1-L Transcription of SARS-CoV-2 Spike Protein S1 Subunit
Abstract
:1. Introduction

2. Results
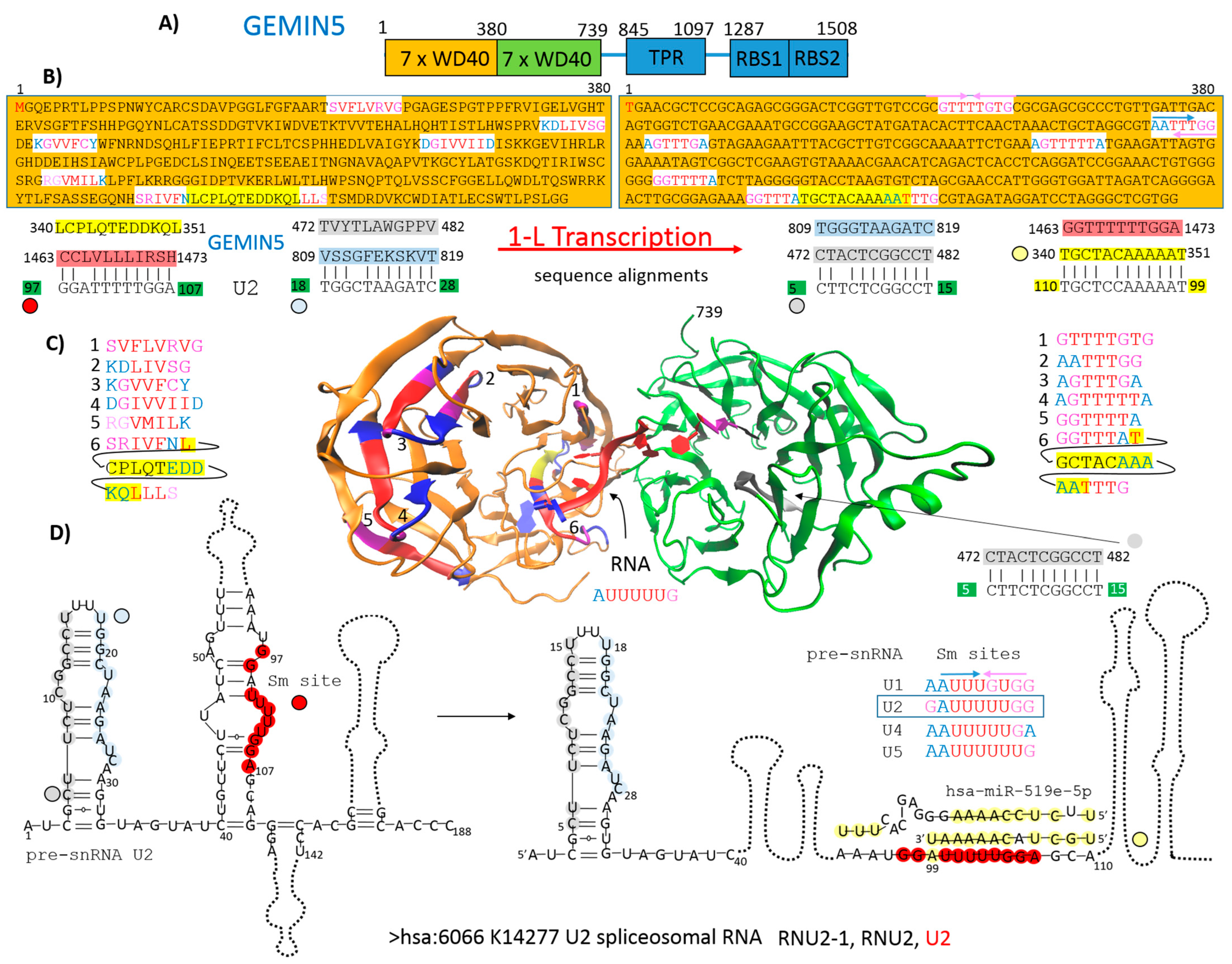
2.1. GEMIN5 as an Example of How Protein Primary Structure Is Involved in Protein–RNA Recognition
2.2. Genes/Proteins Identified by 1-L Transcription of N-(AA)n-C Sequences
2.3. Genes/Proteins Identified by 1-L Transcription of C-(AA)n-N Sequences
3. Discussion
3.1. GEMIN5 as an Example of How Protein Primary Structure Is Involved in Protein–RNA Recognition
3.2. 1-L Transcription of the SARS-CoV-2 Spike Protein S1 Subunit and Genes/Proteins Identified as Being Relevant to COVID-19
3.2.1. Genes/Proteins Known to Be Related to COVID-19
3.2.2. Genes/Proteins Indirectly Related to COVID-19
3.2.3. Unknown Genes Related to COVID-19
3.3. Immune Responses and Inflammation
3.4. T2D and Cardiac Stress
3.5. Cilia and Lung Injury
4. Materials and Methods
4.1. 1-L Transcription Procedure
4.2. BLASTn Screening Process
4.3. GEMIN5 as an Example
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Callaway, E. The next generation of Coronavirus vaccines. Nature 2023, 614, 22–26. [Google Scholar] [CrossRef]
- Ni, T.; Mendonça, L.; Zhu, Y.; Howe, A.; Radecke, J.; Shah, P.M.; Sheng, Y.; Krebs, A.S.; Duyvesteyn, H.M.E.; Allen, E.; et al. ChAdOx1 COVID vaccines express RBD open prefusion SARS-CoV-2 spikes on the cell surface. iScience 2023, 26, 107882. [Google Scholar] [CrossRef]
- Montgomerie, I.; Bird, T.W.; Palmer, O.R.; Mason, N.C.; Pankhurst, T.E.; Lawley, B.; Hernández, L.C.; Harfoot, R.; Authier-Hall, A.; Anderson, D.E.; et al. Incorporation of SARS-CoV-2 spike NTD to RBD protein vaccine improves immunity against viral variants. iScience 2023, 26, 106256. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.J.; Cines, D.B.; Gernsheimer, T.; Kessler, C.; Michel, M.; Tarantino, M.D.; Semple, J.W.; Arnold, D.M.; Godeau, B.; Lambert, M.P.; et al. Thrombocytopenia following Pfizer and Moderna SARS-CoV-2 vaccination. Am. J. Hematol. 2021, 96, 534–537. [Google Scholar] [CrossRef]
- Simpson, C.R.; Shi, T.; Vasileiou, E.; Katikireddi, S.V.; Kerr, S.; Moore, E.; McCowan, C.; Agrawal, U.; Shah, S.A.; Ritchie, L.D.; et al. First-dose ChAdOx1 and BNT162b2 COVID-19 vaccines and thrombocytopenic, thromboembolic and hemorrhagic events in Scotland. Nat. Med. 2021, 27, 1290–1297. [Google Scholar] [CrossRef]
- Chary, M.; Barbuto, A.F.; Izadmehr, S.; Tarsillo, M.; Fleischer, E.; Burns, M.M. Therapeutics: Use, mechanism of action, and toxicity (vaccines, monoclonal antibodies, and immunotherapeutics). J. Med. Toxicol. 2023, 19, 205–218. [Google Scholar] [CrossRef]
- Paknahad, M.H.; Yancheshmeh, F.B.; Soleimani, A. Cardiovascular complications of COVID-19 vaccines: A review of case-report and case-series studies. Heart Lung 2023, 59, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Chakravarty, A. Neurological complications following COVID-19 vaccination. Curr. Neurol. Neurosci. Rep. 2023, 23, 1–14. [Google Scholar] [CrossRef]
- Parry, P.I.; Lefringhausen, A.; Turni, C.; Neil, C.J.; Cosford, R.; Hudson, N.J.; Gillespie, J. ‘Spikeopathy’: COVID-19 spike protein is pathogenic, from both virus and vaccine mRNA. Biomedicines 2023, 11, 2287. [Google Scholar] [CrossRef]
- Nahalka, J. Theoretical analysis of S, M and N structural proteins by the protein–RNA recognition code leads to genes/proteins that are relevant to the SARS-CoV-2 life cycle and pathogenesis. Front. Genet. 2021, 12, 763995. [Google Scholar] [CrossRef]
- Su, J.; Zheng, J.; Huang, W.; Zhang, Y.; Lv, C.; Zhang, B.; Jiang, L.; Cheng, T.; Yuan, Q.; Xia, N.; et al. PIKfyve inhibitors against SARS-CoV-2 and its variants including Omicron. Signal Transduct. Target. Ther. 2022, 7, 167. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, C.; Yang, B.; Zhang, H.; Jiao, J.; Zhang, R.; Liu, S.; Xiao, S.; Chen, Y.; Liu, B.; et al. SARS-CoV-2 ORF10 impairs cilia by enhancing CUL2ZYG11B activity. J. Cell Biol. 2022, 221, e202108015. [Google Scholar] [CrossRef] [PubMed]
- Nahalka, J. Transcription of the envelope protein by 1-L protein–RNA recognition code leads to genes/proteins that are relevant to the SARS-CoV-2 life cycle and pathogenesis. Curr. Issues Mol. Biol. 2022, 44, 791–816. [Google Scholar] [CrossRef] [PubMed]
- Nahalka, J. 1-L Transcription in Alzheimer’s Disease. Curr. Issues Mol. Biol. 2022, 44, 3533–3551. [Google Scholar] [CrossRef] [PubMed]
- Nahalka, J. 1-L Transcription in Parkinson’s Disease. Front. Biosci. (Landmark Ed.) 2023, 28, 292. [Google Scholar] [CrossRef] [PubMed]
- Nahalka, J. The role of the protein–RNA recognition code in neurodegeneration. Cell. Mol. Life Sci. 2019, 76, 2043–2058. [Google Scholar] [CrossRef] [PubMed]
- Oronsky, B.; Larson, C.; Hammond, T.C.; Oronsky, A.; Kesari, S.; Lybeck, M.; Reid, T.R. A review of persistent post-COVID syndrome (PPCS). Clin. Rev. Allergy Immunol. 2023, 64, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Pánek, J.; Roithová, A.; Radivojević, N.; Sýkora, M.; Prusty, A.B.; Huston, N.; Wan, H.; Pyle, A.M.; Fischer, U.; Staněk, D. The SMN complex drives structural changes in human snRNAs to enable snRNP assembly. Nat. Commun. 2023, 14, 6580. [Google Scholar] [CrossRef] [PubMed]
- Faravelli, I.; Riboldi, G.M.; Rinchetti, P.; Lotti, F. The SMN complex at the crossroad between RNA metabolism and neurodegeneration. Int. J. Mol. Sci. 2023, 24, 2247. [Google Scholar] [CrossRef]
- Xu, C.; Ishikawa, H.; Izumikawa, K.; Li, L.; He, H.; Nobe, Y.; Yamauchi, Y.; Shahjee, H.M.; Xian-Hui, W.; Yi-Tao, Y.; et al. Structural insights into Gemin5-guided selection of pre-snRNAs for snRNP assembly. Genes Dev. 2016, 30, 2376–2390. [Google Scholar] [CrossRef]
- Guo, Q.; Zhao, S.; Francisco-Velilla, R.; Zhang, J.; Embarc-Buh, A.; Abellan, S.; Lv, M.; Tang, P.; Gong, Q.; Shen, H.; et al. Structural basis for Gemin5 decamer-mediated mRNA binding. Nat. Commun. 2022, 13, 5166. [Google Scholar] [CrossRef] [PubMed]
- Lechtreck, K. Cargo adapters expand the transport range of intraflagellar transport. J. Cell Sci. 2022, 135, jcs260408. [Google Scholar] [CrossRef] [PubMed]
- Bearce, E.A.; Irons, Z.H.; Craig, S.B.; Kuhns, C.J.; Sabazali, C.; Farnsworth, D.R.; Miller, A.C.; Grimes, D.T. Daw1 regulates the timely onset of cilia motility during development. Development 2022, 149, dev200017. [Google Scholar] [CrossRef] [PubMed]
- Harrision, D.; Gravells, P.; Thompson, R.; Bryant, H.E. Glycohydrolase (PARG) vs. Poly(ADP-ribose) polymerase (PARP)—Function in genome maintenance and relevance of inhibitors for anti-cancer therapy. Front. Mol. Biosci. 2020, 7, 191. [Google Scholar] [CrossRef]
- Alhammad, Y.M.O.; Kashipathy, M.M.; Roy, A.; Gagné, J.-P.; McDonald, P.; Gao, P.; Nonfoux, L.; Battaile, K.P.; Johnson, D.K.; Holmstrom, E.D.; et al. The SARS-CoV-2 conserved macrodomain is a mono-ADP-ribosylhydrolase. J. Virol. 2021, 95, e01969-20. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, J.J.; Ferraris, D.; Fehr, A.R. An update on the current state of SARS-CoV-2 Mac1 inhibitors. Pathogens 2023, 12, 1221. [Google Scholar] [CrossRef]
- Alhammad, Y.M.; Parthasarathy, S.; Ghimire, R.; Kerr, C.M.; O’Connor, J.J.; Pfannenstiel, J.J.; Chanda, D.; Miller, C.A.; Baumlin, N.; Salathe, M.; et al. SARS-CoV-2 Mac1 is required for IFN antagonism and efficient virus replication in cell culture and in mice. Proc. Natl. Acad. Sci. USA 2023, 120, e2302083120. [Google Scholar] [CrossRef] [PubMed]
- Kouhpayeh, S.; Shariati, L.; Boshtam, M.; Rahimmanesh, I.; Mirian, M.; Esmaeili, Y.; Najaflu, M.; Khanahmad, N.; Zeinalian, M.; Trovato, M.; et al. The molecular basis of COVID-19 pathogenesis, conventional and nanomedicine therapy. Int. J. Mol. Sci. 2021, 22, 5438. [Google Scholar] [CrossRef] [PubMed]
- Quistgaard, E.M. BAP31: Physiological functions and roles in disease. Biochimie 2021, 186, 105–129. [Google Scholar] [CrossRef]
- Ran, S.; Su, K.; Zhang, S.; Liu, B. The association between Coronavirus disease 2019 infection and blood constituents: A mendelia constituents: A mendelian randomization analysis. J. Infect. Dis. 2021, 224, 922–924. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, Y.; Yan, X.; Ye, C.; Weirich, S.; Zhang, B.; Wang, X.; Song, L.; Jiang, C.; Jeltsch, A.; et al. CRL2ZER1/ZYG11B recognizes small N-terminal residues for degradation. Nat. Commun. 2022, 13, 7636. [Google Scholar] [CrossRef]
- Fonseca, B.F.; Chakrabarti, L.A. A close shave: How SARS-CoV-2 induces the loss of cilia. J. Cell Biol. 2022, 221, e202206023. [Google Scholar] [CrossRef]
- Ng, V.H.; Spencer, Z.; Neitzel, L.R.; Nayak, A.; Loberg, M.A.; Shen, C.; Kassel, S.N.; Kroh, H.K.; An, Z.; Anthony, C.C.; et al. The USP46 complex deubiquitylates LRP6 to promote Wnt/β-catenin signaling. Nat. Commun. 2023, 14, 6173. [Google Scholar] [CrossRef]
- Koval, A.; Xu, J.; Williams, N.; Schmolke, M.; Krause, K.-H.; Katanaev, V.L. Wnt-independent SARS-CoV-2 infection in pulmonary epithelial cells. Microbiol. Spectr. 2023, 11, e04827-22. [Google Scholar] [CrossRef]
- Li, J.; Ren, J.X.; Liao, H.P.; Guo, W.; Feng, K.Y.; Huang, T.; Cai, Y.D. Identification of dynamic gene expression profiles during sequential vaccination with ChAdOx1/BNT162b2 using machine learning methods. Front. Microbiol. 2023, 14, 1138674. [Google Scholar] [CrossRef]
- Chen, J.Y.; Tan, X.; Wang, Z.H.; Liu, Y.Z.; Zhou, J.F.; Rong, X.Z.; Lu, L.; Li, Y. The ribosome biogenesis protein Esf1 is essential for pharyngeal cartilage formation in zebrafish. FEBS J. 2018, 285, 3464–3484. [Google Scholar] [CrossRef]
- Pan, X.; Chen, S.; Chen, X.; Ren, Q.; Yue, L.; Niu, S.; Li, Z.; Zhu, R.; Chen, X.; Jia, Z.; et al. UTP14A, DKC1, DDX10, PinX1, and ESF1 modulate cardiac angiogenesis leading to obesity-induced cardiac injury. J. Diabetes Res. 2022, 2022, 2923291. [Google Scholar] [CrossRef]
- de Rooij, L.P.M.H.; Becker, L.M.; Teuwen, L.A.; Boeckx, B.; Jansen, S.; Feys, S.; Verleden, S.; Liesenborghs, L.; Stalder, A.K.; Libbrecht, S.; et al. The pulmonary vasculature in lethal COVID-19 and idiopathic pulmonary fibrosis at single-cell resolution. Cardiovasc. Res. 2023, 119, 520–535. [Google Scholar] [CrossRef]
- Xin, Z.; Li, Y.; Meng, L.; Dong, L.; Ren, J.; Men, J. Elevated expression of the MYB proto-oncogene like 2 (MYBL2)-encoding gene as a prognostic and predictive biomarker in human cancers. Math. Biosci. Eng. 2022, 19, 1825–1842. [Google Scholar] [CrossRef]
- Wu, H.; Han, F. Investigation of shared genes and regulatory mechanisms associated with coronavirus disease 2019 and ischemic stroke. Front. Neurol. 2023, 14, 1151946. [Google Scholar] [CrossRef]
- Zhu, J.; Li, X.; Lv, F.; Zhou, W. Bioinformatics approach to identify the influences of COVID-19 on ischemic stroke. Biochem. Genet. 2023, 61, 2222–2241. [Google Scholar] [CrossRef]
- Wang, Q.S.; Edahiro, R.; Namkoong, H.; Hasegawa, T.; Shirai, Y.; Sonehara, K.; Tanaka, H.; Lee, H.; Saiki, R.; Hyugaji, T.; et al. The whole blood transcriptional regulation landscape in 465 COVID-19 infected samples from Japan COVID-19 Task Force. Nat. Commun. 2022, 13, 4830. [Google Scholar] [CrossRef]
- Castro de Moura, M.; Davalos, V.; Planas-Serra, L.; Alvarez-Errico, D.; Arribas, C.; Ruiz, M.; Aguilera-Albesa, S.; Troya, J.; Valencia-Ramos, J.; Vélez-Santamaria, V.; et al. Epigenome-wide association study of COVID-19 severity with respiratory failure. EBioMedicine 2021, 66, 103339. [Google Scholar] [CrossRef]
- Sloan, D.C.; Cryan, C.E.; Muntean, B.S. Multiple potassium channel tetramerization domain (KCTD) family members interact with Gβγ, with effects on cAMP signaling. J. Biol. Chem. 2023, 299, 102924. [Google Scholar] [CrossRef]
- Vatner, S.F.; Park, M.; Yan, L.; Lee, G.J.; Lai, L.; Iwatsubo, K.; Ishikawa, Y.; Pessin, J.; Vatner, D.E. Adenylyl cyclase type 5 in cardiac disease, metabolism, and aging. Am. J. Physiol. Heart Circ. Physiol. 2013, 305, H1–H8. [Google Scholar] [CrossRef]
- Kim, E.J.; Kim, S.H.; Jin, X.; Jin, X.; Kim, H. KCTD2, an adaptor of Cullin3 E3 ubiquitin ligase, suppresses gliomagenesis by destabilizing c-Myc. Cell Death Differ. 2017, 24, 649–659. [Google Scholar] [CrossRef]
- Millarte, V.; Spiess, M. RABEP1/Rabaptin5: A link between autophagy and early endosome homeostasis. Autophagy 2022, 18, 698–699. [Google Scholar] [CrossRef]
- Atik, N.; Wirawan, F.; Amalia, R.; Khairani, A.F.; Pradini, G.W. Differences in endosomal Rab gene expression between positive and negative COVID-19 patients. BMC Res. Notes 2022, 15, 252. [Google Scholar] [CrossRef]
- Hou, W.; Wang, S.; Wu, H.; Xue, L.; Wang, B.; Wang, S.; Wang, H. Small GTPase—A key role in host cell for Coronavirus infection and a potential target for Coronavirus vaccine adjuvant discovery. Viruses 2022, 14, 2044. [Google Scholar] [CrossRef]
- Qiu, S.; Champagne, D.L.; Peters, M.; Catania, E.H.; Weeber, E.J.; Levitt, P.; Pimenta, A.F. Loss of limbic system-associated membrane protein leads to reduced hippocampal mineralocorticoid receptor expression, impaired synaptic plasticity, and spatial memory deficit. Biol. Psychiatry 2010, 68, 197–204. [Google Scholar] [CrossRef]
- Hoppmann, J.; Perwitz, N.; Meier, B.; Fasshauer, M.; Hadaschik, D.; Lehnert, H.; Klein, J. The balance between gluco- and mineralo-corticoid action critically determines inflammatory adipocyte responses. J. Endocrinol. 2010, 204, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Hauser, E.R.; Shah, S.H.; Seo, D.; Sivashanmugam, P.; Exum, S.T.; Gregory, S.G.; Granger, C.B.; Haines, J.L.; Jones, C.J.H.; et al. Polymorphisms of the tumor suppressor gene LSAMP are associated with left main coronary artery disease. Ann. Hum. Genet. 2008, 72, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Xu, Y.; Cheung, A.K.; Tomlinson, R.C.; Alcázar-Román, A.; Murphy, L.; Billich, A.; Zhang, B.; Feng, Y.; Klumpp, M.; et al. PIKfyve, a class III PI Kinase, is the target of the small molecular IL-12/IL-23 inhibitor apilimod and a player in toll-like receptor signaling. Chem. Biol. 2013, 20, 912–921. [Google Scholar] [CrossRef] [PubMed]
- Logue, J.; Chakraborty, A.R.; Johnson, R.; Goyal, G.; Rodas, M.; Taylor, L.J.; Baracco, L.; McGrath, M.E.; Haupt, R.; Furlong, B.A.; et al. PIKfyve-specific inhibitors restrict replication of multiple coronaviruses in vitro but not in a murine model of COVID-19. Commun. Biol. 2022, 5, 808. [Google Scholar] [CrossRef] [PubMed]
- Kant, R.; Kareinen, L.; Ojha, R.; Strandin, T.; Hassan Saber, S.; Kuivanen, S.; Sirnonen, T.; Joensuu, M.; Vapalahti, O.; Kipar, A.; et al. Complete protection from SARS-CoV-2 lung infection in mice through combined intranasal delivery of PIKfyve kinase and TMPRSS2 protease inhibitors. bioRxiv 2023. [Google Scholar] [CrossRef]
- Baker, J.; Ombredane, H.; Daly, L.; Knowles, I.; Rapeport, G.; Ito, K. Pan-antiviral effects of a PIKfyve inhibitor on respiratory virus infection in human nasal epithelium and mice. Antimicrob. Agents Chemother. 2024, 68, e0105023. [Google Scholar] [CrossRef]
- Sevastre, A.S.; Buzatu, I.M.; Baloi, C.; Oprita, A.; Dragoi, A.; Tataranu, L.G.; Alexandru, O.; Tudorache, S.; Dricu, A. ELTD1—An emerging silent actor in cancer drama play. Int. J. Mol. Sci. 2021, 22, 5151. [Google Scholar] [CrossRef] [PubMed]
- Favara, D.M.; Liebscher, I.; Jazayeri, A.; Nambiar, M.; Sheldon, H.; Banham, A.H.; Harris, A.L. Elevated expression of the adhesion GPCR ADGRL4/ELTD1 promotes endothelial sprouting angiogenesis without activating canonical GPCR signalling. Sci. Rep. 2021, 11, 8870. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, H.; Zhang, W.; Bridges, E.; Ang, K.H.; Lin, S.; Masiero, M.; Li, D.; Handford, P.A.; Whiteman, P.; Fischer, R.; et al. ELTD1 is present in extracellular vesicles derived from endothelial cells as a cleaved extracellular domain which induces in vivo angiogenesis. J. Extracell. Biol. 2022, 1, e52. [Google Scholar] [CrossRef]
- Yahalom-Ronen, Y.; Tamir, H.; Melamed, S.; Politi, B.; Achdout, H.; Erez, N.; Israeli, O.; Cohen-Gihon, I.; Chery Mimran, L.; Barlev-Gross, M.; et al. VSV-ΔG-Spike Candidate Vaccine Induces Protective Immunity and Protects K18-hACE2 Mice against SARS-CoV-2 Variants. Viruses 2023, 15, 1364. [Google Scholar] [CrossRef]
- Mao, X.; Nagler, A.R.; Farber, S.A.; Choi, E.J.; Jackson, L.H.; Leiferman, K.M.; Ishii, N.; Hashimoto, T.; Amagai, M.; Zone, J.J.; et al. Autoimmunity to desmocollin 3 in pemphigus vulgaris. Am. J. Pathol. 2010, 177, 2724–2730. [Google Scholar] [CrossRef]
- Muhammad, J.S.; Saheb Sharif-Askari, N.; Cui, Z.G.; Hamad, M.; Halwani, R. SARS-CoV-2 infection-induced promoter hypomethylation as an epigenetic modulator of heat shock protein A1L (HSPA1L) gene. Front. Genet. 2021, 12, 622271. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, W.; Li, M.; Gao, Y.; Zhang, W.; Huang, Y.; Zhuo, W.; Yan, X.; Liu, W.; Wang, F.; et al. NudCL2 is an Hsp90 cochaperone to regulate sister chromatid cohesion by stabilizing cohesin subunits. Cell. Mol. Life Sci. 2019, 76, 381–395. [Google Scholar] [CrossRef]
- Li, M.; Xu, X.; Zhang, J.; Liu, M.; Wang, W.; Gao, Y.; Sun, Q.; Zhang, J.; Lu, Y.; Wang, F.; et al. NudC-like protein 2 restrains centriole amplification by stabilizing HERC2. Cell Death Dis. 2019, 10, 628. [Google Scholar] [CrossRef]
- Galligan, J.T.; Martinez-Noël, G.; Arndt, V.; Hayes, S.; Chittenden, T.W.; Harper, J.W.; Howley, P.M. Proteomic analysis and identification of cellular interactors of the giant ubiquitin ligase HERC2. J. Proteome Res. 2015, 14, 953–966. [Google Scholar] [CrossRef]
- Xu, G.; Wu, Y.; Xiao, T.; Qi, F.; Fan, L.; Zhang, S.; Zhou, J.; He, Y.; Gao, X.; Zeng, H.; et al. Multiomics approach reveals the ubiquitination-specific processes hijacked by SARS-CoV-2. Signal Transduct. Target. Ther. 2022, 7, 312. [Google Scholar] [CrossRef]
- Lee, M.J.; Blish, C.A. Defining the role of natural killer cells in COVID-19. Nat. Immunol. 2023, 24, 1628–1638. [Google Scholar] [CrossRef]
- Hsieh, W.C.; Lai, E.Y.; Liu, Y.T.; Wang, Y.F.; Tzeng, Y.S.; Cui, L.; Lai, Y.J.; Huang, H.C.; Huang, J.H.; Ni, H.C.; et al. NK cell receptor and ligand composition influences the clearance of SARS-CoV-2. J. Clin. Investig. 2021, 131, e146408. [Google Scholar] [CrossRef]
- Pandey, A.; Li-Kroeger, D.; Sethi, M.K.; Lee, T.V.; Buettner, F.F.R.; Bakker, H.; Jafar-Nejad, H. Sensitized genetic backgrounds reveal differential roles for EGF repeat xylosyltransferases in Drosophila Notch signaling. Glycobiology 2018, 28, 849–859. [Google Scholar] [CrossRef]
- Zeng, H.; Wang, Y.; Wang, Y.; Zhang, Y. XXYLT1 methylation contributes to the occurrence of lung adenocarcinoma: Methylation and lung adenocarcinoma. Medicine 2021, 100, E24150. [Google Scholar] [CrossRef]
- Assar, S.; Dastbaz, M.; Amini, K.; Roghani, S.A.; Lotfi, R.; Taghadosi, M.; Kafi, H.; Abdan, Z.; Allahyari, H.; Rostampour, R.; et al. Assessing the gene expression of the adenosine 5′-monophosphate-activated protein kinase (AMPK) and its relation with the IL-6 and IL-10 plasma levels in COVID-19 patients. Mol. Biol. Rep. 2023, 50, 9925–9933. [Google Scholar] [CrossRef]
- Zhang, J.; Dong, J.; Martin, M.; He, M.; Gongol, B.; Marin, T.L.; Chen, L.; Shi, X.; Yin, Y.; Shang, F.; et al. AMP-activated protein kinase phosphorylation of angiotensin-converting enzyme 2 in endothelium mitigates pulmonary hypertension. Am. J. Respir. Crit. Care Med. 2018, 198, 509–520. [Google Scholar] [CrossRef]
- Kohyanagi, N.; Ohama, T. The impact of SETBP1 mutations in neurological diseases and cancer. Genes Cells 2023, 28, 629–641. [Google Scholar] [CrossRef]
- Yadav, H.; Devalaraja, S.; Chung, S.T.; Rane, S.G. TGF-β1/Smad3 pathway targets PP2A-AMPK-FoxO1 signaling to regulate hepatic gluconeogenesis. J. Biol. Chem. 2017, 292, 3420–3432. [Google Scholar] [CrossRef]
- Galbo, T.; Perry, R.J.; Nishimura, E.; Samuel, V.T.; Quistorff, B.; Shulman, G.I. PP2A inhibition results in hepatic insulin resistance despite Akt2 activation. Aging 2013, 5, 770–781. [Google Scholar] [CrossRef]
- LoPresti, M.; Beck, D.B.; Duggal, P.; Cummings, D.A.T.; Solomon, B.D. The role of host genetic factors in Coronavirus susceptibility: Review of animal and systematic review of human literature. Am. J. Hum. Genet. 2020, 107, 381–402. [Google Scholar] [CrossRef]
- Yang, Y.; Ren, P.; Liu, X.; Sun, X.; Zhang, C.; Du, X.; Xing, B. PPP1R26 drives hepatocellular carcinoma progression by controlling glycolysis and epithelial-mesenchymal transition. J. Exp. Clin. Cancer Res. 2022, 41, 101. [Google Scholar] [CrossRef]
- Damasceno, L.E.A.; Prado, D.S.; Veras, F.P.; Fonseca, M.M.; Toller-Kawahisa, J.E.; Rosa, M.H.; Públio, G.A.; Martins, T.V.; Ramalho, F.S.; Waismaz, A.; et al. PKM2 promotes Th17 cell differentiation and autoimmune inflammation by fine-tuning STAT3 activation. J. Exp. Med. 2020, 217, e20190613. [Google Scholar] [CrossRef]
- McElvaney, O.J.; McEvoy, N.L.; McElvaney, O.F.; Carroll, T.P.; Murphy, M.P.; Dunlea, D.M.; Choileáin, O.N.; Clarke, J.; O’Connor, E.; Hogan, G.; et al. Characterization of the inflammatory response to severe COVID-19 illness. Am. J. Respir. Crit. Care Med. 2020, 202, 812–821. [Google Scholar] [CrossRef]
- Misra, A.; Green, M.R. From polyadenylation to splicing: Dual role for mRNA 3’ end formation factors. RNA Biol. 2016, 13, 259–264. [Google Scholar] [CrossRef]
- An, S.; Li, Y.; Lin, Y.; Chu, J.; Su, J.; Chen, Q.; Wang, H.; Pan, P.; Zheng, R.; Li, J.; et al. Genome-wide profiling reveals alternative polyadenylation of innate immune-related mRNA in patients with COVID-19. Front. Immunol. 2021, 12, 756288. [Google Scholar] [CrossRef]
- You, H.; Zhao, Q.; Dong, M. The key genes underlying pathophysiology correlation between the acute myocardial infarction and COVID-19. Int. J. Gen. Med. 2022, 15, 2479–2490. [Google Scholar] [CrossRef]
- Kabashima, T.; Kawaguchi, T.; Wadzinski, B.E.; Uyeda, K. Xylulose 5-phosphate mediates glucose-induced lipogenesis by xylulose 5-phosphate-activated protein phosphatase in rat liver. Proc. Natl. Acad. Sci. USA 2003, 100, 5107–5112. [Google Scholar] [CrossRef]
- Chen, P.; Wu, M.; He, Y.; Jiang, B.; He, M.L. Metabolic alterations upon SARS-CoV-2 infection and potential therapeutic targets against Coronavirus infection. Signal Transduct. Target. Ther. 2023, 8, 237. [Google Scholar] [CrossRef]
- De Silva, I.W.; Nayek, S.; Singh, V.; Reddy, J.; Granger, J.K.; Verbeck, G.F. Paper spray mass spectrometry utilizing Teslin® substrate for rapid detection of lipid metabolite changes during COVID-19 infection. Analyst 2020, 145, 5725–5732. [Google Scholar] [CrossRef]
- Wu, D.; Shu, T.; Yang, X.; Song, J.X.; Zhang, M.; Yao, C.; Liu, W.; Huang, M.; Yu, Y.; Yang, Q.; et al. Plasma metabolomic and lipidomic alterations associated with COVID-19. Natl. Sci. Rev. 2020, 7, 1157–1168. [Google Scholar] [CrossRef]
- Urday, P.; Gayen nee’ Betal, S.; Sequeira Gomes, R.; Al-Kouatly, H.B.; Solarin, K.; Chan, J.S.Y.; Li, D.; Rahman, I.; Addya, S.; Boelig, R.C.; et al. SARS-CoV-2 COVID-19 infection during pregnancy and differential DNA methylation in human cord blood cells from term neonates. Epigenet. Insights 2023, 16, 25168657231184665. [Google Scholar] [CrossRef]
- Hu, Y.; Mivechi, N.F. Promotion of heat shock factor Hsf1 degradation via adaptor protein filamin A-interacting protein 1-like (FILIP-1L). J. Biol. Chem. 2011, 286, 31397–31408. [Google Scholar] [CrossRef]
- Prodromou, C. Mechanisms of Hsp90 regulation. Biochem. J. 2016, 473, 2439–2452. [Google Scholar] [CrossRef]
- Wyler, E.; Mösbauer, K.; Franke, V.; Diag, A.; Gottula, L.T.; Arsiè, R.; Klironomos, F.; Koppstein, D.; Hönzke, K.; Ayoub, S.; et al. Transcriptomic profiling of SARS-CoV-2 infected human cell lines identifies HSP90 as target for COVID-19 therapy. iScience 2021, 24, 102151. [Google Scholar] [CrossRef]
- Pauciull, S.; Riccio, A.; Rossi, A.; Santopolo, S.; Piacentini, S.; Santoro, M.G. Human coronaviruses activate and hijack the proteostasis guardian HSF1 to enhance viral replication. bioRxiv 2022. [Google Scholar] [CrossRef]
- Snigdha, M.; Akter, A.; Amin, M.A.; Islam, M.Z. Bioinformatics approach to analyse COVID-19 biomarkers accountable for generation of intracranial aneurysm in COVID-19 patients. Inform. Med. Unlocked 2023, 39, 101247. [Google Scholar] [CrossRef]
- Zhang, N.; Zhao, C.; Zhang, X.; Cui, X.; Zhao, Y.; Yang, J.; Gao, X. Growth arrest–specific 2 protein family: Structure and function. Cell Prolif. 2021, 54, e12934. [Google Scholar] [CrossRef]
- García-Hidalgo, M.C.; Peláez, R.; González, J.; Santisteve, S.; Benítez, I.D.; Molinero, M.; Perez-Pons, M.; Belmonte, T.; Torres, G.; Moncusí-Moix, A.; et al. Genome-wide transcriptional profiling of pulmonary functional sequelae in ARDS-secondary to SARS-CoV-2 infection. Biomed. Pharmacother. 2022, 154, 113617. [Google Scholar] [CrossRef]
- Kataoka, T. Biological properties of the BCL-2 family protein BCL-RAMBO, which regulates apoptosis, mitochondrial fragmentation, and mitophagy. Front. Cell Dev. Biol. 2022, 10, 1065702. [Google Scholar] [CrossRef]
- Kawabe, Y.; Mori, J.; Morimoto, H.; Yamaguchi, M.; Miyagaki, S.; Ota, T.; Tsuma, Y.; Fukuhara, S.; Nakajima, H.; Oudit, G.Y.; et al. ACE2 exerts anti-obesity effect via stimulating brown adipose tissue and induction of browning in white adipose tissue. Am. J. Physiol. Endocrinol. Metab. 2019, 317, 1140–1149. [Google Scholar] [CrossRef]
- Ju, L.; Chen, S.; Alimujiang, M.; Bai, N.; Yan, H.; Fang, Q.; Han, J.; Ma, X.; Yang, Y.; Jia, W. A novel role for Bcl2l13 in promoting beige adipocyte biogenesis. Biochem. Biophys. Res. Commun. 2018, 506, 485–491. [Google Scholar] [CrossRef]
- Jing, X.; Wu, J.; Dong, C.; Gao, J.; Seki, T.; Kim, C.; Urgard, E.; Hosaka, K.; Yang, Y.; Long, S.; et al. COVID-19 instigates adipose browning and atrophy through VEGF in small mammals. Nat. Metab. 2022, 4, 1674–1683. [Google Scholar] [CrossRef]
- Zaffagni, M.; Harris, J.M.; Patop, I.L.; Reddy Pamudurti, N.; Nguyen, S.; Kadener, S. SARS-CoV-2 Nsp14 mediates the effects of viral infection on the host cell transcriptome. eLife 2022, 11, e71945. [Google Scholar] [CrossRef]
- Ding, D.X.; Wang, Y.; Yan, W.; Fu, W.N. MYCT1 alters the glycogen shunt by regulating selective translation of RACK1-mediated enzymes. iScience 2022, 25, 103955. [Google Scholar] [CrossRef]
- Li, X.; Fan, Q.L.; Ma, T.K.; Liu, C.; Shi, H.; Sun, Y.Y.; Wang, Y.; Ding, D.X.; Tang, A.; Qin, Y.; et al. MYCT1 attenuates renal fibrosis and tubular injury in diabetic kidney disease. iScience 2023, 26, 107609. [Google Scholar] [CrossRef]
- Kabir, A.U.; Subramanian, M.; Lee, D.H.; Wang, X.; Krchma, K.; Wu, J.; Naismith, T.; Halabi, C.M.; Kim, J.Y.; Pulous, F.E.; et al. Dual role of endothelial Myct1 in tumor angiogenesis and tumor immunity. Sci. Transl. Med. 2021, 13, eabb6731. [Google Scholar] [CrossRef]
- Tu, X.; Li, C.; Sun, W.; Tian, X.; Li, Q.; Wang, S.; Ding, X.; Huang, Z. Suppression of cancer cell stemness and drug resistance via MYC destabilization by deubiquitinase USP45 inhibition with a natural small molecule. Cancers 2023, 15, 930. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Wang, Q.; Bian, H.; Chen, Z.; He, H.; Zhao, X.; Gong, P. Comprehensive analysis reveals USP45 as a novel putative oncogene in pan-cancer. Front. Mol. Biosci. 2022, 9, 886904. [Google Scholar] [CrossRef] [PubMed]
- Asano, A.; Nelson-Harrington, J.L.; Travis, A.J. Phospholipase B is activated in response to sterol removal and stimulates acrosome exocytosis in murine sperm. J. Biol. Chem. 2013, 288, 28104–28115. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Zhao, L.; Larsson, A.; Venge, P. The identification of a phospholipase B precursor in human neutrophils. FEBS J. 2009, 276, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Pang, S.W.; Lahiri, C.; Poh, C.L.; Tan, K.O. PNMA family: Protein interaction network and cell signalling pathways implicated in cancer and apoptosis. Cell Signal. 2018, 45, 54–62. [Google Scholar] [CrossRef]
- Kuwae, Y.; Kakehashi, A.; Wakasa, K.; Wei, M.; Yamano, S.; Ishii, N.; Ohsawa, M.; Wanibuchi, H. Paraneoplastic Ma antigen-like 1 as a potential prognostic biomarker in Human pancreatic ductal adenocarcinoma. Pancreas 2015, 44, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Kondybayeva, A.; Akimniyazova, A.; Kamenova, S.; Duchshanova, G.; Aisina, D.; Goncharova, A.; Ivashchenko, A. Prediction of miRNA interaction with mRNA of stroke candidate genes. Neurol. Sci. 2020, 41, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Bediaga, N.G.; Garnham, A.L.; Naselli, G.; Bandala-Sanchez, E.; Stone, N.L.; Cobb, J.; Harbison, J.E.; Wentworth, J.M.; Ziegler, A.G.; Coupe, J.J.; et al. Cytotoxicity-related gene expression and chromatin accessibility define a subset of CD4+ T cells that mark progression to type 1 diabetes. Diabetes 2022, 71, 556–577. [Google Scholar] [CrossRef]
- Zeniya, M.; Sohara, E.; Kita, S.; Iwamoto, T.; Susa, K.; Mori, T.; Oi, K.; Chiga, M.; Takahashi, D.; Yang, S.-S.; et al. Dietary salt intake regulates WNK3-SPAK-NKCC1 phosphorylation cascade in mouse aorta through angiotensin II. Hypertension 2013, 62, 872–878. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, Q.; Jiang, G.; Zhang, C. Dysfunction of Cullin 3 RING E3 ubiquitin ligase causes vasoconstriction and increased sodium reabsorption in diabetes. Arch. Biochem. Biophys. 2021, 710, 109000. [Google Scholar] [CrossRef] [PubMed]
- Giambruno, R.; Zacco, E.; Ugolini, C.; Vandelli, A.; Mulroney, L.; D’Onghia, M.; Giuliani, B.; Criscuolo, E.; Castelli, M.; Clementi, N.; et al. Unveiling the role of PUS7-mediated pseudouridylation in host protein interactions specific for the SARS-CoV-2 RNA genome. Mol. Ther. Nucleic Acids 2023, 34, 102052. [Google Scholar] [CrossRef] [PubMed]
- Mousa, M.; Vurivi, H.; Kannout, H.; Uddin, M.; Alkaabi, N.; Mahboub, B.; Tay, G.K.; Alsafar, H.S. Genome-wide association study of hospitalized COVID-19 patients in the United Arab Emirates. EBioMedicine 2021, 74, 103695. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; O’connell, J.R.; Mcardle, P.F.; Wade, J.B.; Dorff, S.E.; Shah, S.J.; Shi, X.; Pan, L.; Rampersaud, E.; Shen, H.; et al. Whole-genome association study identifies STK39 as a hypertension susceptibility gene. Proc. Natl. Acad. Sci. USA 2009, 106, 226–231. [Google Scholar] [CrossRef] [PubMed]
- He, N.; Tan, H.; Deng, X.; Shu, L.; Qing, B.; Liang, H. MiR-223-3p-loaded exosomes from bronchoalveolar lavage fluid promote alveolar macrophage autophagy and reduce acute lung injury by inhibiting the expression of STK39. Hum. Cell 2023, 35, 1736–1751. [Google Scholar] [CrossRef] [PubMed]
- Taira, J.; Kida, Y.; Inatomi, K.; Komatsu, H.; Higashimoto, Y.; Sakamoto, H. Phosphorylation of clustered serine residues in the N-terminus of BPS domain negatively regulates formation of the complex between human Grb14 and insulin receptor. J. Biochem. 2017, 162, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Iyer, R.; Novotny, C.; Metzger, D.; Zhou, H.H.; Smith, G.I.; Yoshino, M.; Yoshino, J.; Klein, S.; Swaminath, G.; et al. Inhibition of Grb14, a negative modulator of insulin signaling, improves glucose homeostasis without causing cardiac dysfunction. Sci. Rep. 2020, 10, 3417. [Google Scholar] [CrossRef]
- Hiepen, C.; Jatzlau, J.; Hildebrandt, S.; Kampfrath, B.; Goktas, M.; Murgai, A.; Cuellar Camacho, J.L.; Haag, R.; Ruppert, C.; Sengle, G.; et al. BMPR2 acts as a gatekeeper to protect endothelial cells from increased TGFβ responses and altered cell mechanics. PLoS Biol. 2019, 17, e3000557. [Google Scholar] [CrossRef]
- Andruska, A.; Spiekerkoetter, E. Consequences of BMPR2 deficiency in the pulmonary vasculature and beyond: Contributions to pulmonary arterial hypertension. Int. J. Mol. Sci. 2018, 19, 2499. [Google Scholar] [CrossRef]
- Brock, M.; Trenkmann, M.; Gay, R.E.; Michel, B.A.; Gay, S.; Fischler, M.; Ulrich, S.; Speich, R.; Huber, L.C. Interleukin-6 modulates the expression of the bone morphogenic protein receptor type II through a novel STAT3-microRNA cluster 17/92 pathway. Circ. Res. 2009, 104, 1184–1191. [Google Scholar] [CrossRef]
- Hilton, L.R.; Rätsep, M.T.; VandenBroek, M.M.; Jafri, S.; Laverty, K.J.; Mitchell, M.; Theilmann, A.L.; Smart, J.A.; Hawke, L.G.; Moore, S.D.; et al. Impaired interleukin-15 signaling via BMPR2 loss drives natural killer cell deficiency and pulmonary hypertension. Hypertension 2022, 79, 2493–2504. [Google Scholar] [CrossRef]
- Karmouty-Quintana, H.; Thandavarayan, R.A.; Keller, S.P.; Sahay, S.; Pandit, L.M.; Akkanti, B. Emerging mechanisms of pulmonary vasoconstriction in SARS-CoV-2-induced acute respiratory distress syndrome (ARDS) and potential therapeutic targets. Int. J. Mol. Sci. 2020, 21, 8081. [Google Scholar] [CrossRef]
- Fujita, Y.; Hoshina, T.; Matsuzaki, J.; Yoshioka, Y.; Kadota, T.; Hosaka, Y.; Fujimoto, S.; Kawamoto, H.; Watanabe, N.; Sawaki, K.; et al. Early prediction of COVID-19 severity using extracellular vesicle COPB2. J. Extracell. Vesicles 2021, 10, e12092. [Google Scholar] [CrossRef]
- Cattin-Ortolá, J.; Welch, L.G.; Maslen, S.L.; Papa, G.; James, L.C.; Munro, S. Sequences in the cytoplasmic tail of SARS-CoV-2 Spike facilitate expression at the cell surface and syncytia formation. Nat. Commun. 2021, 12, 5333. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Lei, X.; Zhang, L.; Wan, H.; Pan, H.; Wu, J.; Zou, M.; Zhu, L.; Mi, Y. COPB2: A transport protein with multifaceted roles in cancer development and progression. Clin. Transl. Oncol. 2021, 23, 2195–2205. [Google Scholar] [CrossRef]
- Salipante, S.J.; Rojas, M.E.B.; Korkmaz, B.; Duan, Z.; Wechsler, J.; Benson, K.F.; Person, R.E.; Grimes, H.L.; Horwitz, M.S. Contributions to neutropenia from PFAAP5 (N4BP2L2), a novel protein mediating transcriptional repressor cooperation between Gfi1 and neutrophil elastase. Mol. Cell. Biol. 2009, 29, 4394–4405. [Google Scholar] [CrossRef]
- Rydzynska, Z.; Pawlik, B.; Krzyzanowski, D.; Mlynarski, W.; Madzio, J. Neutrophil elastase defects in congenital neutropenia. Front. Immunol. 2021, 12, 653932. [Google Scholar] [CrossRef]
- Kim, D.-K.; Weller, B.; Lin, C.-W.; Sheykhkarimli, D.; Knapp, J.; Kishore, N.; Sauer, M.; Rayhan, A.; Young, V.; Marin-De, N.; et al. A map of binary SARS-CoV-2 protein interactions implicates host immune regulation and ubiquitination. bioRxiv 2021. [Google Scholar] [CrossRef]
- Nikpay, M.; Soubeyrand, S.; Tahmasbi, R.; McPherson, R. Multiomics screening identifies molecular biomarkers causally associated with the risk of coronary artery disease. Circ. Genom. Precis. Med. 2020, 13, e002876. [Google Scholar] [CrossRef]
- Ma, L.; Liu, Y.; Wang, Y.; Yang, J.; Lu, J.; Feng, H.; Ye, S.; Liu, Y. Identification of PTPN20 as an innate immunity-related gene in gastric cancer with Helicobacter pylori infection. Front. Immunol. 2023, 14, 1212692. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Miyajima, M.; Nakajima, M.; Ogino, I.; Kawamura, K.; Akiba, C.; Kamohara, C.; Sakamoto, K.; Karagiozov, K.; Nakamura, E.; et al. Ptpn20 deletion in H-Tx rats enhances phosphorylation of the NKCC1 cotransporter in the choroid plexus: An evidence of genetic risk for hydrocephalus in an experimental study. Fluids Barriers CNS 2022, 19, 39. [Google Scholar] [CrossRef]
- Schmidt, F.; Abdesselem, H.B.; Suhre, K.; Vaikath, N.N.; Sohail, M.U.; Al-Nesf, M.; Bensmail, I.; Mashod, F.; Sarwath, H.; Bernhardt, J.; et al. Auto-immunoproteomics analysis of COVID-19 ICU patients revealed increased levels of autoantibodies related to the male reproductive system. Front. Physiol. 2023, 14, 1203723. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Shang, L.; Zhang, J.; Liu, Y.; Jin, C.; Zhao, Y.; Lei, X.; Wang, W.; Xiao, X.; Zhang, X.; et al. An antibody-based proximity labeling map reveals mechanisms of SARS-CoV-2 inhibition of antiviral immunity. Cell Chem. Biol. 2022, 29, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Kausalya, P.J.; Ong, A.G.M.; Goh, C.M.F.; Mohamed Ali, S.; Hunziker, W. ZO-2/Tjp2 suppresses Yap and Wwtr1/Taz-mediated hepatocyte to cholangiocyte transdifferentiation in the mouse liver. NPJ Regen. Med. 2022, 7, 55. [Google Scholar] [CrossRef]
- Wang, S.; Zhou, L.; Ling, L.; Meng, X.; Chu, F.; Zhang, S.; Zhou, F. The crosstalk between Hippo-YAP pathway and innate immunity. Front. Immunol. 2020, 11, 323. [Google Scholar] [CrossRef]
- Kalashnikova, E.; Lorca, R.A.; Kaur, I.; Barisone, G.A.; Li, B.; Ishimaru, T.; Trimmer, J.S.; Mohapatra, D.P.; Díaz, E. SynDIG1: An activity-regulated, AMPA- receptor-interacting transmembrane protein that regulates excitatory synapse development. Neuron 2010, 65, 80–93. [Google Scholar] [CrossRef] [PubMed]
- Marziali, F.; Cimarelli, A. Membrane interference against HIV-1 by intrinsic antiviral factors: The case of IFITMs. Cells 2021, 10, 1171. [Google Scholar] [CrossRef] [PubMed]
- Scott, C.L.; T’Jonck, W.; Martens, L.; Todorov, H.; Sichien, D.; Soen, B.; Bonnardel, J.; de Prijck, S.; Vandamme, N.; Cannoodt, R.; et al. The transcription factor ZEB2 is required to maintain the tissue-specific identities of macrophages. Immunity 2018, 49, 312–325. [Google Scholar] [CrossRef]
- de Coninck, S.; Berx, G.; Taghon, T.; van Vlierberghe, P.; Goossens, S. ZEB2 in T-cells and T-ALL. Adv. Biol. Regul. 2019, 74, 100639. [Google Scholar] [CrossRef]
- Omilusik, K.D.; Adam Best, J.; Yu, B.; Goossens, S.; Weidemann, A.; Nguyen, J.V.; Seuntjens, E.; Stryjewska, A.; Zweier, C.; Roychoudhuri, R.; et al. Transcriptional repressor ZEB2 promotes terminal differentiation of CD8+ effector and memory T cell populations during infection. J. Exp. Med. 2015, 212, 2027–2039. [Google Scholar] [CrossRef]
- DaSilva-Arnold, S.C.; Kuo, C.Y.; Davra, V.; Remache, Y.; Kim, P.C.W.; Fisher, J.P.; Zamudio, S.; Al-Khan, A.; Birge, R.B.; Illsley, N.P. ZEB2, a master regulator of the epithelial-mesenchymal transition, mediates trophoblast differentiation. Mol. Hum. Reprod. 2018, 25, 61–75. [Google Scholar] [CrossRef] [PubMed]
- Fardi, M.; Alivand, M.; Baradaran, B.; Farshdousti Hagh, M.; Solali, S. The crucial role of ZEB2: From development to epithelial-to-mesenchymal transition and cancer complexity. J. Cell. Physiol. 2019, 234, 14783–14799. [Google Scholar] [CrossRef]
- Novelli, G.; Liu, J.; Biancolella, M.; Alonzi, T.; Novelli, A.; Patten, J.J.; Cocciadiferro, D.; Agolini, E.; Colona, V.L.; Rizzacasa, B.; et al. Inhibition of HECT E3 ligases as potential therapy for COVID-19. Cell Death Dis. 2021, 12, 310. [Google Scholar] [CrossRef] [PubMed]
- Centofanti, F.; Alonzi, T.; Latini, A.; Spitalieri, P.; Murdocca, M.; Chen, X.; Cui, W.; Shang, Q.; Goletti, D.; Shi, Y.; et al. Indole-3-carbinol in vitro antiviral activity against SARS-CoV-2 virus and in vivo toxicity. Cell Death Discov. 2022, 8, 491. [Google Scholar] [CrossRef]
- Lv, B.; Stuck, M.W.; Desai, P.B.; Cabrera, O.A.; Pazour, G.J. E3 ubiquitin ligase wwp1 regulates ciliary dynamics of the hedgehog receptor smoothened. J. Cell Biol. 2021, 220, e202010177. [Google Scholar] [CrossRef] [PubMed]
- Pi, P.; Zeng, Z.; Zeng, L.; Han, B.; Bai, X.; Xu, S. Molecular mechanisms of COVID-19-induced pulmonary fibrosis and epithelial-mesenchymal transition. Front. Pharmacol. 2023, 14, 1218059. [Google Scholar] [CrossRef]
- Lu, X.; Yang, B.; Qi, R.; Xie, Q.; Li, T.; Yang, J.; Tong, T.; Niu, K.; Li, M.; Pan, W.; et al. Targeting WWP1 ameliorates cardiac ischemic injury by suppressing KLF15-ubiquitination mediated myocardial inflammation. Theranostics 2023, 13, 417–437. [Google Scholar] [CrossRef]
- Zhao, D.; Zhong, G.; Li, J.; Pan, J.; Zhao, Y.; Song, H.; Sun, W.; Jin, X.; Li, Y.; Du, R.; et al. Targeting E3 ubiquitin ligase WWP1 prevents cardiac hypertrophy through destabilizing DVL2 via inhibition of K27-linked ubiquitination. Circulation 2021, 144, 694–711. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.M.; Lim, J.C.; Kim, G.; Levine, R.L. Myristoylated methionine sulfoxide reductase A is a late endosomal protein. J. Biol. Chem. 2018, 293, 7355–7366. [Google Scholar] [CrossRef]
- Davis, D.A.; Bulut, H.; Shrestha, P.; Mitsuya, H.; Yarchoan, R. Regulation of retroviral and SARS-CoV-2 protease dimerization and activity through reversible oxidation. Antioxidants 2022, 11, 2054. [Google Scholar] [CrossRef]
- Bennet, S.; Kaufmann, M.; Takami, K.; Sjaarda, C.; Douchant, K.; Moslinger, E.; Wong, H.; Reed, D.E.; Ellis, A.K.; Vanner, S.; et al. Small-molecule metabolome identifies potential therapeutic targets against COVID-19. Sci. Rep. 2022, 12, 10029. [Google Scholar] [CrossRef] [PubMed]
- Saheb Sharif-Askari, N.; Saheb Sharif-Askari, F.; Mdkhana, B.; Hussain Alsayed, H.A.; Alsafar, H.; Alrais, Z.F.; Hamid, Q.; Halwani, R. Upregulation of oxidative stress gene markers during SARS-CoV-2 viral infection. Free Radic. Biol. Med. 2021, 172, 688–698. [Google Scholar] [CrossRef]
- Kschonsak, M.; Chua, H.C.; Weidling, C.; Chakouri, N.; Noland, C.L.; Schott, K.; Chang, T.; Tam, C.; Patel, N.; Arthur, C.P.; et al. Structural architecture of the human NALCN channelosome. Nature 2022, 603, 180–186. [Google Scholar] [CrossRef]
- Cochet-Bissuel, M.; Lory, P.; Monteil, A. The sodium leak channel, NALCN, in health and disease. Front. Cell. Neurosci. 2014, 8, 132. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Feng, T. Network-based data analysis reveals ion channel-related gene features in COVID-19: A bioinformatic approach. Biochem. Genet. 2023, 61, 471–505. [Google Scholar] [CrossRef] [PubMed]
- Siira, S.J.; Spåhr, H.; Shearwood, A.M.J.; Ruzzenente, B.; Larsson, N.G.; Rackham, O.; Filipovska, A. LRPPRC-mediated folding of the mitochondrial transcriptome. Nat. Commun. 2017, 8, 1532. [Google Scholar] [CrossRef]
- Guha, S.; Bhaumik, S.R. Viral regulation of mRNA export with potentials for targeted therapy. Biochim. Biophys. Acta Gene Regul. Mech. 2021, 1864, 194655. [Google Scholar] [CrossRef]
- Zhang, N.; Wang, S.; Wong, C.C.L. Proteomics research of SARS-CoV-2 and COVID-19 disease. Med. Rev. 2022, 2, 427–445. [Google Scholar] [CrossRef]
- Wang, H.; Tang, A.; Cui, Y.; Gong, H.; Li, H. LRPPRC facilitates tumor progression and immune evasion through upregulation of m6A modification of PD-L1 mRNA in hepatocellular carcinoma. Front. Immunol. 2023, 14, 1144774. [Google Scholar] [CrossRef]
- Qian, W.; Zhou, J.; Duan, L.; Wang, H.; Xu, S.; Cao, Y. m6A methylation: A potential key player in understanding and treating COVID-2019 infection. Cell Death Discov. 2023, 9, 300. [Google Scholar] [CrossRef]
- Zang, L.; Gu, J.; Yang, X.; Yuan, Y.; Guo, H.; Zhou, W.; Ma, J.; Chen, Y.; Wu, Y.; Zheng, H.; et al. Ubiquitin-specific protease 24 promotes EV71 infection by restricting K63-linked polyubiquitination of TBK1. Virol. Sin. 2023, 38, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Ullah, T.R.; Johansen, M.D.; Balka, K.R.; Ambrose, R.L.; Gearing, L.J.; Roest, J.; Vivian, J.P.; Sapkota, S.; Jayasekara, W.S.N.; Wenholz, D.S.; et al. Pharmacological inhibition of TBK1/IKKε blunts immunopathology in a murine model of SARS-CoV-2 infection. Nat. Commun. 2023, 14, 5666. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.C.; Wu, Y.S.; Hung, C.Y.; Wang, S.A.; Young, M.J.; Hsu, T.I.; Hung, J.J. USP24 induces IL-6 in tumor-associated microenvironment by stabilizing p300 and β-TrCP and promotes cancer malignancy. Nat. Commun. 2018, 9, 3996. [Google Scholar] [CrossRef] [PubMed]
- Thayer, J.A.; Awad, O.; Hegdekar, N.; Sarkar, C.; Tesfay, H.; Burt, C.; Zeng, X.; Feldman, R.A.; Lipinski, M.M. The PARK10 gene USP24 is a negative regulator of autophagy and ULK1 protein stability. Autophagy 2020, 16, 140–153. [Google Scholar] [CrossRef]
- Gao, T.; McKenna, B.; Li, C.; Reichert, M.; Nguyen, J.; Singh, T.; Yang, C.; Pannikar, A.; Doliba, N.; Zhang, T.; et al. Pdx1 maintains β cell identity and function by repressing an α cell program. Cell Metab. 2014, 19, 259–271. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Ma, Q.; Li, C.; Liu, R.; Zhao, L.; Wang, W.; Zhang, P.; Liu, X.; Gao, G.; Liu, F.; et al. Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg. Microbes Infect. 2020, 9, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Xu, L.D.; Zhang, F.; Liang, Q.Z.; Jiao, Y.; Shi, F.S.; He, B.; Xu, P.; Huang, Y.W. Heat shock protein 90 facilitates SARS-CoV-2 structural protein-mediated virion assembly and promotes virus-induced pyroptosis. J. Biol. Chem. 2023, 299, 104668. [Google Scholar] [CrossRef]
- Yu, H.; Takeuchi, M.; LeBarron, J.; Kantharia, J.; London, E.; Bakker, H.; Haltiwanger, R.S.; Li, H.; Takeuchi, H. Notch-modifying xylosyltransferase structures support an SNi-like retaining mechanism. Nat. Chem. Biol. 2015, 11, 847–854. [Google Scholar] [CrossRef]
- Benamar, M.; Chen, Q.; Chou, J.; Julé, A.M.; Boudra, R.; Contini, P.; Crestani, E.; Lai, P.S.; Wang, M.; Fong, J.; et al. The Notch1/CD22 signaling axis disrupts Treg function in SARS-CoV-2–associated multisystem inflammatory syndrome in children. J. Clin. Investig. 2023, 133, e163235. [Google Scholar] [CrossRef]
- Liu, X.; Verma, A.; Jr, G.G.; Ramage, H.; Lucas, A.; Myers, R.L.; Michaelson, J.J.; Coryell, W.; Kumar, A.; Charney, A.W. Targeting the coronavirus nucleocapsid protein through GSK-3 inhibition. Proc. Natl. Acad. Sci. USA 2021, 118, e2113401118. [Google Scholar] [CrossRef]
- Urazov, S.; Chernov, A.; Popov, O.; Klenkova, N.; Sushentseva, N.; Polkovnikova, I.; Apalko, S.; Kislyuk, K.; Pavlovich, D.; Ivanov, A.; et al. Secretory phospholipase A2 and interleukin-6 levels as predictive markers of the severity and outcome of patients with COVID-19 infections. Int. J. Mol. Sci. 2023, 24, 5540. [Google Scholar] [CrossRef]
- Govender, N.; Khaliq, O.P.; Moodley, J.; Naicker, T. Insulin resistance in COVID-19 and diabetes. Prim. Care Diabetes 2021, 15, 629–634. [Google Scholar] [CrossRef]
- Adamo, S.; Michler, J.; Zurbuchen, Y.; Cervia, C.; Taeschler, P.; Raeber, M.E.; Baghai Sain, S.; Nilsson, J.; Moor, A.E.; Boyman, O. Signature of long-lived memory CD8+ T cells in acute SARS-CoV-2 infection. Nature 2022, 602, 148–155. [Google Scholar] [CrossRef]
- Ortillon, J.; le Bail, J.C.; Villard, E.; Léger, B.; Poirier, B.; Girardot, C.; Beeske, S.; Ledein, L.; Blanchard, V.; Brieu, P.; et al. High glucose activates YAP signaling to promote vascular inflammation. Front. Physiol. 2021, 12, 665994. [Google Scholar] [CrossRef]
- Zhu, L.; She, Z.G.; Cheng, X.; Qin, J.J.; Zhang, X.J.; Cai, J.; Lei, F.; Wang, H.; Xie, J.; Wang, W.; et al. Association of blood glucose control and outcomes in patients with COVID-19 and pre-existing type 2 diabetes. Cell Metab. 2020, 31, 1068–1077. [Google Scholar] [CrossRef]
- Lim, S.; Bae, J.H.; Kwon, H.S.; Nauck, M.A. COVID-19 and diabetes mellitus: From pathophysiology to clinical management. Nat. Rev. Endocrinol. 2021, 17, 11–30. [Google Scholar] [CrossRef]
- Zhang, Y.; Xin, B.; Liu, Y.; Jiang, W.; Han, W.; Deng, J.; Wang, P.; Hong, X.; Yan, D. SARS-CoV-2 protein NSP9 promotes cytokine production by targeting TBK1. Front. Immunol. 2023, 14, 1211816. [Google Scholar] [CrossRef]
- Kimura, A.; Kishimoto, T. Regulator of Treg/Th17 balance. Eur. J. Immunol. 2010, 40, 1830–1835. [Google Scholar] [CrossRef]
- Galván-Peña, S.; Leon, J.; Chowdhary, K.; Michelson, D.A.; Vijaykumar, B.; Yang, L.; Magnuson, A.M.; Chen, F.; Manickas-Hill, Z.; Piechocka-Trocha, A.; et al. Profound Treg perturbations correlate with COVID-19 severity. Proc. Natl. Acad. Sci. USA 2021, 118, e2111315118. [Google Scholar] [CrossRef]
- Murakami, M.; Sato, H.; Taketomi, Y. Modulation of immunity by the secreted phospholipase A2 family. Immunol. Rev. 2023, 317, 42–70. [Google Scholar] [CrossRef]
- Ripon, M.A.R.; Bhowmik, D.R.; Amin, M.T.; Hossain, M.S. Role of arachidonic cascade in COVID-19 infection: A review. Prostaglandins Other Lipid Mediat. 2021, 154, 106539. [Google Scholar] [CrossRef]
- Dennis, E.A.; Norris, P.C. Eicosanoid storm in infection and inflammation. Nat. Rev. Immunol. 2015, 15, 511–523. [Google Scholar] [CrossRef]
- Ricke-Hoch, M.; Stelling, E.; Lasswitz, L.; Gunesch, A.P.; Kasten, M.; Zapatero-Belinchón, F.J.; Brogden, G.; Gerold, G.; Pietschmann, T.; Montiel, V.; et al. Impaired immune response mediated by prostaglandin E2 promotes severe COVID-19 disease. PLoS ONE 2021, 16, e0255335. [Google Scholar] [CrossRef]
- Ding, Q.; Wang, Y.; Zhang, A.-l.; Xu, T.; Zhou, D.-d.; Li, X.-F.; Yang, J.-F.; Zhang, L.; Wang, X. ZEB2 attenuates LPS-induced inflammation by the NF-κB pathway in HK-2 cells. Inflammation 2018, 41, 722–731. [Google Scholar] [CrossRef]
- Hiwasa, T.; Wang, H.; Goto, K.-i.; Mine, S.; Machida, T.; Kobayashi, E.; Yoshida, Y.; Adachi, A.; Matsutani, T.; Sata, M.; et al. Serum anti-DIDO1, anti-CPSF2, and anti-FOXJ2 antibodies as predictive risk markers for acute ischemic stroke. BMC Med. 2021, 19, 131. [Google Scholar] [CrossRef]
- Zheng, Q.W.; Ni, Q.Z.; Zhu, B.; Liang, X.; Ma, N.; Wang, Y.K.; Xu, S.; Cao, H.J.; Xia, J.; Zhang, F.K.; et al. PPDPF promotes lung adenocarcinoma progression via inhibiting apoptosis and NK cell-mediated cytotoxicity through STAT3. Oncogene 2022, 41, 4244–4256. [Google Scholar] [CrossRef]
- Wang, F.; Zhou, Y.; Fu, B.; Wu, Y.; Zhang, R.; Sun, R.; Tian, Z.; Wei, H. Molecular signatures and transcriptional regulatory networks of human immature decidual NK and mature peripheral NK cells. Eur. J. Immunol. 2014, 44, 2771–2784. [Google Scholar] [CrossRef]
- Lee, A.; Seo, J.; Park, S.; Cho, Y.; Kim, G.; Li, J.; Liang, L.; Park, T.; Chung, W. Type 2 diabetes and its genetic susceptibility are associated with increased severity and mortality of COVID-19 in UK Biobank. Commun. Biol. 2024, 7, 122. [Google Scholar] [CrossRef]
- Xiao, Y.; Wang, Y.; Ryu, J.; Liu, W.; Zou, H.; Zhang, R.; Yan, Y.; Dai, Z.; Zhang, D.; Sun, L.-Z.; et al. Upregulated TGF-β1 contributes to hyperglycaemia in type 2 diabetes by potentiating glucagon signalling. Diabetologia 2023, 66, 1142–1155. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nahalka, J. 1-L Transcription of SARS-CoV-2 Spike Protein S1 Subunit. Int. J. Mol. Sci. 2024, 25, 4440. https://doi.org/10.3390/ijms25084440
Nahalka J. 1-L Transcription of SARS-CoV-2 Spike Protein S1 Subunit. International Journal of Molecular Sciences. 2024; 25(8):4440. https://doi.org/10.3390/ijms25084440
Chicago/Turabian StyleNahalka, Jozef. 2024. "1-L Transcription of SARS-CoV-2 Spike Protein S1 Subunit" International Journal of Molecular Sciences 25, no. 8: 4440. https://doi.org/10.3390/ijms25084440




