LP) |
| 36 | TTN | c.8320G>A | p.Glu2774Lys | rs763666119 | LB | 0.0003 | VUS (2017) | VUS (VUS-LP) |
| 37 | TNNI3 | c.-8G>A | NA | rs773513015 | VUSc | 0.001 | VUS (2017) | VUS |
| 37 | TTN | c.40796G>A | p.Arg13599Gln | rs778774812 | VUS | 0.001 | VUS (2017) | VUS (VUS-LP) |
| 38 | TTN | c.57709G>T | p.Val19237Leu | rs1397460981 | NA | NA | VUS (2017) | VUS (VUS-LP) |
| 39 | TTN | c.95967dup | p.Arg31990ThrfsTer10 | NA | NA | NA | VUS (2017) | VUS (VUS-LP) |
| 40 | TTN | c.12389G>A | p.Cys4130Tyr | rs375577529 | VUSc | 0.008 | VUS (2017) | VUS (VUS-LB) |
| 40 | VCL | c.2905G>A | p.Ala969Thr | rs199751261 | VUS | 0.002 | VUS (2017) | VUS (VUS-LP) |
| 41 | LMNA | c.253C>G | p.Leu85Val | NA | NA | NA | VUS (2017) | VUS (VUS-LP) |
| 41 | TTN | c.78602G>A | p.Gly26201Asp | rs756222422 | NA | 0.0004 | VUS (2017) | VUS (VUS-LP) |
| 41 | TTN | c.64137G>C | p.Lys21379Asn | rs56019808 | VUSc | 0.014 | VUS (2017) | VUS (VUS-LB) |
| 41 | TTN | c.36844+9A>G | NA | rs372725070 | LB | 0.013 | VUS (2017) | VUS (VUS-LB) |
| 41 | TTN | c.21355G>T | p.Ala7119Ser | rs200972189 | VUSc | 0.02 | VUS (2017) | VUS (VUS-LB) |
| 42 | TTN | c.92472G>C | p.Lys30824Asn | NA | NA | NA | VUS (2017) | VUS (VUS-LP) |
| 42 | TTN | c.25810C>T | p.Arg8604Ter | NA | NA | NA | VUS (2017) | VUS (VUS-LP) |
| 42 | TTN | c.28738+7T>G | NA | NA | NA | NA | VUS (2017) | VUS (VUS-LP) |
| 42 | TTN | c.14656A>T | p.Thr4886Ser | rs794727816 | VUS | NA | VUS (2017) | VUS (VUS-LP) |
| 43 | TNNC1 | c.400G>A | p.Glu134Lys | rs1553651640 | VUS | NA | VUS (2017) | VUS (VUS-LP) |
| 43 | TTN | c.95173A>G | p.Lys31725Glu | rs72629783 | VUSc | 0.021 | VUS (2017) | VUS (VUS-LB) |
| 43 | TTN | c.31709T>C | p.Ile10570Thr | rs72650057 | VUSc | 0.013 | VUS (2017) | VUS (VUS-LB) |
| 44 | TTN | c.56541G>A | p.Trp18847Ter | NA | LP | NA | VUS (2017) | VUS (VUS-LP) |
| 45 | FLNC | c.3612del | p.His1205ThrfsTer65 | NA | NA | NA | VUS (2017) | VUS (VUS-LP) |
| 46 | TTN | c.19570G>A | p.Asp6524Asn | rs72648973 | VUSc | 0.075 | VUS (2017) | VUS (VUS-LB) |
| 47 | TNNT2 | c.476G>A | p.Arg159Gln | rs45501500 | VUSc | NA | VUS (2017) | VUS |
| 48 | TTN | c.51369_51384del | p.Asp17123GlufsTer4 | NA | NA | NA | VUS (2017) | VUS (VUS-LP) |
| 49 | JPH2 | c.572C>G | p.Pro191Arg | rs554853074 | LB | 0.045 | VUS (2017) | VUS (VUS-LB) |
| 49 | TNNT2 | c.230C>T | p.Pro77Leu | rs144900708 | VUS | 0.003 | VUS (2017) | VUS (VUS-LP) |
| 50 | TTN | c.86270C>A | p.Ser28757Ter | NA | LP | NA | VUS (2017) | VUS (VUS-LP) |
| 50 | TTN | c.62666A>T | p.Asp20889Val | rs535816123 | NA | 0.002 | VUS (2017) | VUS (VUS-LP) |
| 50 | TTN | c.47435T>C | p.Ile15812Thr | rs72646819 | VUSc | 0.007 | VUS (2017) | VUS (VUS-LB) |
| 50 | TTN | c.16066A>G | p.Thr5356Ala | rs530353051 | VUS | 0.002 | VUS (2017) | VUS |
| 51 | BAG3 | c.903del | p.Arg301SerfsTer6 | NA | NA | NA | VUS (2018) | VUS (VUS-LP) |
| 51 | TTN | c.29327A>G | p.Tyr9776Cys | rs72650035 | VUSc | 0.02 | VUS (2018) | VUS (VUS-LB) |
| 52 | FLNC | c.1414del | p.Cys472ValfsTer20 | NA | NA | NA | VUS (2018) | VUS (VUS-LP) |
| 53 | MYH7 | c.5452C>T | p.Arg1818Trp | rs763073072 | VUS | 0.0005 | VUS (2018) | VUS (VUS-LP) |
| 54 | TNNC1 | c.394G>A | p.Asp132Asn | rs397516846 | VUS | 0.0003 | VUS (2018) | VUS (VUS-LP) |
| 55 | TTN | c.24617A>G | p.Asn8206Ser | NA | NA | NA | VUS (2018) | VUS |
| 55 | TTN | c.12889+7A>T | NA | rs10200398 | VUSc | 0.07 | VUS (2018) | VUS (VUS-LB) |
| 56 | TTN | c.20839G>A | p.Glu6947Lys | rs201326258 | VUS | 0.003 | VUS (2018) | VUS |
| 56 | VCL | c.1620T>G | p.Asp540Glu | rs533622785 | NA | 0.0006 | VUS (2018) | VUS (VUS-LP) |
| 57 | DSP | c.5673_5674dup | p.Lys1892ArgfsTer38 | NA | NA | NA | VUS (2018) | VUS (VUS-LP) |
| 57 | SCN5A | c.2924G>A | p.Arg975Gln | rs753149586 | VUS | 0.005 | VUS (2018) | VUS (VUS-LP) |
| 58 | TNNT2 | c.835G>A | p.Gly279Arg | rs757664792 | VUS | 0.0004 | VUS (2018) | VUS (VUS-LP) |
| 59 | TTN | c.9220C>T | p.Arg3074Ter | rs780706937 | VUSc | NA | VUS (2018) | VUS |
| 60 | TTN | c.81493+1G>T | NA | NA | NA | NA | VUS (2018) | VUS (VUS-LP) |
| 61 | TNNT2 | c.391C>G | p.Arg131Gly | rs74315380 | LP | NA | VUS (2019) | VUS (VUS-LP) |
| 62 | TTN | c.78412C>T | p.Arg26138Ter | rs794729384 | LP | 0.0004 | VUS (2019) | LP |
| 63 | TTN | c.67528G>T | p.Glu22510Ter | NA | NA | NA | VUS (2019) | VUS (VUS-LP) |
| 64 | TTN | c.39790C>T | p.Arg13264Ter | rs751746401 | LP | 0.0004 | VUS (2019) | LP |
| 65 | TTN | c.59787G>A | p.Trp19929Ter | NA | NA | NA | VUS (2019) | VUS (VUS-LP) |
| 65 | VCL | c.158A>G | p.Asn53Ser | rs751938777 | VUS | 0.001 | VUS (2019) | VUS (VUS-LP) |
| 2023 | Intronic | Indels | Nonsense | Missense | Total | ||
|---|---|---|---|---|---|---|---|
| B | 0 | 0 | 0 | 0 | 0 | ||
| LB | 0 | 0 | 0 | 5 (4%) | 5 (4%) | ||
| VUS | VUS-LB | 3 (2.4%) | 1 (0.8%) | 0 | 23 (18.4%) | 27 (21.6%) | 110 (88%) |
| VUS | 2 (1.6%) | 0 | 1 (0.8%) | 13 (10.4%) | 16 (12.8%) | ||
| VUS-LP | 2 (1.6%) | 10 (8%) | 7 (5.6%) | 48 (38.4%) | 67 (53.6%) | ||
| LP | 0 | 0 | 4 (3.2%) | 6 (4.8%) | 10 (8%) | ||
| P | 0 | 0 | 0 | 0 | 0 | ||
| Total | 7 (5.6%) | 11 (8.8%) | 12 (9.6%) | 95 (76%) | 125 (100%) | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Serra, A.; Toro, R.; Martinez-Barrios, E.; Iglesias, A.; Fernandez-Falgueras, A.; Alcalde, M.; Coll, M.; Puigmulé, M.; del Olmo, B.; Picó, F.; et al. Implementing a New Algorithm for Reinterpretation of Ambiguous Variants in Genetic Dilated Cardiomyopathy. Int. J. Mol. Sci. 2024, 25, 3807. https://doi.org/10.3390/ijms25073807
Pérez-Serra A, Toro R, Martinez-Barrios E, Iglesias A, Fernandez-Falgueras A, Alcalde M, Coll M, Puigmulé M, del Olmo B, Picó F, et al. Implementing a New Algorithm for Reinterpretation of Ambiguous Variants in Genetic Dilated Cardiomyopathy. International Journal of Molecular Sciences. 2024; 25(7):3807. https://doi.org/10.3390/ijms25073807
Chicago/Turabian StylePérez-Serra, Alexandra, Rocío Toro, Estefanía Martinez-Barrios, Anna Iglesias, Anna Fernandez-Falgueras, Mireia Alcalde, Mónica Coll, Marta Puigmulé, Bernat del Olmo, Ferran Picó, and et al. 2024. "Implementing a New Algorithm for Reinterpretation of Ambiguous Variants in Genetic Dilated Cardiomyopathy" International Journal of Molecular Sciences 25, no. 7: 3807. https://doi.org/10.3390/ijms25073807
APA StylePérez-Serra, A., Toro, R., Martinez-Barrios, E., Iglesias, A., Fernandez-Falgueras, A., Alcalde, M., Coll, M., Puigmulé, M., del Olmo, B., Picó, F., Lopez, L., Arbelo, E., Cesar, S., Llano, C. T. d., Mangas, A., Brugada, J., Sarquella-Brugada, G., Brugada, R., & Campuzano, O. (2024). Implementing a New Algorithm for Reinterpretation of Ambiguous Variants in Genetic Dilated Cardiomyopathy. International Journal of Molecular Sciences, 25(7), 3807. https://doi.org/10.3390/ijms25073807








