Genome-Wide Characterization and Functional Validation of the ACS Gene Family in the Chestnut Reveals Its Regulatory Role in Ovule Development
Abstract
:1. Introduction
2. Results
2.1. Genome-Wide Analysis of the Chestnut ACS Gene (CmACS) Family
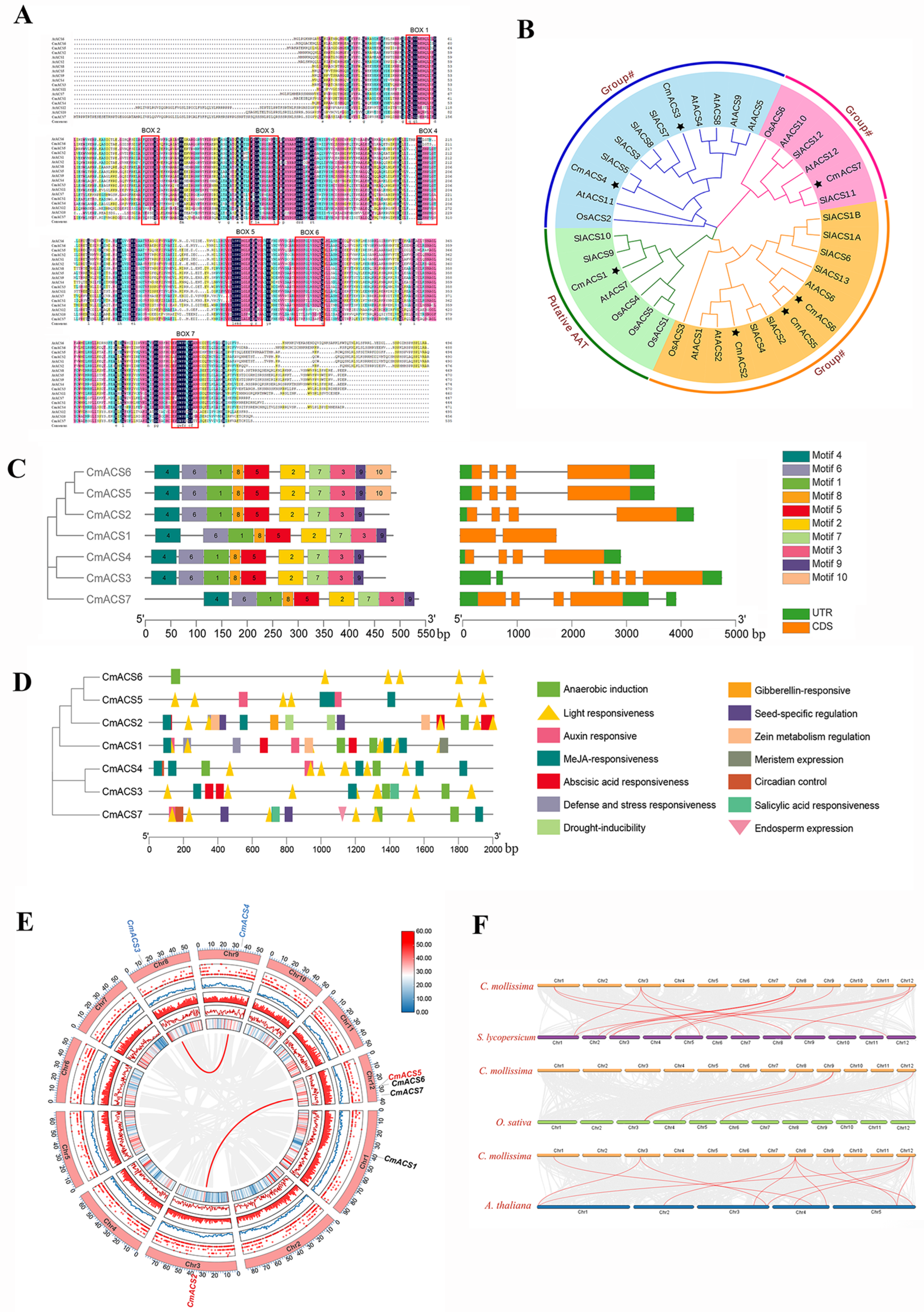
2.2. Multiple-Sequence Alignment and Phylogenetic Analysis of the CmACS Gene Family
2.3. Structural Characterization of CmACS Proteins
2.4. Analysis of Cis-Acting Elements in CmACS Promoters
2.5. Chromosome Localization and Covariance Analysis of CmACSs
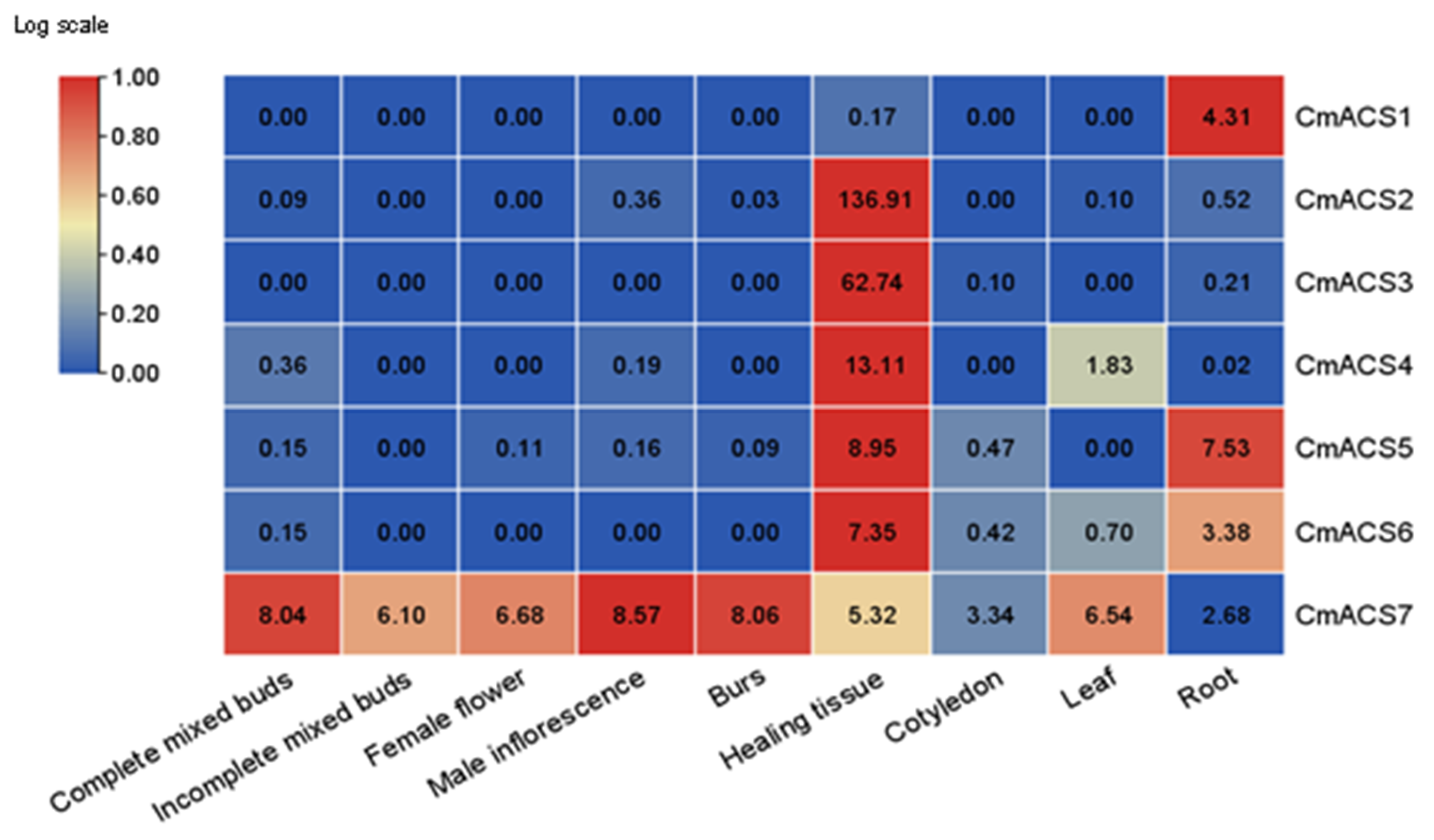
2.6. Tissue Expression Pattern Analysis of CmACSs
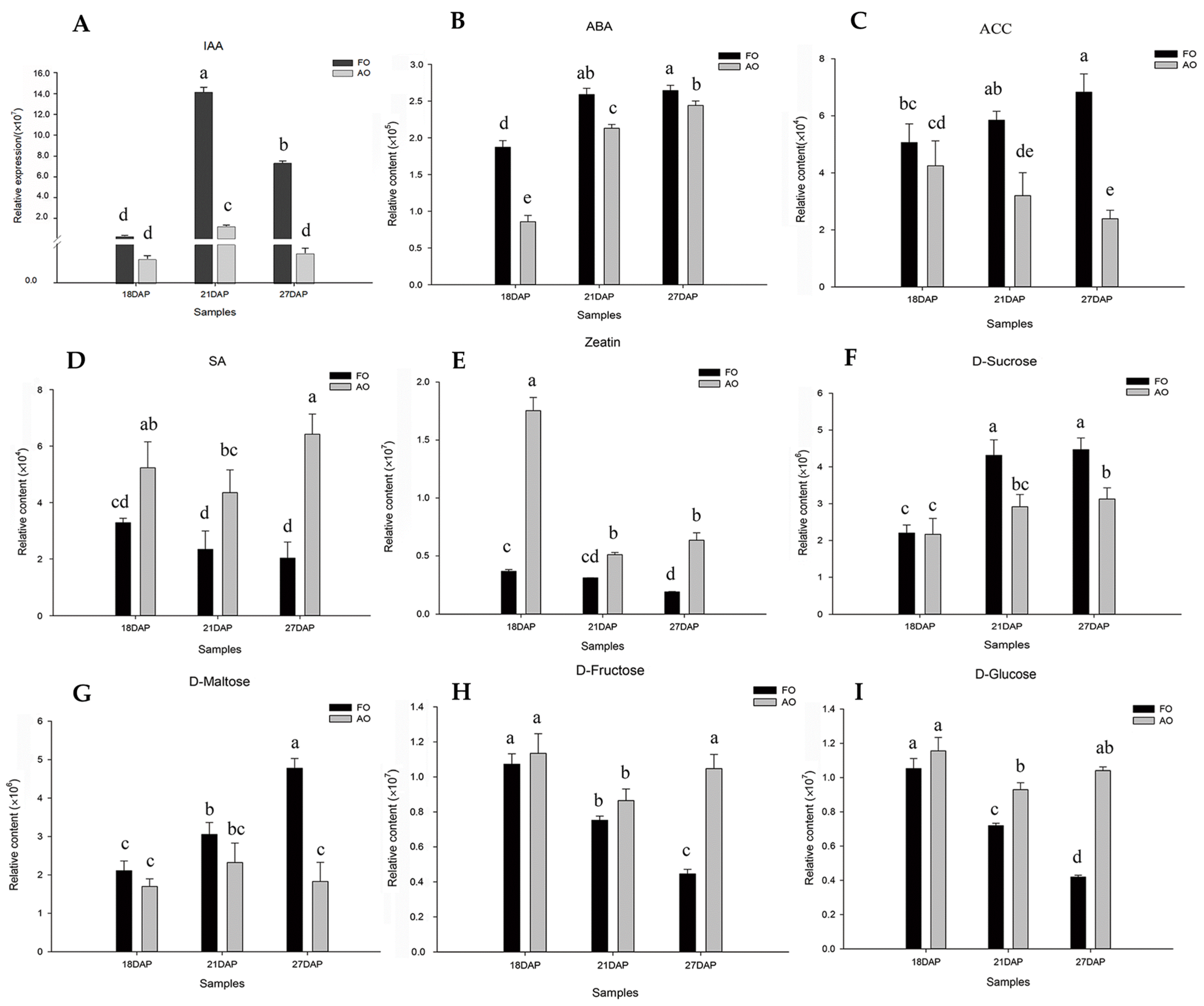
2.7. Expression Analysis of CmACS Genes during Ovule Development
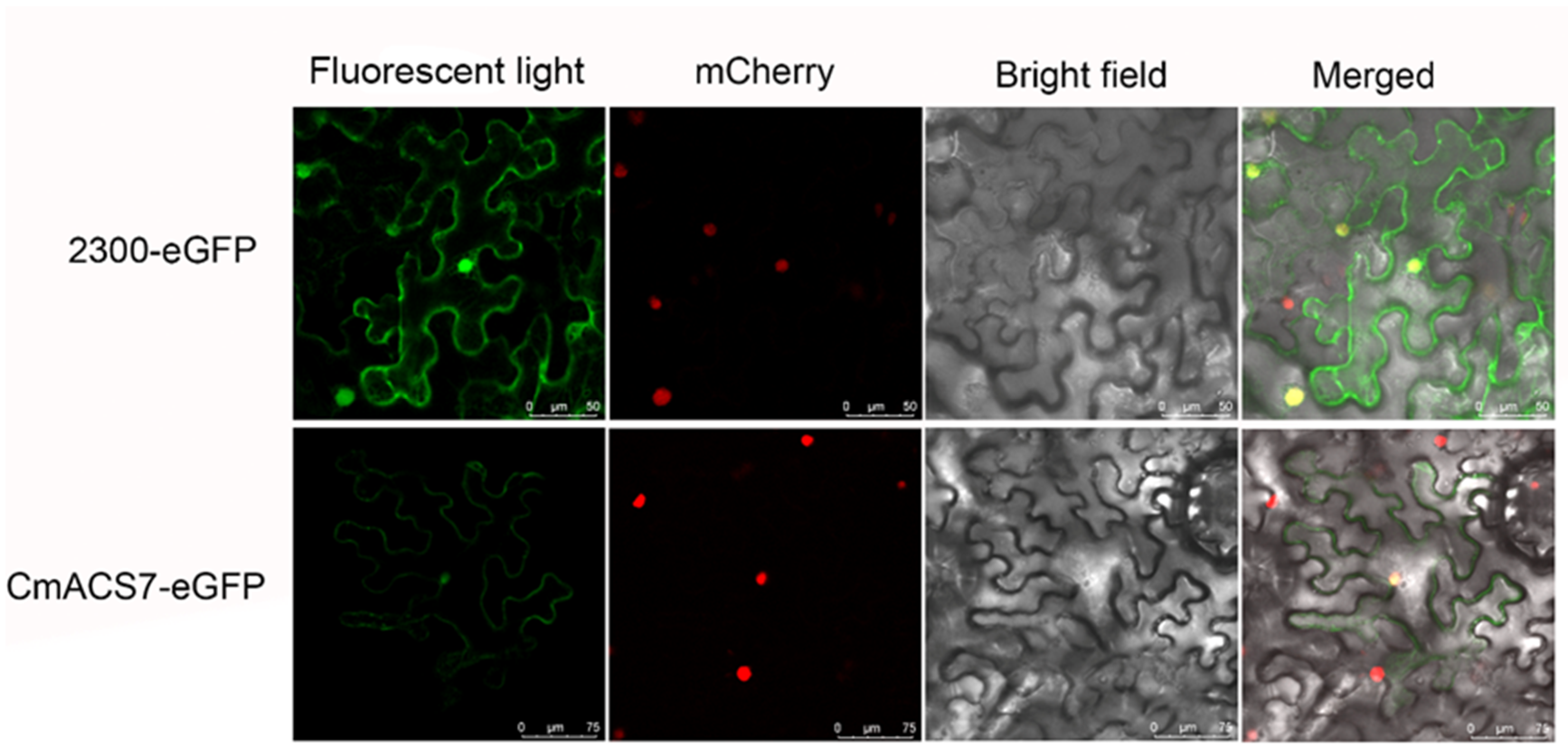
2.8. Subcellular Localization
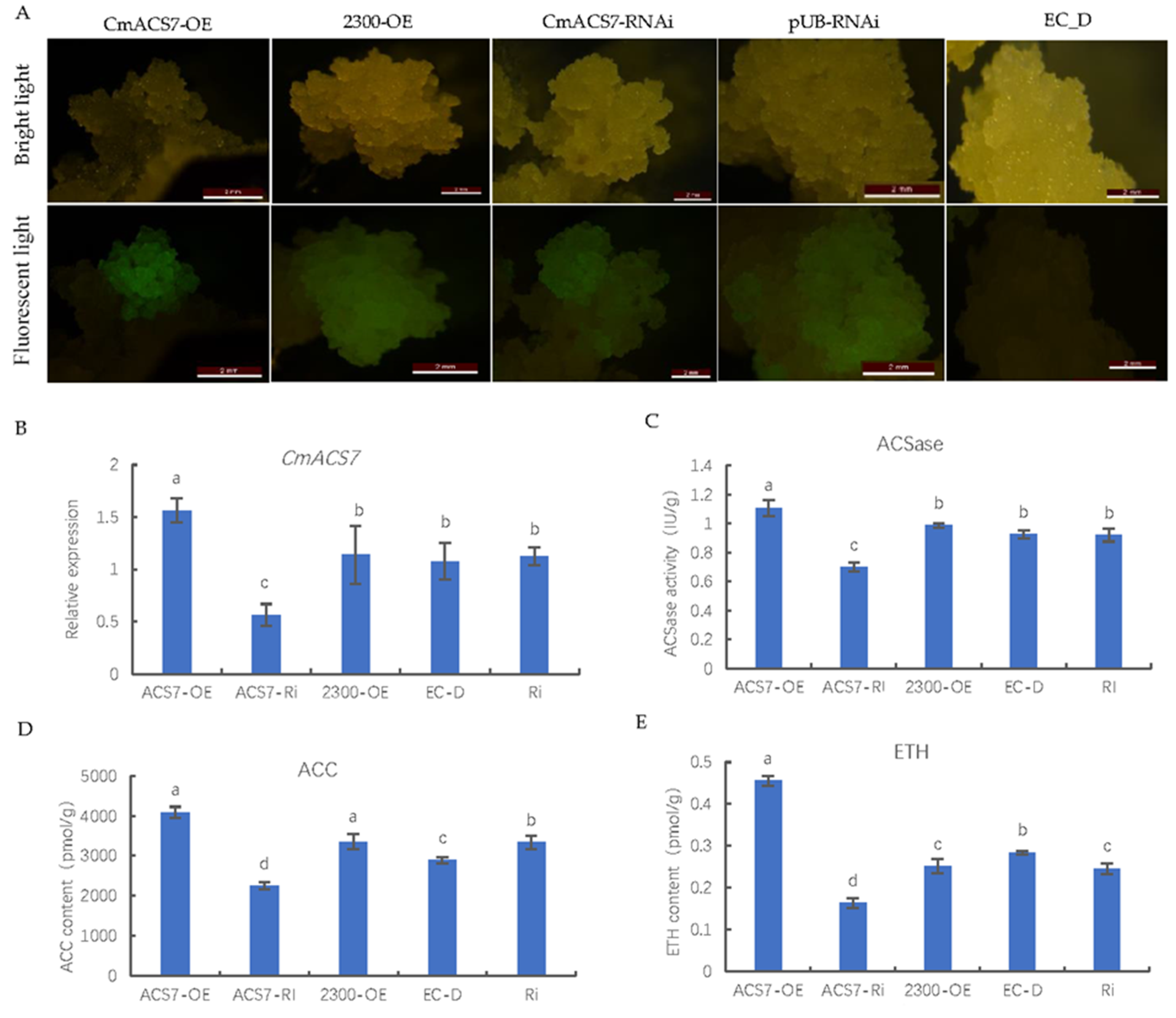
2.9. Genetic Transformation of Chestnut Healing Tissues
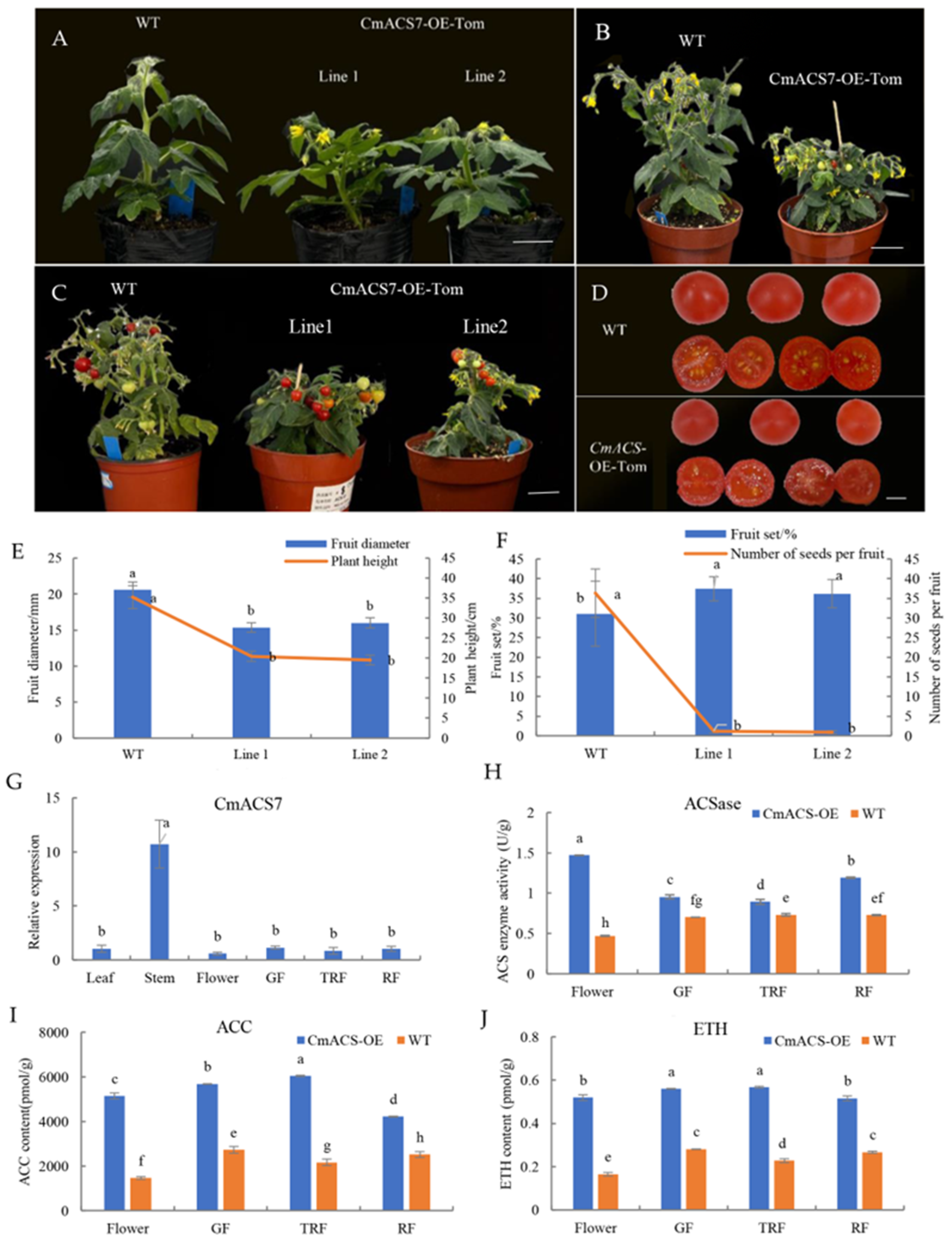
2.10. Genetic Transformation of Micro S. lycoperisicum
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Identification and Protein Structure Analysis of the CmACSs
4.3. Analysis of Cis-Acting Elements of ACS Promoters
4.4. Multiple-Sequence Comparison and Construction of the Phylogenetic Evolutionary Tree of CmACSs
4.5. Chromosome Location and Collinearity Analysis
4.6. Analysis of the Expression of CmACSs in Different Tissues
4.7. Expression Analysis of CmACSs during Ovule Development
4.8. Construction of CmACS7 Gene Overexpression and Interference Vectors
4.9. Subcellular Localization
4.10. Genetic Transformation of Chestnut Healing Tissues
4.11. Transformation of S. lycoperisicum by A. tumefaciens Containing CmACS7
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xu, C.; Hao, B.; Sun, G.; Mei, Y.; Sun, L.; Sun, Y.; Wang, Y.; Zhang, Y.Y.; Zhang, W.; Zhang, M.; et al. Dual activities of ACC synthase: Novel clues regarding the molecular evolution of ACS genes. Sci. Adv. 2021, 7, eabg8752. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Zhong, S.; Grierson, D. Recent advances in ethylene research. J. Exp. Bot. 2009, 60, 3311–3336. [Google Scholar] [CrossRef] [PubMed]
- Zeng, C.C. Identification of ACS Genes Regulating Peanut Pod Development. Master’s Thesis, Jiangxi Agricultural University, Nanchang, China, 2021. [Google Scholar] [CrossRef]
- Liu, L. Advances in ACC synthase gene research in higher plants. Tianjin Agric. Sci. 2013, 19, 22–25. [Google Scholar]
- Zhang, T.-C.; Qiao, Q.; Zhong, Y. Detecting adaptive evolution and functional divergence in aminocyclopropane-1-Carboxylate synthase (ACS) gene family. Comput. Biol. Chem. 2012, 38, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Lv, S.F.; Jiang, J. Advances in the Arabidopsis thaliana ethylene synthase ACS gene family. Biotechnol. Bull. 2014, 11, 7–13. [Google Scholar] [CrossRef]
- Gamalero, E.; Lingua, G.; Glick, B.R. Ethylene, ACC, and the plant growth-promoting enzyme ACC deaminase. Biology 2023, 12, 1043. [Google Scholar] [CrossRef] [PubMed]
- Arteca, J.M.; Arteca, R.N. A multi-responsive gene encoding 1-aminocyclopropane-1-carboxylate synthase (ACS6) in mature Arabidopsis leaves. Plant Mol. Biol. 1999, 39, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.J.; Chang, C.L.; Wang, P.H.; Tsai, M.C.; Hsu, P.H.; Chang, I.F. A type III ACC synthase, ACS7, is involved in root gravitropism in Arabidopsis thaliana. J. Exp. Bot. 2013, 64, 4343–4360. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Liu, H.; Ding, X.; Qiu, J.; Zhang, M.; Chu, Z. Arabidopsis thaliana ACS8 plays a crucial role in the early biosynthesis of ethylene elicited by Cu2+ ions. J. Cell Sci. 2018, 131, jcs202424. [Google Scholar] [CrossRef]
- Lincoln, J.E.; Campbell, A.D.; Oetiker, J.; Rottmann, W.H.; Oeller, P.W.; Shen, N.F.; Theologis, A. LE-ACS4, a Fruit ripening and wound-induced 1-aminocyclopropane-1-carboxylate synthase gene of tomato (Lycopersicon Esculentum). Expression in escherichia coli, structural characterization, expression characteristics, and phylogenetic analysis. J. Biol. Chem. 1993, 268, 19422–19430. [Google Scholar] [CrossRef]
- Wang, C.; Li, W.; Chen, F.; Cheng, Y.; Huang, X.; Zou, B.; Wang, Y.; Xu, W.; Qu, S. Genome-wide Identification and characterization of members of the ACS gene family in Cucurbita Maxima and their transcriptional responses to the specific treatments. Int. J. Mol. Sci. 2022, 23, 8476. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Oh, J.; Kim, B.; Choi, E.K.; Hwang, U.S.; Staub, J.E.; Chung, S.-M.; Park, Y. The CmaACS-7 gene provides sequence variation for development of DNA markers associated with monoecious sex expression in melon (Cucumis Melo L.). Hortic. Environ. Biotechnol. 2015, 56, 535–545. [Google Scholar] [CrossRef]
- Plett, J.M.; Williams, M.; LeClair, G.; Regan, S.; Beardmore, T. Heterologous over-expression of ACC SYNTHASE8 (ACS8) in Populus tremula x P. alba clone 717-1B4 results in elevated levels of ethylene and induces stem dwarfism and reduced leaf size through separate genetic pathways. Front. Plant Sci. 2014, 5, 514. [Google Scholar] [CrossRef] [PubMed]
- Aguado, E.; García, A.; Manzano, S.; Valenzuela, J.L.; Cuevas, J.; Pinillos, V.; Jamilena, M. The sex-determining gene CitACS4 is a pleiotropic regulator of flower and fruit development in watermelon (Citrullus lanatus). Plant Reprod. 2018, 31, 411–426. [Google Scholar] [CrossRef] [PubMed]
- Tsuchisaka, A.; Theologis, A. Unique and overlapping expression patterns among the Arabidopsis 1-amino-cyclopropane-1-carboxylate synthase gene family members. Plant Physiol. 2004, 136, 2982–3000. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Flores-Sandoval, E.; Ahtesham, U.; Coleman, A.; Clay, J.M.; Bowman, J.L.; Chang, C. Ethylene-independent functions of the ethylene precursor ACC in marchantia polymorpha. Nat. Plants 2020, 6, 1335–1344. [Google Scholar] [CrossRef]
- Xu, S.L.; Rahman, A.; Baskin, T.I.; Kieber, J.J. Two leucine-rich repeat receptor kinases mediate signaling, linking cell wall biosynthesis and ACC synthase in Arabidopsis. Plant Cell 2008, 20, 3065–3079. [Google Scholar] [CrossRef]
- Basu, D.; Tian, L.; Debrosse, T.; Poirier, E.; Emch, K.; Herock, H.; Travers, A.; Showalter, A.M. Glycosylation of a Fasciclin-Like Arabinogalactan-Protein (SOS5) mediates root growth and seed mucilage adherence via a cell wall Receptor-Like Kinase (FEI1/FEI2) pathway in Arabidopsis. PLoS ONE 2016, 11, e0145092. [Google Scholar] [CrossRef]
- Tsang, D.L.; Edmond, C.; Harrington, J.L.; Nühse, T.S. Cell wall integrity controls root elongation via a general 1-aminocyclopropane-1-carboxylic acid-dependent, ethylene-independent pathway. Plant Physiol. 2011, 156, 596–604. [Google Scholar] [CrossRef]
- Mou, W.; Kao, Y.-T.; Michard, E.; Simon, A.A.; Li, D.; Wudick, M.M.; Lizzio, M.A.; Feijó, J.A.; Chang, C. Ethylene-independent signaling by the ethylene precursor ACC in Arabidopsis ovular pollen tube attraction. Nat. Commun. 2020, 11, 4082. [Google Scholar] [CrossRef]
- Cui, Y.; Ji, X.; Zhang, Y.; Liu, Y.; Bai, Q.; Su, S. Transcriptomic and metabolic profiling reveal the mechanism of ovule development in Castanea mollissima. Int. J. Mol. Sci. 2024, 25, 1974. [Google Scholar] [CrossRef]
- Cui, Y.H.; Li, L.; Bai, Q.; Yang, Q.; Su, S. Chai. Identification of the Hsp90 gene family in chestnut and analysis of its expression during ovule development. J. Beijing For. Univ. 2022, 44, 10–19. [Google Scholar] [CrossRef]
- Kawamoto, N.; Del Carpio, D.P.; Hofmann, A.; Mizuta, Y.; Kurihara, D.; Higashiyama, T.; Uchida, N.; Torii, K.U.; Colombo, L.; Groth, G.; et al. A Peptide pair coordinates regular ovule initiation patterns with seed number and fruit size. Curr. Biol. 2020, 30, 4352–4361.e4. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Guo, W.; Chen, B.; Pan, F.; Yang, H.; Zhou, J.; Wang, G.; Li, X. Transcriptional and hormonal responses in ethephon-induced promotion of femaleness in Pumpkin. Front. Plant Sci. 2021, 12, 715487. [Google Scholar] [CrossRef]
- Xing, Y.; Liu, Y.; Zhang, Q.; Nie, X.; Sun, Y.; Zhang, Z.; Li, H.; Fang, K.; Wang, G.; Huang, H.; et al. Hybrid de novo genome assembly of Chinese chestnut (Castanea mollissima). Gigascience 2019, 8, giz112. [Google Scholar] [CrossRef] [PubMed]
- Wittkopp, P.J.; Kalay, G. Cis-regulatory elements: Molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 2012, 13, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Vanderstraeten, L.; Depaepe, T.; Bertrand, S.; Van Der Straeten, D. The ethylene precursor ACC affects early vegetative development independently of ethylene signaling. Front. Plant Sci. 2019, 10, 1591. [Google Scholar] [CrossRef]
- Rowe, J.H.; Topping, J.F.; Liu, J.; Lindsey, K. Abscisic acid regulates root growth under osmotic stress conditions via an interacting hormonal network with cytokinin, ethylene and auxin. New Phytol. 2016, 211, 225–239. [Google Scholar] [CrossRef]
- Zhao, Y.; Song, C.; Brummell, D.A.; Qi, S.; Lin, Q.; Duan, Y. Jasmonic acid treatment alleviates chilling injury in peach fruit by promoting sugar and ethylene metabolism. Food Chem. 2021, 338, 128005. [Google Scholar] [CrossRef]
- Liu, S.; Lei, C.; Zhu, Z.; Li, M.; Chen, Z.; He, W.; Liu, B.; Chen, L.; Li, X.; Xie, Y. Genome-wide analysis and identification of 1-aminocyclopropane-1-carboxylate synthase (ACS) gene family in wheat (Triticum aestivum L.). Int. J. Mol. Sci. 2023, 24, 11158. [Google Scholar] [CrossRef]
- Sharma, K.; Gupta, S.; Sarma, S.; Rai, M.; Sreelakshmi, Y.; Sharma, R. Mutations in tomato 1-aminocyclopropane carboxylic acid synthase2 uncover its role in development beside fruit ripening. Plant J. Cell Mol. Biol. 2021, 106, 95–112. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, S.; Tao, Q.; Pan, J.; Si, L.; Gong, Z.; Cai, R. A putative positive feedback regulation mechanism in CsACS2 expression suggests a modified model for sex determination in cucumber (Cucumis Sativus L.). J. Exp. Bot. 2012, 63, 4475–4484. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Su, S.; Kou, Y.R. Relationship between fruit set and early fruit development of chestnut and endogenous hormone mass fraction. J. Northeast. For. Univ. 2020, 48, 55–61. [Google Scholar] [CrossRef]
- Li, L. Anatomical Observation and Preliminary Study of Molecular Mechanism of Fertilization and Embryo Development in Chestnut. Master’s Thesis, Beijing Forestry University, Beijing, China, 2020. [Google Scholar] [CrossRef]
- Oeller, P.; Lu, M.; Taylor, L.; Pike, D.; Theologis, A. Reversible inhibition of tomato fruit senescence by antisense RNA. Science 1991, 254, 437–439. [Google Scholar] [CrossRef] [PubMed]
- Trusov, Y.; Botella, J.R. Silencing of the ACC Synthase Gene ACACS2 Causes Delayed Flowering in Pineapple [Ananas Comosus (L.) Merr.]. J. Exp. Bot. 2006, 57, 3953–3960. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Sparkes, I.A.; Runions, J.; Kearns, A.; Hawes, C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006, 1, 2019–2025. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.J.; Chu, T.S.; Song, L.; Ge, J.Y.; Qin, L.; Xing, Y. Optimization and application of a transient transformation system for chestnut healing tissues. Northwest J. Bot. 2022, 42, 154–161. [Google Scholar] [CrossRef]
- Cortina, C.; Culiáñez-Macià, F.A. Tomato transformation and transgenic plant production. Plant Cell Tissue Organ Cult. 2004, 76, 269–275. [Google Scholar] [CrossRef]
| Gene Name | Gene ID | Subcellular Localization | CDS Length/bp | No. of Exons | Protein Characteristics | Signal Peptide Sites | |||
|---|---|---|---|---|---|---|---|---|---|
| Amino Acid Length/aa | Instability Coefficient | Molecular Weight/kDa | Isoelectric Point | ||||||
| CmACS1 | Cm01G01961 | Cytosol | 1459 | 2 | 485 | 50.07 | 54.60 | 6.06 | No |
| CmACS2 | Cm03G01038 | Cytosol | 1435 | 4 | 477 | 45.94 | 53.82 | 8.14 | No |
| CmACS3 | Cm08G00844 | Nucleus | 1414 | 6 | 470 | 41.07 | 53.25 | 6.90 | No |
| CmACS4 | Cm09G01545 | Cytosol | 1417 | 4 | 471 | 43.91 | 53.14 | 5.54 | No |
| CmACS5 | Cm12G01476 | Cytosol | 1477 | 4 | 491 | 43.43 | 55.17 | 6.74 | No |
| CmACS6 | Cm12G01477 | Cytosol | 1477 | 4 | 491 | 43.43 | 55.17 | 6.74 | No |
| CmACS7 | Cm12G01910 | Nucleus | 1607 | 5 | 535 | 50.79 | 59.25 | 7.02 | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, Y.; Ji, X.; Yu, W.; Liu, Y.; Bai, Q.; Su, S. Genome-Wide Characterization and Functional Validation of the ACS Gene Family in the Chestnut Reveals Its Regulatory Role in Ovule Development. Int. J. Mol. Sci. 2024, 25, 4454. https://doi.org/10.3390/ijms25084454
Cui Y, Ji X, Yu W, Liu Y, Bai Q, Su S. Genome-Wide Characterization and Functional Validation of the ACS Gene Family in the Chestnut Reveals Its Regulatory Role in Ovule Development. International Journal of Molecular Sciences. 2024; 25(8):4454. https://doi.org/10.3390/ijms25084454
Chicago/Turabian StyleCui, Yanhong, Xingzhou Ji, Wenjie Yu, Yang Liu, Qian Bai, and Shuchai Su. 2024. "Genome-Wide Characterization and Functional Validation of the ACS Gene Family in the Chestnut Reveals Its Regulatory Role in Ovule Development" International Journal of Molecular Sciences 25, no. 8: 4454. https://doi.org/10.3390/ijms25084454




