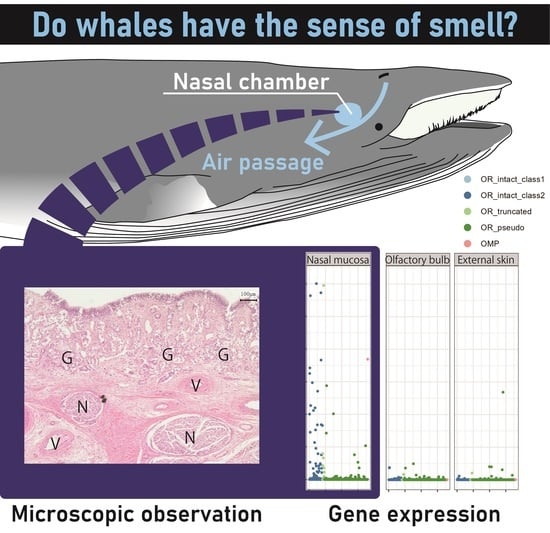
Localized Expression of Olfactory Receptor Genes in the Olfactory Organ of Common Minke Whales
Abstract
:1. Introduction
2. Results
2.1. Gross Observation
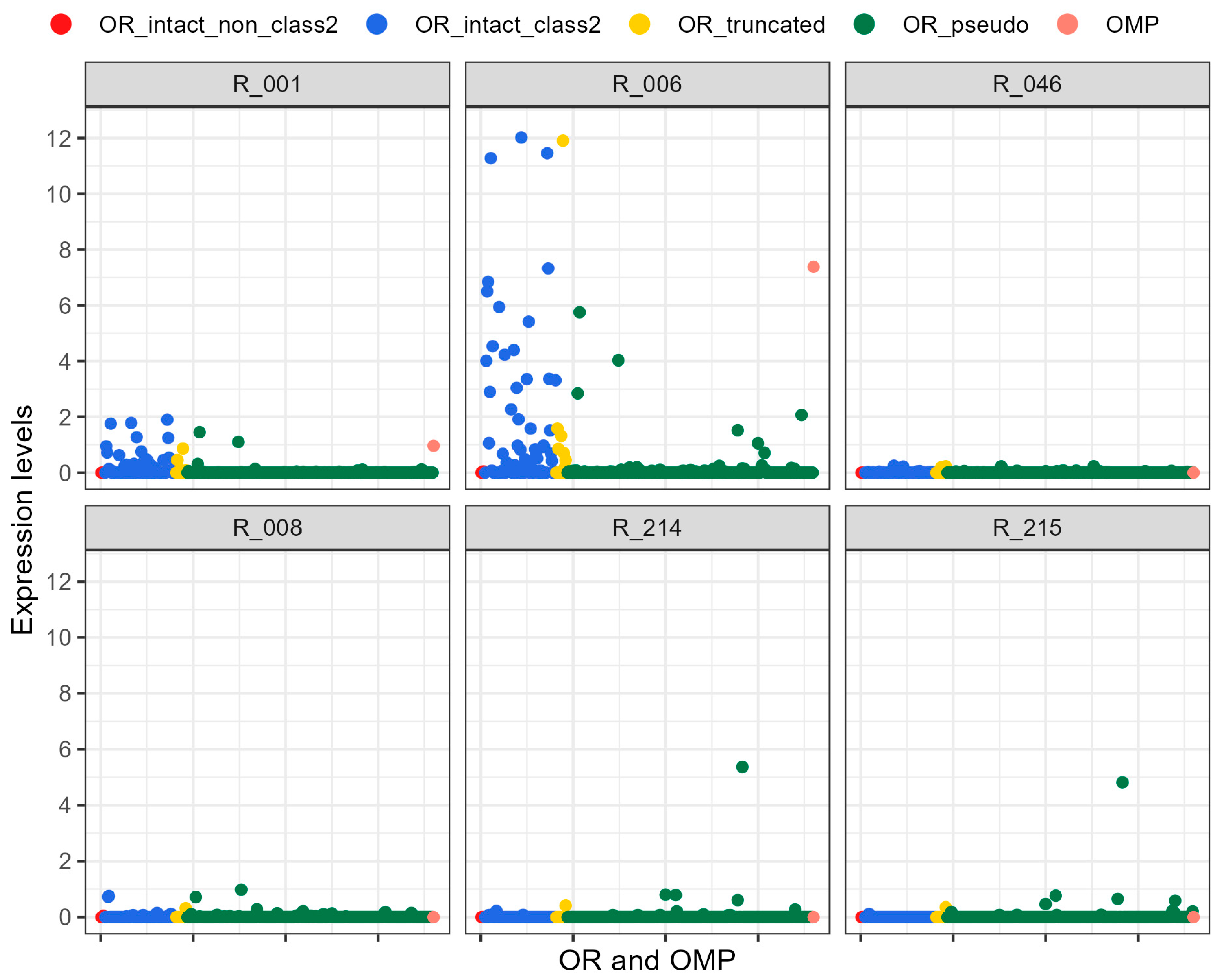
2.2. Expression of the Olfactory Receptor Genes
2.3. Histological Staining
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Gross Observation
4.3. RNA Expression
4.4. Histology Staining
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Doty, R.L. Odor-guided behavior in mammals. Experientia 1986, 42, 257–271. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Niimura, Y.; Nozawa, M. The evolution of animal chemosensory receptor gene repertoires: Roles of chance and necessity. Nat. Rev. Genet. 2008, 9, 951–963. [Google Scholar] [CrossRef]
- Corona, R.; Lévy, F. Chemical olfactory signals and parenthood in mammals. Horm. Behav. 2015, 68, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Buck, L.; Axel, R. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell 1991, 65, 175–187. [Google Scholar] [CrossRef] [PubMed]
- Niimura, Y.; Nei, M. Evolutionary dynamics of olfactory receptor genes in fishes and tetrapods. Proc. Natl. Acad. Sci. USA 2005, 102, 6039–6044. [Google Scholar] [CrossRef] [PubMed]
- Niimura, Y. On the origin and evolution of vertebrate olfactory receptor genes: Comparative genome analysis among 23 chordate species. Genome Biol. Evol. 2009, 1, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Malnic, B.; Hirono, J.; Sato, T.; Buck, L.B. Combinatorial receptor codes for odors. Cell 1999, 96, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Saito, H.; Chi, Q.; Zhuang, H.; Matsunami, H.; Mainland, J.D. Odor coding by a Mammalian receptor repertoire. Sci. Signal. 2009, 2, ra9. [Google Scholar] [CrossRef]
- Pihlström, H.; Fortelius, M.; Hemilä, S.; Forsman, R.; Reuter, T. Scaling of mammalian ethmoid bones can predict olfactory organ size and performance. Proc. R. Soc. B Biol. Sci. 2005, 272, 957–962. [Google Scholar] [CrossRef] [PubMed]
- Niimura, Y. Evolutionary dynamics of olfactory receptor genes in chordates: Interaction between environments and genomic contents. Hum. Genom. 2009, 4, 107. [Google Scholar] [CrossRef]
- Zhou, Y.; Shearwin-Whyatt, L.; Li, J.; Song, Z.; Hayakawa, T.; Stevens, D.; Fenelon, J.C.; Peel, E.; Cheng, Y.; Pajpach, F.; et al. Platypus and echidna genomes reveal mammalian biology and evolution. Nature 2021, 592, 756–762. [Google Scholar] [CrossRef]
- Moran, D.T.; Rowley, J.C.; Jafek, B.W.; Lovell, M.A. The fine structure of the olfactory mucosa in man. J. Neurocytol. 1982, 11, 721–746. [Google Scholar] [CrossRef] [PubMed]
- Bird, D.J.; Murphy, W.J.; Fox-Rosales, L.; Hamid, I.; Eagle, R.A.; Van Valkenburgh, B. Olfaction written in bone: Cribriform plate size parallels olfactory receptor gene repertoires in Mammalia. Proc. R. Soc. B Biol. Sci. 2018, 285, 20180100. [Google Scholar] [CrossRef] [PubMed]
- Niimura, Y.; Nei, M. Evolution of olfactory receptor genes in the human genome. Proc. Natl. Acad. Sci. USA 2003, 100, 12235–12240. [Google Scholar] [CrossRef] [PubMed]
- Niimura, Y.; Nei, M. Extensive gains and losses of olfactory receptor genes in mammalian evolution. PLoS ONE 2007, 2, e708. [Google Scholar] [CrossRef] [PubMed]
- Roe, L.J.; Thewissen, J.G.M.; Quade, J.; O’Neil, J.R.; Bajpai, S.; Sahni, A.; Hussain, S.T. Isotopic Approaches to Understanding the Terrestrial-to-Marine Transition of the Earliest Cetaceans. In The Emergence of Whales: Evolutionary Patterns in the Origin of Cetacea; Thewissen, J.G.M., Ed.; Springer: Boston, MA, USA, 1998; pp. 399–422. [Google Scholar] [CrossRef]
- Clementz, M.T.; Goswami, A.; Gingerich, P.D.; Koch, P.L. Isotopic records from early whales and sea cows: Contrasting patterns of ecological transition. J. Vertebr. Paleontol. 2006, 26, 355–370. [Google Scholar] [CrossRef]
- Gatesy, J.; Geisler, J.H.; Chang, J.; Buell, C.; Berta, A.; Meredith, R.W.; Springer, M.S.; McGowen, M.R. A phylogenetic blueprint for a modern whale. Mol. Phylogenet. Evol. 2013, 66, 479–506. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, M.; Rooney, A.P.; Okada, N. Phylogenetic relationships among cetartiodactyls based on insertions of short and long interpersed elements: Hippopotamuses are the closest extant relatives of whales. Proc. Natl. Acad. Sci. USA 1999, 96, 10261–10266. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, M.; Matsuno, F.; Abe, H.; Shimamura, M.; Hamilton, H.; Matsubayashi, H.; Okada, N. Evolution of CHR-2 SINEs in cetartiodactyl genomes: Possible evidence for the monophyletic origin of toothed whales. Mamm. Genome 2001, 12, 909–915. [Google Scholar] [CrossRef]
- Miller, P.J.O.; Roos, M.M.H. Breathing. In Encyclopedia of Marine Mammals; Würsig, B., Thewissen, J.G.M., Kovacs, K.M., Eds.; Academic Press: San Diego, CA, USA, 2018; pp. 140–143. [Google Scholar] [CrossRef]
- Chu, K.C. Dive times and ventilation patterns of singing humpback whales (Megaptera novaeangliae). Can. J. Zool. 1988, 66, 1322–1327. [Google Scholar] [CrossRef]
- Hedley, S.L.; Bannister, J.L.; Dunlop, R.A. Abundance estimates of Southern Hemisphere Breeding Stock ‘D’ humpback whales from aerial and land-based surveys off Shark Bay, Western Australia, 2008. J. Cetacean Res. Manag. 2011, 3, 209–221. [Google Scholar] [CrossRef]
- Godfrey, S.J.; Geisler, J.; Fitzgerald, E.M.G. On the olfactory anatomy in an archaic whale (Protocetidae, Cetacea) and the minke whale Balaenoptera acutorostrata (Balaenopteridae, Cetacea). Anat. Rec. 2013, 296, 257–272. [Google Scholar] [CrossRef]
- Ichishima, H. The ethmoid and presphenoid of cetaceans. J. Morphol. 2016, 277, 1661–1674. [Google Scholar] [CrossRef] [PubMed]
- Hirose, A.; Kishida, T.; Nakamura, G. Nasal mucosa resembling an olfactory system in the common minke whale (Balaenoptera acutorostrata). Cetacean Popul. Stud. 2018, 1, 25–28. [Google Scholar] [CrossRef]
- Thewissen, J.G.M.; George, J.; Rosa, C.; Kishida, T. Olfaction and brain size in the bowhead whale (Balaena mysticetus). Mar. Mammal Sci. 2011, 27, 282–294. [Google Scholar] [CrossRef]
- Kishida, T.; Thewissen, J.; Usip, S.; Suydam, R.S.; George, J.C. Organization and distribution of glomeruli in the bowhead whale olfactory bulb. Peerj 2015, 3, e897. [Google Scholar] [CrossRef] [PubMed]
- Farnkopf, I.C.; George, J.C.; Kishida, T.; Hillmann, D.J.; Suydam, R.S.; Thewissen, J.G.M. Olfactory epithelium and ontogeny of the nasal chambers in the bowhead whale (Balaena mysticetus). Anat. Rec. 2022, 305, 643–667. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Wu, Y.; Zeng, L.; Zhao, S. Building the Chordata Olfactory Receptor Database using more than 400,000 receptors annotated by Genome2OR. Sci. China Life Sci. 2022, 65, 2539–2551. [Google Scholar] [CrossRef] [PubMed]
- Kishida, T. Olfaction of aquatic amniotes. Cell Tissue Res. 2021, 383, 353–365. [Google Scholar] [CrossRef]
- Kishida, T.; Thewissen, J.; Hayakawa, T.; Imai, H.; Agata, K. Aquatic adaptation and the evolution of smell and taste in whales. Zool. Lett. 2015, 1, 9. [Google Scholar] [CrossRef]
- Christmas, M.J.; Kaplow, I.M.; Genereux, D.P.; Dong, M.X.; Hughes, G.M.; Li, X.; Sullivan, P.F.; Hindle, A.G.; Andrews, G.; Armstrong, J.C.; et al. Evolutionary constraint and innovation across hundreds of placental mammals. Science 2023, 380, eabn3943. [Google Scholar] [CrossRef]
- Danciger, E.; Mettling, C.; Vidal, M.; Morris, R.; Margolis, F. Olfactory marker protein gene: Its structure and olfactory neuron-specific expression in transgenic mice. Proc. Natl. Acad. Sci. USA 1989, 86, 8565–8569. [Google Scholar] [CrossRef] [PubMed]
- Buiakova, O.I.; Baker, H.; Scott, J.W.; Farbman, A.; Kream, R.; Grillo, M.; Franzen, L.; Richman, M.; Davis, L.M.; Abbondanzo, S.; et al. Olfactory marker protein (OMP) gene deletion causes altered physiological activity of olfactory sensory neurons. Proc. Natl. Acad. Sci. USA 1996, 93, 9858–9863. [Google Scholar] [CrossRef] [PubMed]
- Kishida, T.; Thewissen, J.G.M. Evolutionary changes of the importance of olfaction in cetaceans based on the olfactory marker protein gene. Gene 2012, 492, 349–353. [Google Scholar] [CrossRef] [PubMed]
- Springer, M.; Gatesy, J. Inactivation of the olfactory marker protein (OMP) gene in river dolphins and other odontocete cetaceans. Mol. Phylogenet. Evol. 2017, 109, 375–387. [Google Scholar] [CrossRef] [PubMed]
- Cranford, T.W.; Amundin, M.; Norris, K.S. Functional morphology and homology in the odontocete nasal complex: Implications for sound generation. J. Morphol. 1996, 228, 223–285. [Google Scholar] [CrossRef]
- Berta, A.; Ekdale, E.G.; Cranford, T.W. Review of the cetacean nose: Form, function, and evolution. Anat. Rec. 2014, 297, 2205–2215. [Google Scholar] [CrossRef] [PubMed]
- Hirose, A.; Kodera, R.; Uekusa, Y.; Katsumata, H.; Katsumata, E.; Nakamura, G.; Kato, H. Comparative anatomy of and around the posterior nasofrontal sac of a beluga whale. Mar. Mammal Sci. 2022, 38, 1272–1285. [Google Scholar] [CrossRef]
- Bouchard, B.; Barnagaud, J.-Y.; Poupard, M.; Glotin, H.; Gauffier, P.; Ortiz, S.T.; Lisney, T.J.; Campagna, S.; Rasmussen, M.; Célérier, A. Behavioural responses of humpback whales to food-related chemical stimuli. PLoS ONE 2019, 14, e0212515. [Google Scholar] [CrossRef]
- Bouchard, B.; Barnagaud, J.; Verborgh, P.; Gauffier, P.; Campagna, S.; Célérier, A. A field study of chemical senses in bottlenose dolphins and pilot whales. Anat. Rec. 2022, 305, 668–679. [Google Scholar] [CrossRef]
- Go, Y.; Niimura, Y. Similar numbers but different repertoires of olfactory receptor genes in humans and chimpanzees. Mol. Biol. Evol. 2008, 25, 1897–1907. [Google Scholar] [CrossRef] [PubMed]
- Kishida, T.; Go, Y.; Tatsumoto, S.; Tatsumi, K.; Kuraku, S.; Toda, M. Loss of olfaction in sea snakes provides new perspectives on the aquatic adaptation of amniotes. Proc. R. Soc. B 2019, 286, 20191828. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhao, H.; Fu, N.; Chen, L. The diversified function and potential therapy of ectopic olfactory receptors in non-olfactory tissues. J. Cell. Physiol. 2018, 233, 2104–2115. [Google Scholar] [CrossRef] [PubMed]
- Parmentier, M.; Libert, F.; Schurmans, S.; Schiffmann, S.; Lefort, A.; Eggerickx, D.; Ledent, C.; Mollereau, C.; Gérard, C.; Perret, J.; et al. Expression of members of the putative olfactory receptor gene family in mammalian germ cells. Nature 1992, 355, 453–455. [Google Scholar] [CrossRef] [PubMed]
- Spehr, M.; Gisselmann, G.; Poplawski, A.; Riffell, J.A.; Wetzel, C.H.; Zimmer, R.K.; Hatt, H. Identification of a testicular odorant receptor mediating human sperm chemotaxis. Science 2003, 299, 2054–2058. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, N.; Yomogida, K.; Okabe, M.; Touhara, K. Functional characterization of a mouse testicular olfactory receptor and its role in chemosensing and in regulation of sperm motility. J. Cell Sci. 2004, 117, 5835–5845. [Google Scholar] [CrossRef] [PubMed]
- Rouquier, S.; Giorgi, D. Olfactory receptor gene repertoires in mammals. Mutat. Res./Fundam. Mol. Mech. Mutagen. 2007, 616, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Neuhaus, E.M.; Zhang, W.; Gelis, L.; Deng, Y.; Noldus, J.; Hatt, H. Activation of an olfactory receptor inhibits proliferation of prostate cancer cells. J. Biol. Chem. 2009, 284, 16218–16225. [Google Scholar] [CrossRef] [PubMed]
- Van Valkenburgh, B.; Pang, B.; Bird, D.; Curtis, A.; Yee, K.; Wysocki, C.; Craven, B.A. Respiratory and olfactory turbinals in feliform and caniform carnivorans: The influence of snout length. Anat. Rec. 2014, 297, 2065–2079. [Google Scholar] [CrossRef]
- Ressler, K.J.; Sullivan, S.L.; Buck, L.B. A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell 1993, 73, 597–609. [Google Scholar] [CrossRef]
- Vassar, R.; Ngai, J.; Axel, R. Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell 1993, 74, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Marchand, J.E.; Yang, X.; Chikaraishi, D.; Krieger, J.; Breer, H.; Kauer, J.S. Olfactory receptor gene expression in tiger salamander olfactory epithelium. J. Comp. Neurol. 2004, 474, 453–467. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.D.; Eiting, T.P.; Bhatnagar, K.P. A Quantitative study of olfactory, non-olfactory, and vomeronasal epithelia in the nasal fossa of the bat Megaderma lyra. J. Mamm. Evol. 2012, 19, 27–41. [Google Scholar] [CrossRef]
- Ruf, I. Comparative anatomy and systematic implications of the turbinal skeleton in Lagomorpha (Mammalia). Anat. Rec. 2014, 297, 2031–2046. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.D.; Eiting, T.P.; Bonar, C.J.; Craven, B.A. Nasal morphometry in marmosets: Loss and redistribution of olfactory surface area. Anat. Rec. 2014, 297, 2093–2104. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Tu, V.T.; Eiting, T.P.; Nojiri, T.; Koyabu, D. On the embryonic development of the nasal turbinals and their homology in bats. Front. Cell Dev. Biol. 2021, 9, 613545. [Google Scholar] [CrossRef] [PubMed]
- Van Valkenburgh, B.; Curtis, A.; Samuels, J.X.; Bird, D.; Fulkerson, B.; Meachen-Samuels, J.; Slater, G.J. Aquatic adaptations in the nose of carnivorans: Evidence from the turbinates. J. Anat. 2011, 218, 298–310. [Google Scholar] [CrossRef]
- Martinez, Q.; Clavel, J.; Esselstyn, J.A.; Achmadi, A.S.; Grohé, C.; Pirot, N.; Fabre, P.-H. Convergent evolution of olfactory and thermoregulatory capacities in small amphibious mammals. Proc. Natl. Acad. Sci. USA 2020, 117, 8958–8965. [Google Scholar] [CrossRef] [PubMed]
- Van Valkenburgh, B.; Smith, T.D.; Craven, B.A. Tour of a labyrinth: Exploring the vertebrate nose. Anat. Rec. 2014, 297, 1975–1984. [Google Scholar] [CrossRef] [PubMed]
- Iwata, T.; Niimura, Y.; Kobayashi, C.; Shirakawa, D.; Suzuki, H.; Enomoto, T.; Touhara, K.; Yoshihara, Y.; Hirota, J. A long-range cis-regulatory element for class I odorant receptor genes. Nat. Commun. 2017, 8, 885. [Google Scholar] [CrossRef]
- Kobayakawa, K.; Kobayakawa, R.; Matsumoto, H.; Oka, Y.; Imai, T.; Ikawa, M.; Okabe, M.; Ikeda, T.; Itohara, S.; Kikusui, T.; et al. Innate versus learned odour processing in the mouse olfactory bulb. Nature 2007, 450, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Barboza, M.L.B.; Larkin, I.V. Gross and microscopic anatomy of the nasal cavity, including olfactory epithelium, of the Florida manatee (Trichechus manatus latirostris). Aquat. Mamm. 2020, 46, 274–284. [Google Scholar] [CrossRef]
- Policarpo, M.; Baldwin, M.W.; Casane, D.; Salzburger, W. Diversity and evolution of the vertebrate chemoreceptor gene repertoire. Nat. Commun. 2024, 15, 1421. [Google Scholar] [CrossRef]
- Tyack, P.L.; Miller, E.H. Vocal anatomy, acoustic communication and echolocation. In Marine Mammal Biology: An Evolutionary Approach; Blackwell Science: Hoboken, NJ, USA, 2002; pp. 142–184. [Google Scholar]
- Feng, P.; Zheng, J.; Rossiter, S.J.; Wang, D.; Zhao, H. Massive Losses of Taste Receptor Genes in Toothed and Baleen Whales. Genome Biol. Evol. 2014, 6, 1254–1265. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.; Zhou, X.; Xu, S.; Sun, D.; Ren, W.; Zhou, K.; Yang, G. The loss of taste genes in cetaceans. BMC Evol. Biol. 2014, 14, 218. [Google Scholar] [CrossRef] [PubMed]
- Sonntag, C.F. The Comparative Anatomy of the Tongues of the Mammalia.—VII. Cetaeea, Sirenia, and Ungulata. Proc. Zool. Soc. Lond. 1922, 92, 639–657. [Google Scholar] [CrossRef]
- Ogawa, T.; Shida, T. On the sensory tubercles of lips and oral cavity in the sei and fin whale. Sci. Rep. Whales Res. Inst. 1950, 3, 1–16. [Google Scholar]
- Tarpley, R.J. Gross and Microscopic Anatomy of the Tongue and Gastrointestinal Tract of the Bowhead Whale (Balaena mysticetus). Ph.D. Thesis, Texas A&M University, College Station, TX, USA, December 1985. [Google Scholar]
- Schulte, H.v.W. Anatomy of a foetus of Balaenoptara borealis. Monogr. Pac. Cetacea 1916, 1, 389–502. [Google Scholar]
- Nevitt, G.A. Olfactory foraging by Antarctic procellariiform seabirds: Life at high Reynolds numbers. Biol. Bull. 2000, 198, 245–253. [Google Scholar] [CrossRef]
- Dell’Ariccia, G.; Celerier, A.; Gabirot, M.; Palmas, P.; Massa, B.; Bonadonna, F. Olfactory foraging in temperate waters: Sensitivity to dimethylsulphide of shearwaters in the Atlantic Ocean and Mediterranean Sea. J. Exp. Biol. 2014, 217, 1701–1709. [Google Scholar] [CrossRef]
- Owen, K.; Saeki, K.; Warren, J.D.; Bocconcelli, A.; Wiley, D.N.; Ohira, S.-I.; Bombosch, A.; Toda, K.; Zitterbart, D.P. Natural dimethyl sulfide gradients would lead marine predators to higher prey biomass. Commun. Biol. 2021, 4, 149. [Google Scholar] [CrossRef]
- Savoca, M.S.; Wohlfeil, M.E.; Ebeler, S.E.; Nevitt, G.A. Marine plastic debris emits a keystone infochemical for olfactory foraging seabirds. Sci. Adv. 2016, 2, e1600395. [Google Scholar] [CrossRef]
- Klima, M. Development of the Cetacean Nasal Skull; Springer: Berlin/Heidelberg, Germany, 1999. [Google Scholar] [CrossRef]
- Ito, K.; Kodeara, R.; Koyasu, K.; Martinez, Q.; Koyabu, D. The development of nasal turbinal morphology of moles and shrews. Vertebr. Zool. 2022, 72, 857–881. [Google Scholar] [CrossRef]
- Maier, W.; Ruf, I. Morphology of the nasal capsule of primates—With special reference to Daubentonia and Homo. Anat. Rec. 2014, 297, 1985–2006. [Google Scholar] [CrossRef] [PubMed]
- Yim, H.-S.; Cho, Y.S.; Guang, X.; Kang, S.G.; Jeong, J.-Y.; Cha, S.-S.; Oh, H.-M.; Lee, J.-H.; Yang, E.C.; Kwon, K.K.; et al. Minke whale genome and aquatic adaptation in cetaceans. Nat. Genet. 2014, 46, 88–92. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Niimura, Y.; Matsui, A.; Touhara, K. Extreme expansion of the olfactory receptor gene repertoire in African elephants and evolutionary dynamics of orthologous gene groups in 13 placental mammals. Genome Res. 2014, 24, 1485–1496. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Roberts, A.; Trapnell, C.; Donaghey, J.; Rinn, J.L.; Pachter, L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011, 12, R22. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; 182p. [Google Scholar]
- Kuraku, S.; Zmasek, C.M.; Nishimura, O.; Katoh, K. aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 2013, 41, W22–W28. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. 2019, 37, 291–294. [Google Scholar] [CrossRef] [PubMed]
- Flouri, T.; Izquierdo-Carrasco, F.; Darriba, D.; Aberer, A.; Nguyen, L.-T.; Minh, B.; Von Haeseler, A.; Stamatakis, A. The Phylogenetic Likelihood Library. Syst. Biol. 2014, 64, 356–362. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
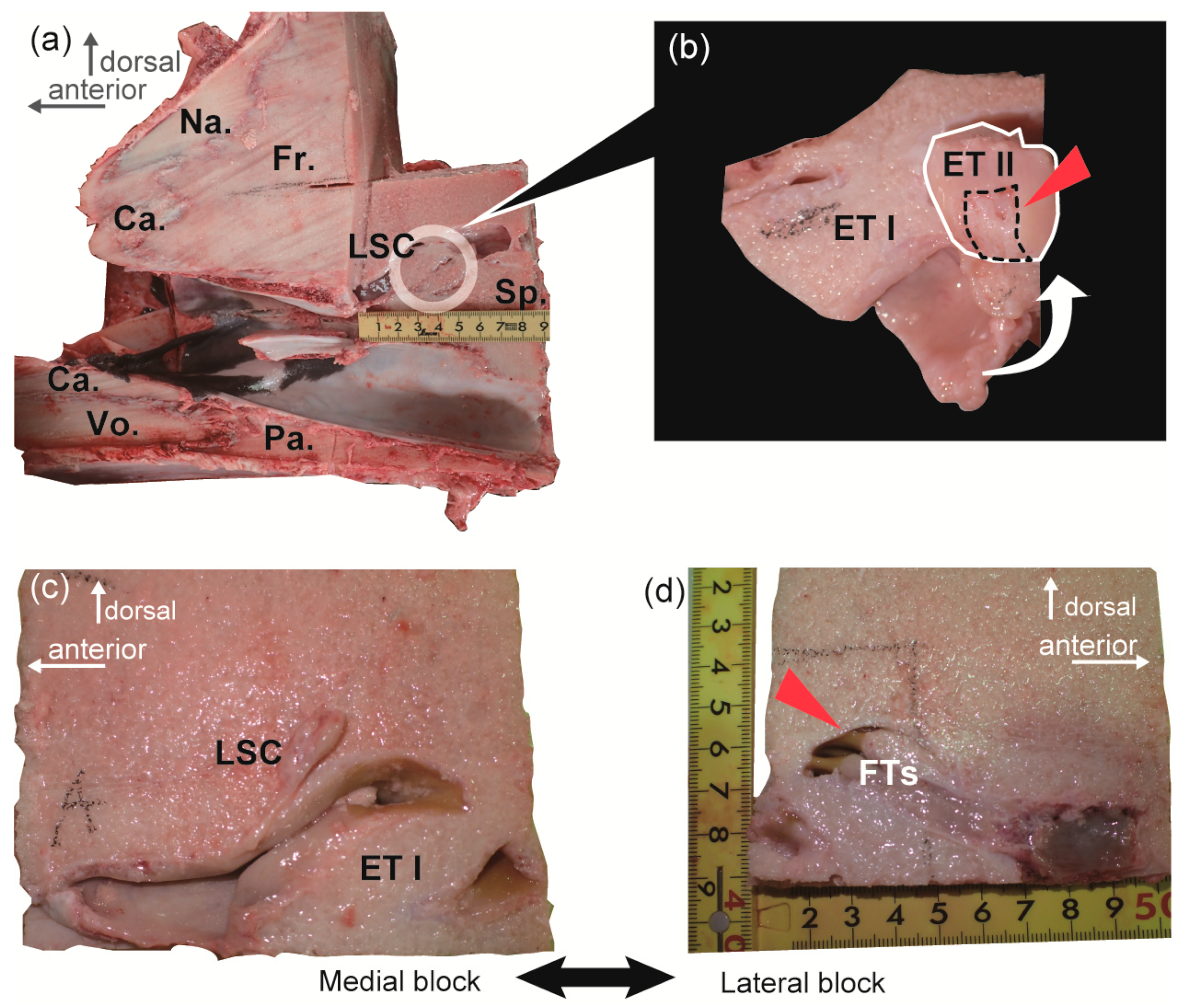
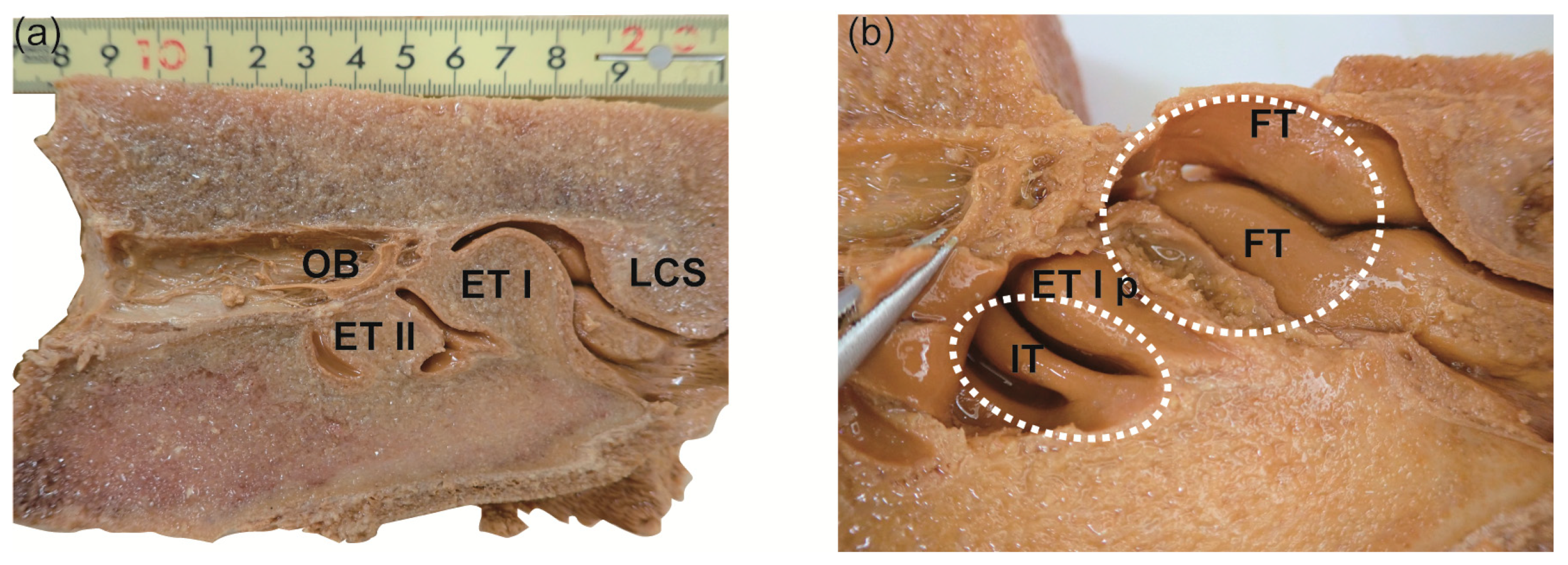


| Location | Year | ID | Sex | Body Length (m) | PMI † (h) | Description of Specimen | Side | Techniques Used | Sample ID |
|---|---|---|---|---|---|---|---|---|---|
| Kushiro, Hokkaido | 2016 | 16NPCK-M009 | M | 7.01 | 4.5 | Nasal mucosa | L | Histology | H-009 |
| Kushiro, Hokkaido | 2018 | 18NPCK-M001 | M | 5.50 | 6 | Nasal mucosa (Ethmoturbinal II) | L | RNA-seq Histology | R-001 H-001 |
| 18NPCK-M006 | M | 7.09 | 6 | Nasal mucosa (Frontoturbinal) | L | RNA-seq | R-006 | ||
| 18NPCK-M008 | F | 4.62 | 6 | Olfactory bulb | R | RNA-seq | R-008 | ||
| Abashiri, Hokkaido | 2018 | 18NPCO-M046 | F | 7.30 | 5 | Nasal mucosa (Anterior portion) | R | RNA-seq Histology | R-046 H-046 |
| Kushiro, Hokkaido | 2019 | 19SK214 | M | 6.96 | 8.5 | External skin | - | RNA-seq | R-214 |
| 19SK215 | M | 7.33 | 5.5 | External skin | - | RNA-seq | R-215 |
| R-001 | R-006 | R-046 | R-008 | R-214 | R-215 | |
|---|---|---|---|---|---|---|
| β actin (FPKM) | 983 | 285 | 549 | 371 | 161 | 143 |
| OMP | ||||||
| FPKM | 9.510 | 21.101 | 0 | 0 | 0 | 0 |
| Expression † | 0.966 | 7.378 | 0 | 0 | 0 | 0 |
| Average of expressing intact ORs | ||||||
| FPKM | 3.640 ± 4.946 | 6.537 ± 8.735 | 0.244 ± 0.372 | 0.543 ± 0.876 | 0.171 ± 0.098 | 0.112 ± 0.054 |
| Expression † | 0.370 ± 0.503 | 2.286 ± 3.054 | 0.045 ± 0.068 | 0.146 ± 0.236 | 0.106 ± 0.061 | 0.078 ± 0.038 |
| Description of Specimen | Sample ID | OR Expression | OMP Expression | Pseudostratified Columnar Epithelium | Bowman’s Glands | Absence of Goblet Cells | Nucleus Free Zone | Peripheral Nerve |
|---|---|---|---|---|---|---|---|---|
| Ethmoturbinal II | H-001 R-001 | low | low | Y | Y | Y | Y | N |
| Frontoturbinal | H-009 R-006 | Y | Y | Y | Y | Y | Y | Y |
| Anterior portion | H-046 R-046 | N | N | Y | Y | Y | Y | N |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hirose, A.; Nakamura, G.; Nikaido, M.; Fujise, Y.; Kato, H.; Kishida, T. Localized Expression of Olfactory Receptor Genes in the Olfactory Organ of Common Minke Whales. Int. J. Mol. Sci. 2024, 25, 3855. https://doi.org/10.3390/ijms25073855
Hirose A, Nakamura G, Nikaido M, Fujise Y, Kato H, Kishida T. Localized Expression of Olfactory Receptor Genes in the Olfactory Organ of Common Minke Whales. International Journal of Molecular Sciences. 2024; 25(7):3855. https://doi.org/10.3390/ijms25073855
Chicago/Turabian StyleHirose, Ayumi, Gen Nakamura, Masato Nikaido, Yoshihiro Fujise, Hidehiro Kato, and Takushi Kishida. 2024. "Localized Expression of Olfactory Receptor Genes in the Olfactory Organ of Common Minke Whales" International Journal of Molecular Sciences 25, no. 7: 3855. https://doi.org/10.3390/ijms25073855