Development of a New Model System to Study Long-Distance Interactions Supported by Architectural Proteins
Abstract
:1. Introduction
2. Results
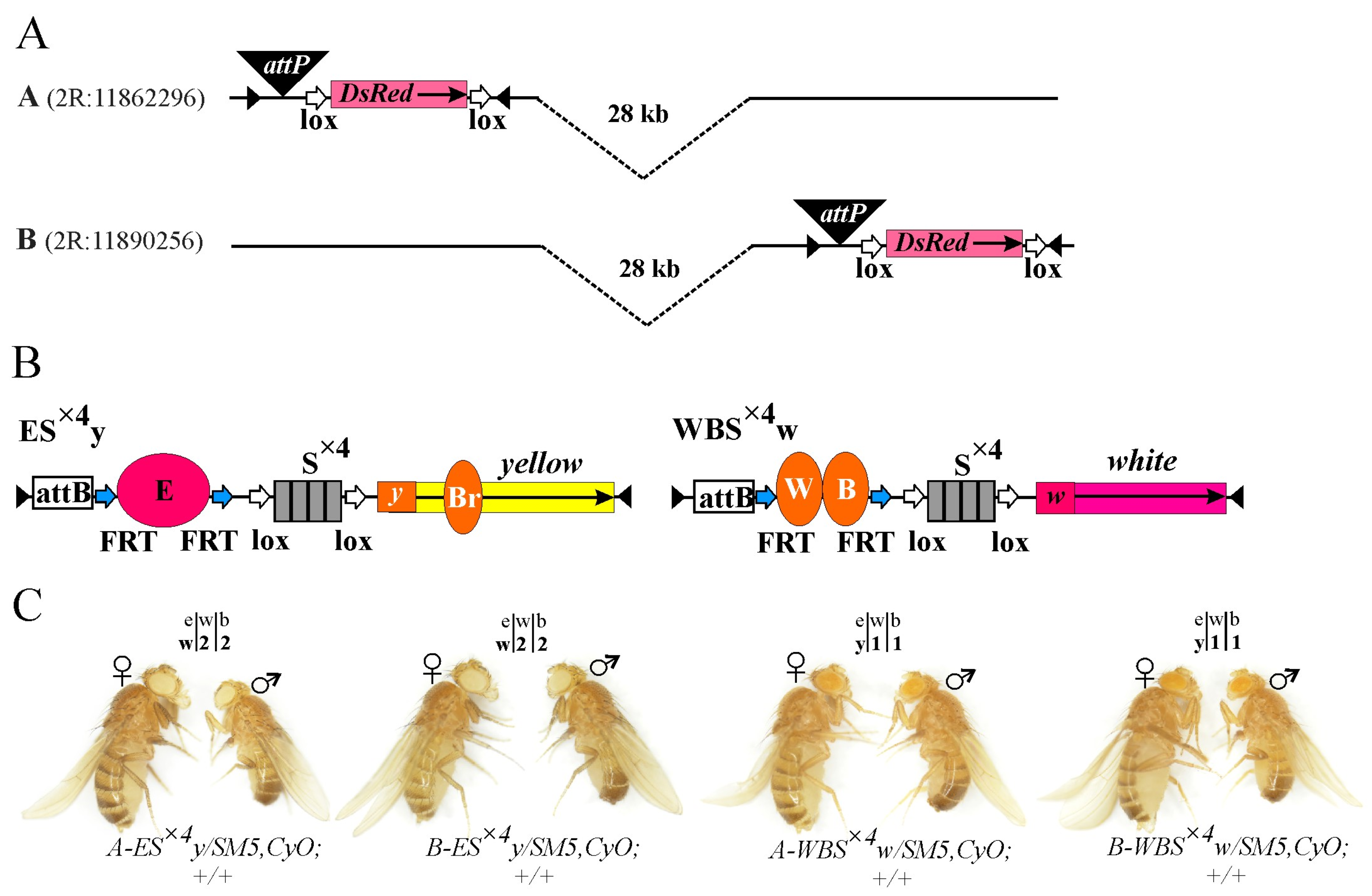
2.1. Generation of a Model System for Testing Long-Distance Interactions between Regulatory Elements
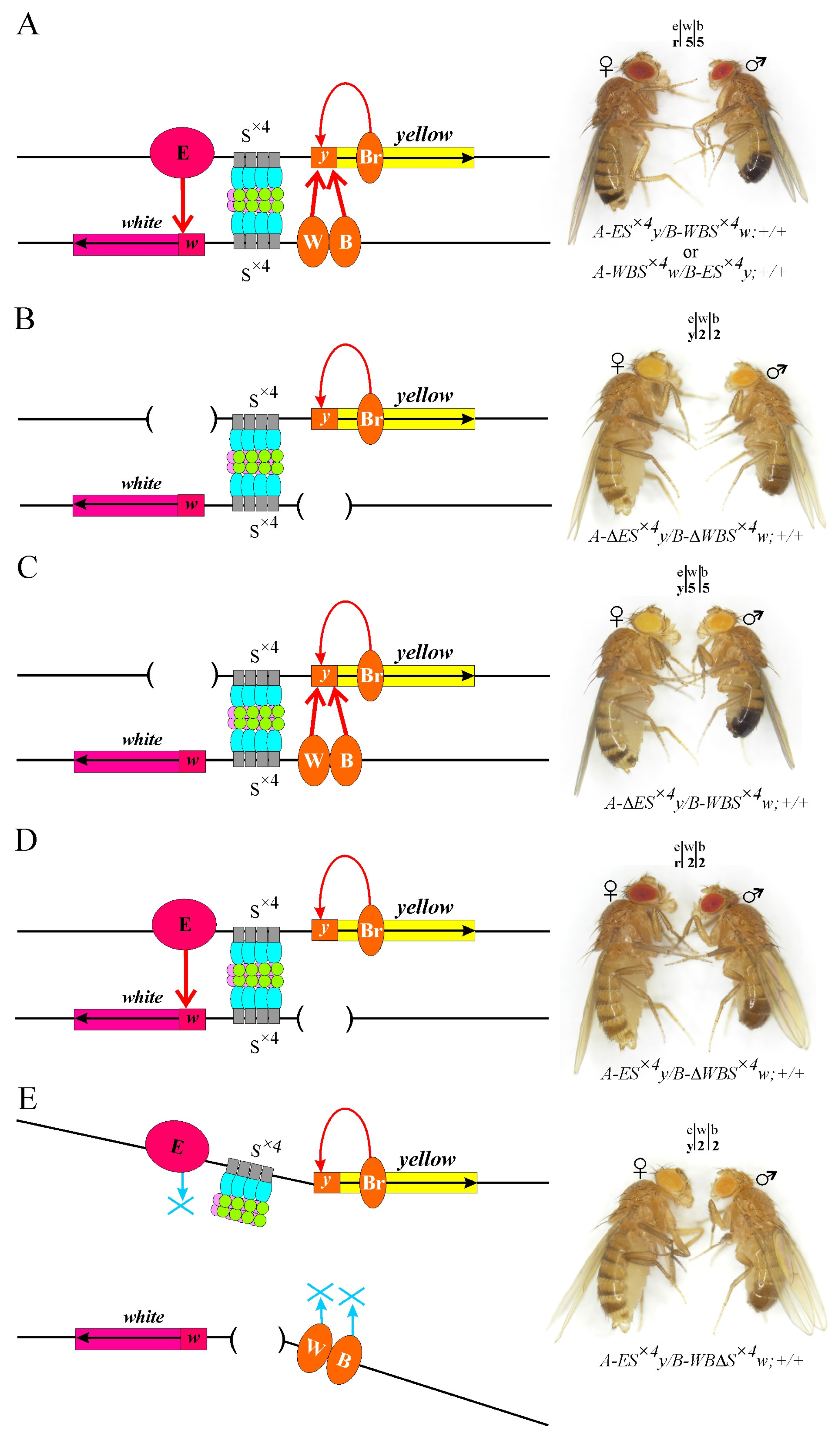
2.2. Artificial Su(Hw) Protein Binding Sites Can Support Long-Distance Interactions between Enhancers and Promoters of Reporter Genes in the Model System
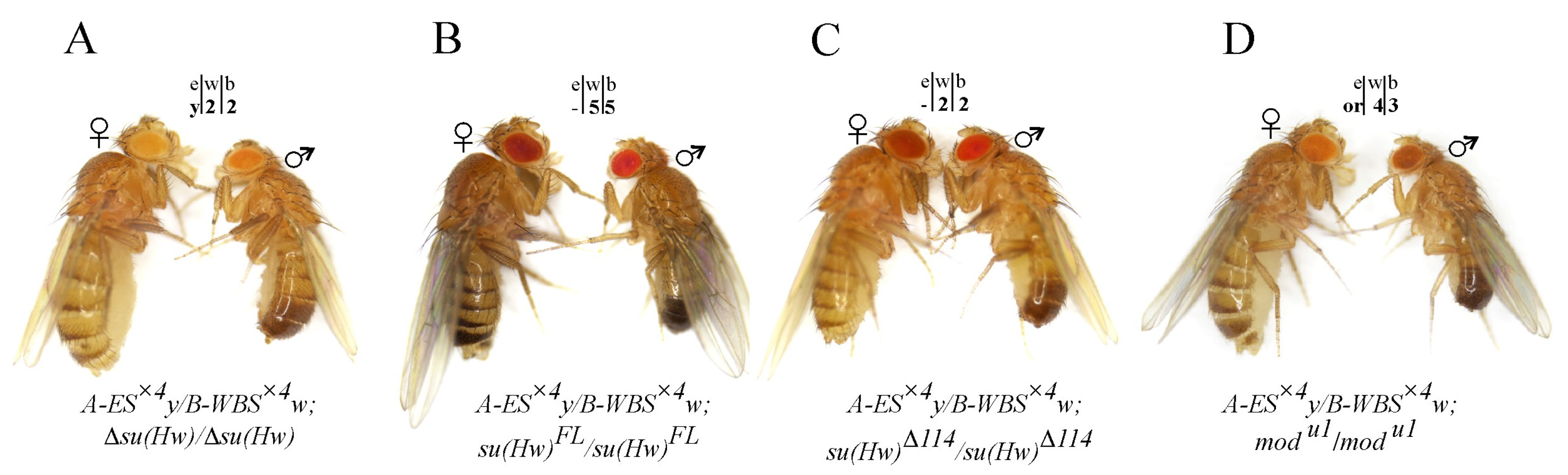
2.3. Testing the Combined Roles of Mod(mdg4)-67.2 and CP190 in Long-Distance Interactions
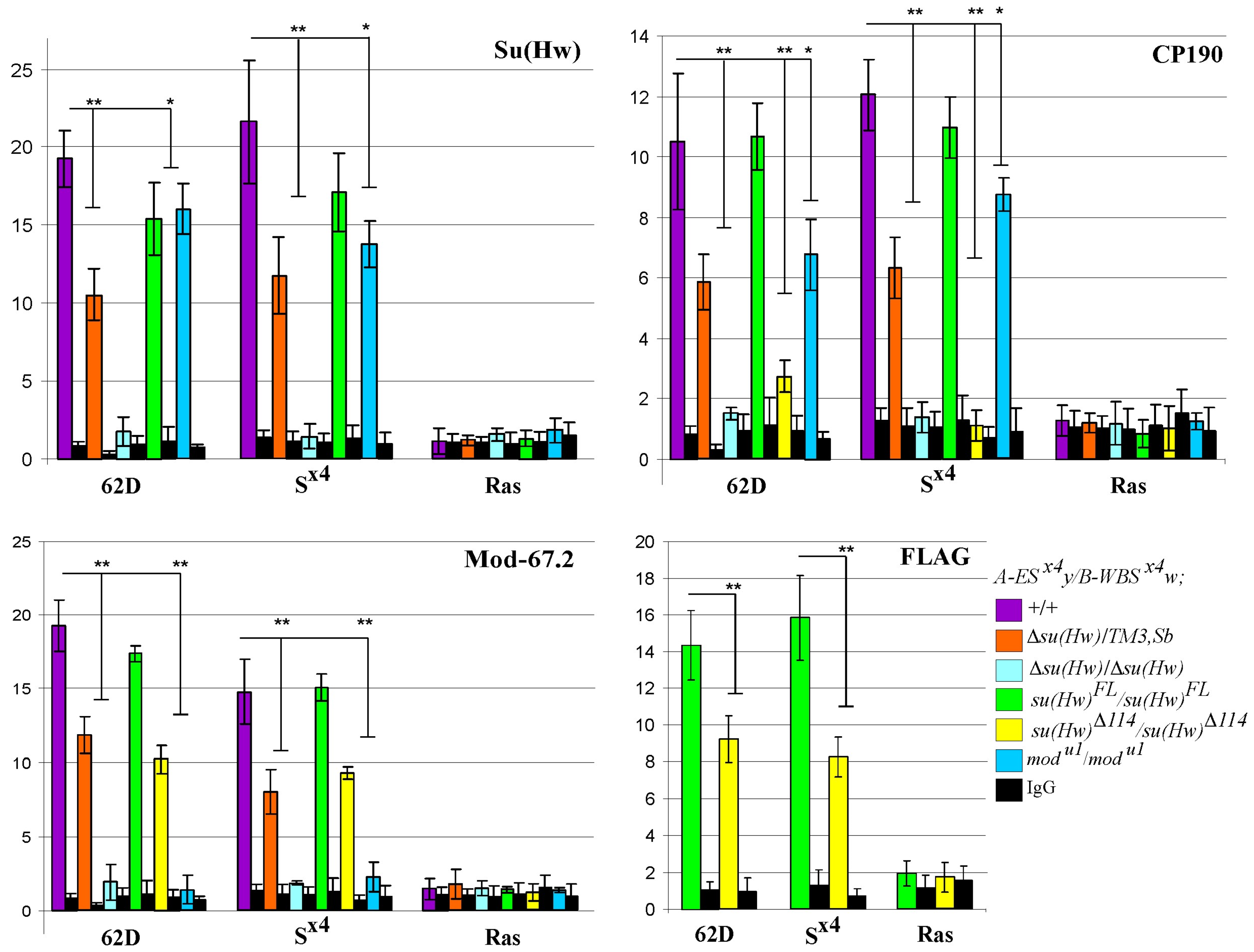
2.4. The Level of Insulator Proteins Binding to S×4 Confirms Their Role in Long-Distance Interactions
2.5. Testing Long-Distance Cis Interactions
3. Discussion
4. Materials and Methods
4.1. Plasmids and Cloning
4.2. Drosophila Strains, Transformation, and Genetic Crosses
4.3. Analysis of Yellow and White Phenotypes
4.4. Chromatin Preparation, ChIP Analysis, and Antibodies
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Van Steensel, B.; Furlong, E.E.M. The Role of Transcription in Shaping the Spatial Organization of the Genome. Nat. Rev. Mol. Cell Biol. 2019, 20, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Lim, B.; Levine, M.S. Enhancer-Promoter Communication: Hubs or Loops? Curr. Opin. Genet. Dev. 2021, 67, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Dean, A. Enhancers Navigate the Three-Dimensional Genome to Direct Cell Fate Decisions. Curr. Opin. Struct. Biol. 2021, 71, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Hafner, A.; Park, M.; Berger, S.E.; Murphy, S.E.; Nora, E.P.; Boettiger, A.N. Loop Stacking Organizes Genome Folding from TADs to Chromosomes. Mol. Cell 2023, 83, 1377–1392.e6. [Google Scholar] [CrossRef] [PubMed]
- Cavalheiro, G.R.; Pollex, T.; Furlong, E.E. To Loop or Not to Loop: What Is the Role of TADs in Enhancer Function and Gene Regulation? Curr. Opin. Genet. Dev. 2021, 67, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Di Stefano, M.; Cavalli, G. Integrative Studies of 3D Genome Organization and Chromatin Structure. Curr. Opin. Struct. Biol. 2022, 77, 102493. [Google Scholar] [CrossRef] [PubMed]
- Batut, P.J.; Bing, X.Y.; Sisco, Z.; Raimundo, J.; Levo, M.; Levine, M.S. Genome Organization Controls Transcriptional Dynamics during Development. Science 2022, 375, 566–570. [Google Scholar] [CrossRef]
- Kyrchanova, O.; Sokolov, V.; Georgiev, P. Mechanisms of Interaction between Enhancers and Promoters in Three Drosophila Model Systems. Int. J. Mol. Sci. 2023, 24, 2855. [Google Scholar] [CrossRef]
- Sigrist, C.J.; Pirrotta, V. Chromatin Insulator Elements Block the Silencing of a Target Gene by the Drosophila Polycomb Response Element (PRE) but Allow Trans Interactions between PREs on Different Chromosomes. Genetics 1997, 147, 209–221. [Google Scholar] [CrossRef]
- Muller, M.; Hagstrom, K.; Gyurkovics, H.; Pirrotta, V.; Schedl, P. The Mcp Element from the Drosophila Melanogaster Bithorax Complex Mediates Long-Distance Regulatory Interactions. Genetics 1999, 153, 1333–1356. [Google Scholar] [CrossRef]
- Li, H.-B.; Müller, M.; Bahechar, I.A.; Kyrchanova, O.; Ohno, K.; Georgiev, P.; Pirrotta, V. Insulators, Not Polycomb Response Elements, Are Required for Long-Range Interactions between Polycomb Targets in Drosophila Melanogaster. Mol. Cell Biol. 2011, 31, 616–625. [Google Scholar] [CrossRef] [PubMed]
- Bantignies, F.; Grimaud, C.; Lavrov, S.; Gabut, M.; Cavalli, G. Inheritance of Polycomb-Dependent Chromosomal Interactions in Drosophila. Genes. Dev. 2003, 17, 2406–2420. [Google Scholar] [CrossRef]
- Fujioka, M.; Emi-Sarker, Y.; Yusibova, G.L.; Goto, T.; Jaynes, J.B. Analysis of an Even-Skipped Rescue Transgene Reveals Both Composite and Discrete Neuronal and Early Blastoderm Enhancers, and Multi-Stripe Positioning by Gap Gene Repressor Gradients. Development 1999, 126, 2527–2538. [Google Scholar] [CrossRef]
- Fujioka, M.; Mistry, H.; Schedl, P.; Jaynes, J.B. Determinants of Chromosome Architecture: Insulator Pairing in Cis and in Trans. PLoS Genet. 2016, 12, e1005889. [Google Scholar] [CrossRef] [PubMed]
- Fujioka, M.; Wu, X.; Jaynes, J.B. A Chromatin Insulator Mediates Transgene Homing and Very Long-Range Enhancer-Promoter Communication. Development 2009, 136, 3077–3087. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Levo, M.; Barinov, L.; Fujioka, M.; Jaynes, J.B.; Gregor, T. Dynamic Interplay between Enhancer-Promoter Topology and Gene Activity. Nat. Genet. 2018, 50, 1296–1303. [Google Scholar] [CrossRef] [PubMed]
- Kyrchanova, O.V.; Bylino, O.V.; Georgiev, P.G. Mechanisms of Enhancer-Promoter Communication and Chromosomal Architecture in Mammals and Drosophila. Front. Genet. 2022, 13, 1081088. [Google Scholar] [CrossRef] [PubMed]
- Mazo, A.M.; Mizrokhi, L.J.; Karavanov, A.A.; Sedkov, Y.A.; Krichevskaja, A.A.; Ilyin, Y.V. Suppression in Drosophila: Su(Hw) and Su(f) Gene Products Interact with a Region of Gypsy (Mdg4) Regulating Its Transcriptional Activity. EMBO J. 1989, 8, 903–911. [Google Scholar] [CrossRef] [PubMed]
- Dorsett, D.; Viglianti, G.A.; Rutledge, B.J.; Meselson, M. Alteration of Hsp82 Gene Expression by the Gypsy Transposon and Suppressor Genes in Drosophila Melanogaster. Genes. Dev. 1989, 3, 454–468. [Google Scholar] [CrossRef]
- Holdridge, C.; Dorsett, D. Repression of Hsp70 Heat Shock Gene Transcription by the Suppressor of Hairy-Wing Protein of Drosophila Melanogaster. Mol. Cell Biol. 1991, 11, 1894–1900. [Google Scholar]
- Geyer, P.K.; Corces, V.G. DNA Position-Specific Repression of Transcription by a Drosophila Zinc Finger Protein. Genes. Dev. 1992, 6, 1865–1873. [Google Scholar] [CrossRef] [PubMed]
- Roseman, R.R.; Swan, J.M.; Geyer, P.K. A Drosophila Insulator Protein Facilitates Dosage Compensation of the X Chromosome Min-White Gene Located at Autosomal Insertion Sites. Development 1995, 121, 3573–3582. [Google Scholar] [CrossRef] [PubMed]
- Baxley, R.M.; Bullard, J.D.; Klein, M.W.; Fell, A.G.; Morales-Rosado, J.A.; Duan, T.; Geyer, P.K. Deciphering the DNA Code for the Function of the Drosophila Polydactyl Zinc Finger Protein Suppressor of Hairy-Wing. Nucleic Acids Res. 2017, 45, 4463–4478. [Google Scholar] [CrossRef] [PubMed]
- Maksimenko, O.G.; Fursenko, D.V.; Belova, E.V.; Georgiev, P.G. CTCF As an Example of DNA-Binding Transcription Factors Containing Clusters of C2H2-Type Zinc Fingers. Acta Naturae 2021, 13, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Sabirov, M.; Popovich, A.; Boyko, K.; Nikolaeva, A.; Kyrchanova, O.; Maksimenko, O.; Popov, V.; Georgiev, P.; Bonchuk, A. Mechanisms of CP190 Interaction with Architectural Proteins in Drosophila Melanogaster. Int. J. Mol. Sci. 2021, 22, 12400. [Google Scholar] [CrossRef] [PubMed]
- Golovnin, A.; Melnikova, L.; Babosha, V.; Pokholkova, G.V.; Slovohotov, I.; Umnova, A.; Maksimenko, O.; Zhimulev, I.F.; Georgiev, P. The N-Terminal Part of Drosophila CP190 Is a Platform for Interaction with Multiple Architectural Proteins. Int. J. Mol. Sci. 2023, 24, 15917. [Google Scholar] [CrossRef] [PubMed]
- Bartkuhn, M.; Straub, T.; Herold, M.; Herrmann, M.; Rathke, C.; Saumweber, H.; Gilfillan, G.D.; Becker, P.B.; Renkawitz, R. Active Promoters and Insulators Are Marked by the Centrosomal Protein 190. EMBO J. 2009, 28, 877–888. [Google Scholar] [CrossRef] [PubMed]
- Ahanger, S.H.; Shouche, Y.S.; Mishra, R.K. Functional Sub-Division of the Drosophila Genome via Chromatin Looping: The Emerging Importance of CP190. Nucleus 2013, 4, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Ahanger, S.H.; Günther, K.; Weth, O.; Bartkuhn, M.; Bhonde, R.R.; Shouche, Y.S.; Renkawitz, R. Ectopically Tethered CP190 Induces Large-Scale Chromatin Decondensation. Sci. Rep. 2014, 4, 3917. [Google Scholar] [CrossRef]
- Wang, Q.; Sun, Q.; Czajkowsky, D.M.; Shao, Z. Sub-Kb Hi-C in D. Melanogaster Reveals Conserved Characteristics of TADs between Insect and Mammalian Cells. Nat. Commun. 2018, 9, 188. [Google Scholar] [CrossRef]
- Bag, I.; Chen, S.; Rosin, L.F.; Chen, Y.; Liu, C.-Y.; Yu, G.-Y.; Lei, E.P. M1BP Cooperates with CP190 to Activate Transcription at TAD Borders and Promote Chromatin Insulator Activity. Nat. Commun. 2021, 12, 4170. [Google Scholar] [CrossRef] [PubMed]
- Bohla, D.; Herold, M.; Panzer, I.; Buxa, M.K.; Ali, T.; Demmers, J.; Krüger, M.; Scharfe, M.; Jarek, M.; Bartkuhn, M.; et al. A Functional Insulator Screen Identifies NURF and dREAM Components to Be Required for Enhancer-Blocking. PLoS ONE 2014, 9, e107765. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.Y.; Grisan, V.; Jang, B.; Herbert, J.; Badenhorst, P. Genome-Wide Mapping Targets of the Metazoan Chromatin Remodeling Factor NURF Reveals Nucleosome Remodeling at Enhancers, Core Promoters and Gene Insulators. PLoS Genet. 2016, 12, e1005969. [Google Scholar] [CrossRef] [PubMed]
- Ali, T.; Krüger, M.; Bhuju, S.; Jarek, M.; Bartkuhn, M.; Renkawitz, R. Chromatin Binding of Gcn5 in Drosophila Is Largely Mediated by CP190. Nucleic Acids Res. 2017, 45, 2384–2395. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Rosin, L.F.; Pegoraro, G.; Moshkovich, N.; Murphy, P.J.; Yu, G.; Lei, E.P. NURF301 Contributes to Gypsy Chromatin Insulator-Mediated Nuclear Organization. Nucleic Acids Res. 2022, 50, 7906–7924. [Google Scholar] [CrossRef]
- Bowman, S.K.; Deaton, A.M.; Domingues, H.; Wang, P.I.; Sadreyev, R.I.; Kingston, R.E.; Bender, W. H3K27 Modifications Define Segmental Regulatory Domains in the Drosophila Bithorax Complex. eLife 2014, 3, e02833. [Google Scholar] [CrossRef] [PubMed]
- Savitsky, M.; Kim, M.; Kravchuk, O.; Schwartz, Y.B. Distinct Roles of Chromatin Insulator Proteins in Control of the Drosophila Bithorax Complex. Genetics 2016, 202, 601–617. [Google Scholar] [CrossRef] [PubMed]
- Sabirov, M.; Kyrchanova, O.; Pokholkova, G.V.; Bonchuk, A.; Klimenko, N.; Belova, E.; Zhimulev, I.F.; Maksimenko, O.; Georgiev, P. Mechanism and Functional Role of the Interaction between CP190 and the Architectural Protein Pita in Drosophila Melanogaster. Epigenetics Chromatin 2021, 14, 16. [Google Scholar] [CrossRef] [PubMed]
- Pai, C.Y.; Lei, E.P.; Ghosh, D.; Corces, V.G. The Centrosomal Protein CP190 Is a Component of the Gypsy Chromatin Insulator. Mol. Cell 2004, 16, 737–748. [Google Scholar] [CrossRef]
- Melnikova, L.; Kostyuchenko, M.; Molodina, V.; Parshikov, A.; Georgiev, P.; Golovnin, A. Interactions between BTB Domain of CP190 and Two Adjacent Regions in Su(Hw) Are Required for the Insulator Complex Formation. Chromosoma 2018, 127, 59–71. [Google Scholar] [CrossRef]
- Buchner, K.; Roth, P.; Schotta, G.; Krauss, V.; Saumweber, H.; Reuter, G.; Dorn, R. Genetic and Molecular Complexity of the Position Effect Variegation Modifier Mod(Mdg4) in Drosophila. Genetics 2000, 155, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, D.; Gerasimova, T.I.; Corces, V.G. Interactions between the Su(Hw) and Mod(Mdg4) Proteins Required for Gypsy Insulator Function. EMBO J. 2001, 20, 2518–2527. [Google Scholar] [CrossRef] [PubMed]
- Gause, M.; Morcillo, P.; Dorsett, D. Insulation of Enhancer-Promoter Communication by a Gypsy Transposon Insert in the Drosophila Cut Gene: Cooperation between Suppressor of Hairy-Wing and Modifier of Mdg4 Proteins. Mol. Cell Biol. 2001, 21, 4807–4817. [Google Scholar] [CrossRef] [PubMed]
- Melnikova, L.; Kostyuchenko, M.; Molodina, V.; Parshikov, A.; Georgiev, P.; Golovnin, A. Multiple Interactions Are Involved in a Highly Specific Association of the Mod(Mdg4)-67.2 Isoform with the Su(Hw) Sites in Drosophila. Open Biol. 2017, 7, 170150. [Google Scholar] [CrossRef] [PubMed]
- Stogios, P.J.; Downs, G.S.; Jauhal, J.J.S.; Nandra, S.K.; Privé, G.G. Sequence and Structural Analysis of BTB Domain Proteins. Genome Biol. 2005, 6, R82. [Google Scholar] [CrossRef] [PubMed]
- Bonchuk, A.; Balagurov, K.; Georgiev, P. BTB Domains: A Structural View of Evolution, Multimerization, and Protein-Protein Interactions. Bioessays 2023, 45, e2200179. [Google Scholar] [CrossRef] [PubMed]
- Alekseyenko, A.A.; Gorchakov, A.A.; Zee, B.M.; Fuchs, S.M.; Kharchenko, P.V.; Kuroda, M.I. Heterochromatin-Associated Interactions of Drosophila HP1a with dADD1, HIPP1, and Repetitive RNAs. Genes. Dev. 2014, 28, 1445–1460. [Google Scholar] [CrossRef] [PubMed]
- Glenn, S.E.; Geyer, P.K. Investigation of the Developmental Requirements of Drosophila HP1 and Insulator Protein Partner, HIPP1. G3 2019, 9, 345–357. [Google Scholar] [CrossRef] [PubMed]
- Melnikova, L.; Molodina, V.; Erokhin, M.; Georgiev, P.; Golovnin, A. HIPP1 Stabilizes the Interaction between CP190 and Su(Hw) in the Drosophila Insulator Complex. Sci. Rep. 2019, 9, 19102. [Google Scholar] [CrossRef]
- Stow, E.C.; Simmons, J.R.; An, R.; Schoborg, T.A.; Davenport, N.M.; Labrador, M. A Drosophila Insulator Interacting Protein Suppresses Enhancer-Blocking Function and Modulates Replication Timing. Gene 2022, 819, 146208. [Google Scholar] [CrossRef]
- Oegema, K.; Marshall, W.F.; Sedat, J.W.; Alberts, B.M. Two Proteins That Cycle Asynchronously between Centrosomes and Nuclear Structures: Drosophila CP60 and CP190. J. Cell Sci. 1997, 110 Pt 14, 1573–1583. [Google Scholar] [CrossRef]
- Melnikova, L.; Molodina, V.; Babosha, V.; Kostyuchenko, M.; Georgiev, P.; Golovnin, A. The MADF-BESS Protein CP60 Is Recruited to Insulators via CP190 and Has Redundant Functions in Drosophila. Int. J. Mol. Sci. 2023, 24, 15029. [Google Scholar] [CrossRef]
- King, M.R.; Matzat, L.H.; Dale, R.K.; Lim, S.J.; Lei, E.P. The RNA-Binding Protein Rumpelstiltskin Antagonizes Gypsy Chromatin Insulator Function in a Tissue-Specific Manner. J. Cell Sci. 2014, 127, 2956–2966. [Google Scholar] [CrossRef]
- Chen, D.; Lei, E.P. Function and Regulation of Chromatin Insulators in Dynamic Genome Organization. Curr. Opin. Cell Biol. 2019, 58, 61–68. [Google Scholar] [CrossRef]
- Bag, I.; Chen, Y.; D’Orazio, K.; Lopez, P.; Wenzel, S.; Takagi, Y.; Lei, E.P. Isha Is a Su(Hw) mRNA-Binding Protein Required for Gypsy Insulator Function. G3 2022, 12, jkac152. [Google Scholar] [CrossRef]
- Gerasimova, T.I.; Byrd, K.; Corces, V.G. A Chromatin Insulator Determines the Nuclear Localization of DNA. Mol. Cell 2000, 6, 1025–1035. [Google Scholar] [CrossRef]
- Capelson, M.; Corces, V.G. The Ubiquitin Ligase dTopors Directs the Nuclear Organization of a Chromatin Insulator. Mol. Cell 2005, 20, 105–116. [Google Scholar] [CrossRef]
- Liang, J.; Lacroix, L.; Gamot, A.; Cuddapah, S.; Queille, S.; Lhoumaud, P.; Lepetit, P.; Martin, P.G.; Vogelmann, J.; Court, F.; et al. Chromatin Immunoprecipitation Indirect Peaks Highlight Long-Range Interactions of Insulator Proteins and Pol II Pausing. Mol. Cell 2014, 53, 672–681. [Google Scholar] [CrossRef]
- Vogelmann, J.; Le Gall, A.; Dejardin, S.; Allemand, F.; Gamot, A.; Labesse, G.; Cuvier, O.; Negre, N.; Cohen-Gonsaud, M.; Margeat, E.; et al. Chromatin Insulator Factors Involved in Long-Range DNA Interactions and Their Role in the Folding of the Drosophila Genome. PLoS Genet. 2014, 10, e1004544. [Google Scholar] [CrossRef]
- Maksimenko, O.; Golovnin, A.; Georgiev, P. Enhancer-Promoter Communication Is Regulated by Insulator Pairing in a Drosophila Model Bigenic Locus. Mol. Cell Biol. 2008, 28, 5469–5477. [Google Scholar] [CrossRef]
- Bell, O.; Wirbelauer, C.; Hild, M.; Scharf, A.N.D.; Schwaiger, M.; MacAlpine, D.M.; Zilbermann, F.; van Leeuwen, F.; Bell, S.P.; Imhof, A.; et al. Localized H3K36 Methylation States Define Histone H4K16 Acetylation during Transcriptional Elongation in Drosophila. EMBO J. 2007, 26, 4974–4984. [Google Scholar] [CrossRef]
- Chory, E.J.; Calarco, J.P.; Hathaway, N.A.; Bell, O.; Neel, D.S.; Crabtree, G.R. Nucleosome Turnover Regulates Histone Methylation Patterns over the Genome. Mol. Cell 2019, 73, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Sauer, B.; Henderson, N. Site-Specific DNA Recombination in Mammalian Cells by the Cre Recombinase of Bacteriophage P1. Proc. Natl. Acad. Sci. USA 1988, 85, 5166–5170. [Google Scholar] [CrossRef]
- Groth, A.C.; Olivares, E.C.; Thyagarajan, B.; Calos, M.P. A Phage Integrase Directs Efficient Site-Specific Integration in Human Cells. Proc. Natl. Acad. Sci. USA 2000, 97, 5995–6000. [Google Scholar] [CrossRef]
- Bischof, J.; Maeda, R.K.; Hediger, M.; Karch, F.; Basler, K. An Optimized Transgenesis System for Drosophila Using Germ-Line-Specific phiC31 Integrases. Proc. Natl. Acad. Sci. USA 2007, 104, 3312–3317. [Google Scholar] [CrossRef]
- Scott, K.C.; Taubman, A.D.; Geyer, P.K. Enhancer Blocking by the Drosophila Gypsy Insulator Depends upon Insulator Anatomy and Enhancer Strength. Genetics 1999, 153, 787–798. [Google Scholar] [CrossRef]
- Geyer, P.K.; Corces, V.G. Separate Regulatory Elements Are Responsible for the Complex Pattern of Tissue-Specific and Developmental Transcription of the Yellow Locus in Drosophila Melanogaster. Genes. Dev. 1987, 1, 996–1004. [Google Scholar] [CrossRef]
- Qian, S.; Varjavand, B.; Pirrotta, V. Molecular Analysis of the Zeste-White Interaction Reveals a Promoter-Proximal Element Essential for Distant Enhancer-Promoter Communication. Genetics 1992, 131, 79–90. [Google Scholar] [CrossRef]
- Geyer, P.K.; Spana, C.; Corces, V.G. On the Molecular Mechanism of Gypsy-Induced Mutations at the Yellow Locus of Drosophila Melanogaster. EMBO J. 1986, 5, 2657–2662. [Google Scholar] [CrossRef]
- McLeod, M.; Craft, S.; Broach, J.R. Identification of the Crossover Site during FLP-Mediated Recombination in the Saccharomyces Cerevisiae Plasmid 2 Microns Circle. Mol. Cell Biol. 1986, 6, 3357–3367. [Google Scholar] [CrossRef]
- Port, F.; Chen, H.-M.; Lee, T.; Bullock, S.L. Optimized CRISPR/Cas Tools for Efficient Germline and Somatic Genome Engineering in Drosophila. Proc. Natl. Acad. Sci. USA 2014, 111, E2967–E2976. [Google Scholar] [CrossRef] [PubMed]
- Harrison, D.A.; Gdula, D.A.; Coyne, R.S.; Corces, V.G. A Leucine Zipper Domain of the Suppressor of Hairy-Wing Protein Mediates Its Repressive Effect on Enhancer Function. Genes. Dev. 1993, 7, 1966–1978. [Google Scholar] [CrossRef] [PubMed]
- Butcher, R.D.; Chodagam, S.; Basto, R.; Wakefield, J.G.; Henderson, D.S.; Raff, J.W.; Whitfield, W.G. The Drosophila Centrosome-Associated Protein CP190 Is Essential for Viability but Not for Cell Division. J. Cell Sci. 2004, 117, 1191–1199. [Google Scholar] [CrossRef] [PubMed]
- Parnell, T.J.; Kuhn, E.J.; Gilmore, B.L.; Helou, C.; Wold, M.S.; Geyer, P.K. Identification of Genomic Sites That Bind the Drosophila Suppressor of Hairy-Wing Insulator Protein. Mol. Cell Biol. 2006, 26, 5983–5993. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.R.; Geyer, P.K.; Wu, C.T. Core Promoter Elements Can Regulate Transcription on a Separate Chromosome in Trans. Genes. Dev. 1999, 13, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Lim, B.; Heist, T.; Levine, M.; Fukaya, T. Visualization of Transvection in Living Drosophila Embryos. Mol. Cell 2018, 70, 287–296.e6. [Google Scholar] [CrossRef] [PubMed]
- Heist, T.; Fukaya, T.; Levine, M. Large Distances Separate Coregulated Genes in Living Drosophila Embryos. Proc. Natl. Acad. Sci. USA 2019, 116, 15062–15067. [Google Scholar] [CrossRef]
- AlHaj Abed, J.; Erceg, J.; Goloborodko, A.; Nguyen, S.C.; McCole, R.B.; Saylor, W.; Fudenberg, G.; Lajoie, B.R.; Dekker, J.; Mirny, L.A.; et al. Highly Structured Homolog Pairing Reflects Functional Organization of the Drosophila Genome. Nat. Commun. 2019, 10, 4485. [Google Scholar] [CrossRef] [PubMed]
- Duncan, I.W. Transvection Effects in Drosophila. Annu. Rev. Genet. 2002, 36, 521–556. [Google Scholar] [CrossRef]
- Kennison, J.A.; Southworth, J.W. Transvection in Drosophila. Adv. Genet. 2002, 46, 399–420. [Google Scholar] [CrossRef]
- Galouzis, C.C.; Prud’homme, B. Transvection Regulates the Sex-Biased Expression of a Fly X-Linked Gene. Science 2021, 371, 396–400. [Google Scholar] [CrossRef]
- Bateman, J.R.; Johnson, J.E. Altering Enhancer-Promoter Linear Distance Impacts Promoter Competition in Cis and in Trans. Genetics 2022, 222, iyac098. [Google Scholar] [CrossRef]
- Chen, J.-L.; Huisinga, K.L.; Viering, M.M.; Ou, S.A.; Wu, C.; Geyer, P.K. Enhancer Action in Trans Is Permitted throughout the Drosophila Genome. Proc. Natl. Acad. Sci. USA 2002, 99, 3723–3728. [Google Scholar] [CrossRef]
- Kravchuk, O.; Kim, M.; Klepikov, P.; Parshikov, A.; Georgiev, P.; Savitsky, M. Transvection in Drosophila: Trans-Interaction between Yellow Enhancers and Promoter Is Strongly Suppressed by a Cis-Promoter Only in Certain Genomic Regions. Chromosoma 2017, 126, 431–441. [Google Scholar] [CrossRef]
- King, T.D.; Johnson, J.E.; Bateman, J.R. Position Effects Influence Transvection in Drosophila Melanogaster. Genetics 2019, 213, 1289–1299. [Google Scholar] [CrossRef]
- Blick, A.J.; Mayer-Hirshfeld, I.; Malibiran, B.R.; Cooper, M.A.; Martino, P.A.; Johnson, J.E.; Bateman, J.R. The Capacity to Act in Trans Varies Among Drosophila Enhancers. Genetics 2016, 203, 203–218. [Google Scholar] [CrossRef]
- Blum, J.A.; Wells, M.; Huxley-Reicher, Z.; Johnson, J.E.; Bateman, J.R. Transvection between Nonallelic Genomic Positions in Drosophila. G3 2024, 14, jkad255. [Google Scholar] [CrossRef]
- Cavalheiro, G.R.; Girardot, C.; Viales, R.R.; Pollex, T.; Cao, T.B.N.; Lacour, P.; Feng, S.; Rabinowitz, A.; Furlong, E.E.M. CTCF, BEAF-32, and CP190 Are Not Required for the Establishment of TADs in Early Drosophila Embryos but Have Locus-Specific Roles. Sci. Adv. 2023, 9, eade1085. [Google Scholar] [CrossRef]
- Kahn, T.G.; Savitsky, M.; Kuong, C.; Jacquier, C.; Cavalli, G.; Chang, J.-M.; Schwartz, Y.B. Topological Screen Identifies Hundreds of Cp190- and CTCF-Dependent Drosophila Chromatin Insulator Elements. Sci. Adv. 2023, 9, eade0090. [Google Scholar] [CrossRef]
- Oliver, D.; Sheehan, B.; South, H.; Akbari, O.; Pai, C.Y. The Chromosomal Association/Dissociation of the Chromatin Insulator Protein Cp190 of Drosophila Melanogaster Is Mediated by the BTB/POZ Domain and Two Acidic Regions. BMC Cell Biol. 2010, 11, 101. [Google Scholar] [CrossRef]
- Plevock, K.M.; Galletta, B.J.; Slep, K.C.; Rusan, N.M. Newly Characterized Region of CP190 Associates with Microtubules and Mediates Proper Spindle Morphology in Drosophila Stem Cells. PLoS ONE 2015, 10, e0144174. [Google Scholar] [CrossRef]
- Mohan, M.; Bartkuhn, M.; Herold, M.; Philippen, A.; Heinl, N.; Bardenhagen, I.; Leers, J.; White, R.A.; Renkawitz-Pohl, R.; Saumweber, H.; et al. The Drosophila Insulator Proteins CTCF and CP190 Link Enhancer Blocking to Body Patterning. EMBO J. 2007, 26, 4203–4214. [Google Scholar] [CrossRef]
- Cuartero, S.; Fresan, U.; Reina, O.; Planet, E.; Espinas, M.L. Ibf1 and Ibf2 Are Novel CP190-Interacting Proteins Required for Insulator Function. EMBO J. 2014, 33, 637–647. [Google Scholar] [CrossRef] [PubMed]
- Zolotarev, N.; Fedotova, A.; Kyrchanova, O.; Bonchuk, A.; Penin, A.A.; Lando, A.S.; Eliseeva, I.A.; Kulakovskiy, I.V.; Maksimenko, O.; Georgiev, P. Architectural Proteins Pita, Zw5, and ZIPIC Contain Homodimerization Domain and Support Specific Long-Range Interactions in Drosophila. Nucleic Acids Res. 2016, 44, 7228–7241. [Google Scholar] [CrossRef]
- Brandão, H.B.; Gabriele, M.; Hansen, A.S. Tracking and Interpreting Long-Range Chromatin Interactions with Super-Resolution Live-Cell Imaging. Curr. Opin. Cell Biol. 2021, 70, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Parteka-Tojek, Z.; Zhu, J.J.; Lee, B.; Jodkowska, K.; Wang, P.; Aaron, J.; Chew, T.-L.; Banecki, K.; Plewczynski, D.; Ruan, Y. Super-Resolution Visualization of Chromatin Loop Folding in Human Lymphoblastoid Cells Using Interferometric Photoactivated Localization Microscopy. Sci. Rep. 2022, 12, 8582. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.H.; Wu, Z.; Rhie, S.K. Characterizing Chromatin Interactions of Regulatory Elements and Nucleosome Positions, Using Hi-C, Micro-C, and Promoter Capture Micro-C. Epigenetics Chromatin 2022, 15, 41. [Google Scholar] [CrossRef]
- Chen, L.-F.; Lee, J.; Boettiger, A. Recent Progress and Challenges in Single-Cell Imaging of Enhancer-Promoter Interaction. Curr. Opin. Genet. Dev. 2023, 79, 102023. [Google Scholar] [CrossRef]
- Loubiere, V.; Delest, A.; Schuettengruber, B.; Martinez, A.-M.; Cavalli, G. Chromatin Immunoprecipitation Experiments from Whole Drosophila Embryos or Larval Imaginal Discs. Bio-Protocol 2017, 7, e2327. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Melnikova, L.; Molodina, V.; Georgiev, P.; Golovnin, A. Development of a New Model System to Study Long-Distance Interactions Supported by Architectural Proteins. Int. J. Mol. Sci. 2024, 25, 4617. https://doi.org/10.3390/ijms25094617
Melnikova L, Molodina V, Georgiev P, Golovnin A. Development of a New Model System to Study Long-Distance Interactions Supported by Architectural Proteins. International Journal of Molecular Sciences. 2024; 25(9):4617. https://doi.org/10.3390/ijms25094617
Chicago/Turabian StyleMelnikova, Larisa, Varvara Molodina, Pavel Georgiev, and Anton Golovnin. 2024. "Development of a New Model System to Study Long-Distance Interactions Supported by Architectural Proteins" International Journal of Molecular Sciences 25, no. 9: 4617. https://doi.org/10.3390/ijms25094617





