The Bifunctional Effects of Lactoferrin (LFcinB11) in Inhibiting Neural Cell Adhesive Molecule (NCAM) Polysialylation and the Release of Neutrophil Extracellular Traps (NETs)
Abstract
:1. Introduction
2. Results
2.1. CD Data
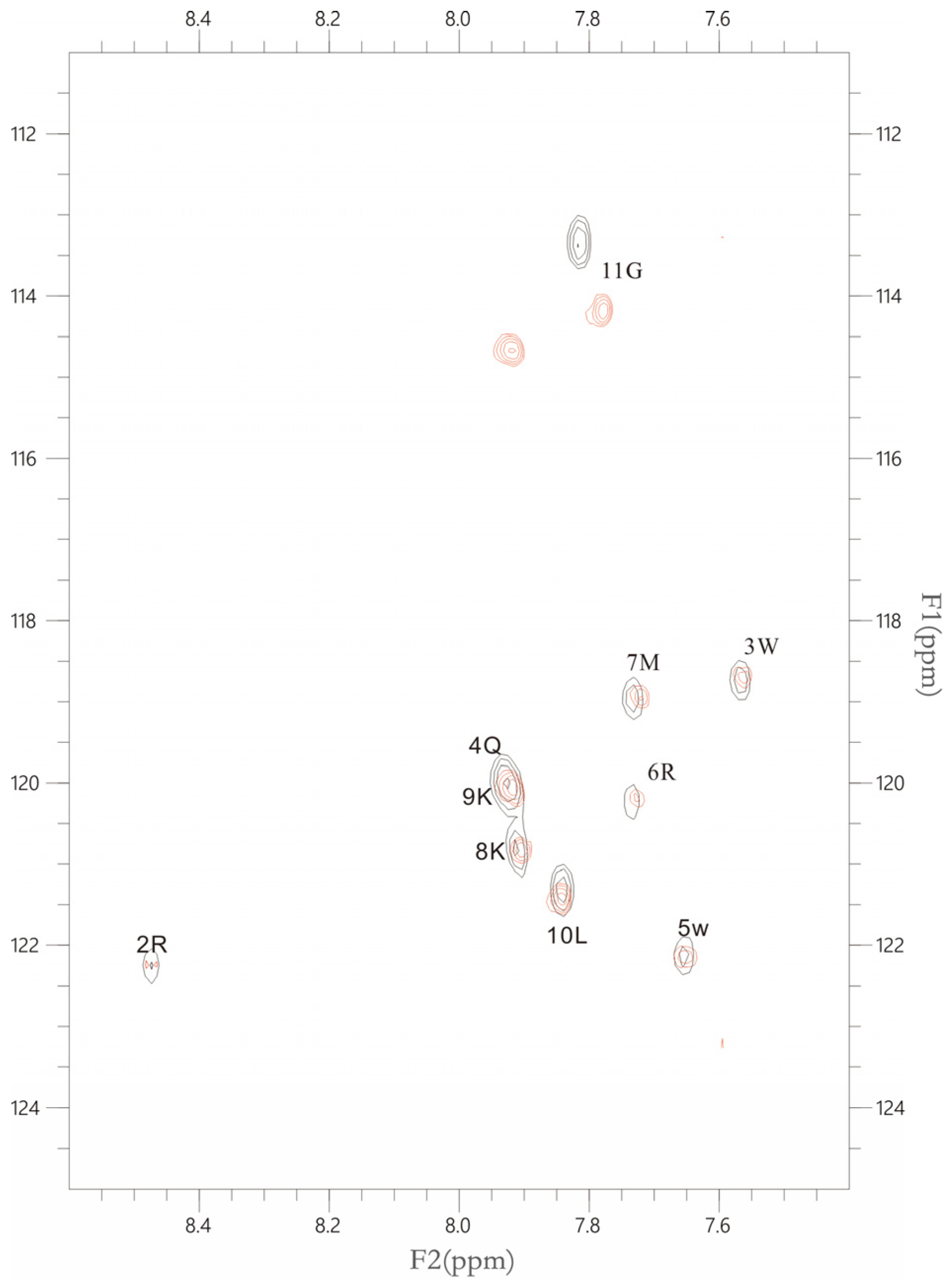
2.2. NMR Results (a)
2.2.1. The Interaction between the PSTD and 20 μM LFcinB11
2.2.2. The Interaction between the PSTD and 40 μM LFcinB11
2.2.3. The Interaction between the PSTD and 60 μM LFcinB11
2.2.4. The Interaction between the PSTD and 80 μM LFcinB11
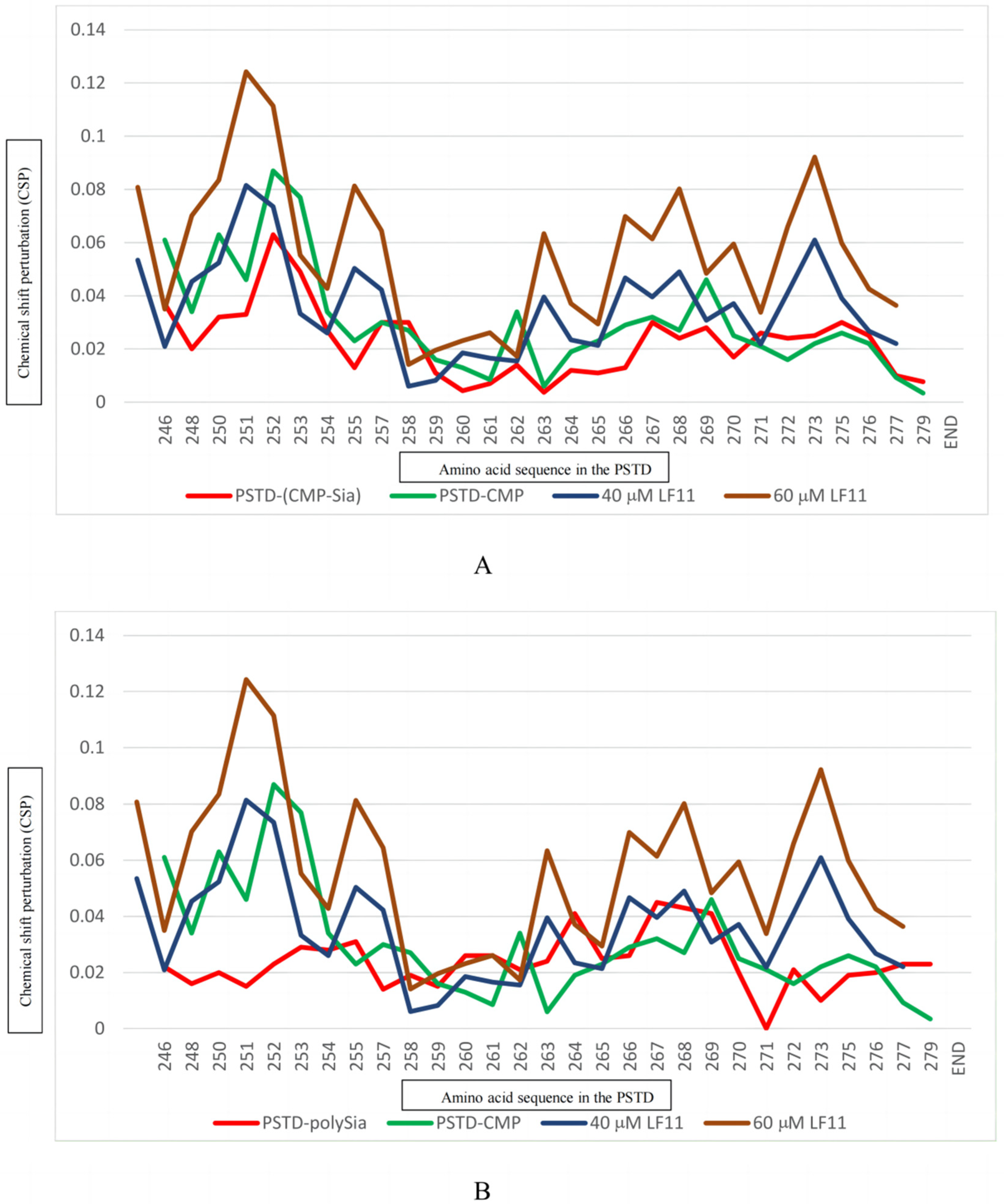
2.2.5. The Interactions between LFcinB11 and polySia
3. Discussion
4. Materials and Methods
4.1. Material Sources
4.2. Circular Dichroism (CD) Spectroscopy
4.3. NMR Sample Preparation
4.4. NMR Spectroscopic Methods
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| polySia | Polysialic acid |
| Sia | Mono-sialic acid |
| CMP-Sia | Cytidine monophosphate-sialic acid |
| NCAMs | Neural cell adhesion molecules |
| polySTs | Polysialyltransferases (ST8Sia II (STX) and ST8Sia IV (PST) |
| PSTD | Polysialyltransferase domain |
| LMWH | Low-molecular-weight heparin |
| CMP | Cytidine monophosphate |
| LF | Lactoferrin |
| CSP | Chemical shift perturbation |
References
- Lepers, A.H.; Petit, D.; Mollicone, R.; Delannoy, P.; Petit, J.M.; Oriol, R. Evolutionary history of the alpha2,8-sialyltransferase (ST8Sia) gene family: Tandem duplications in early deuterostomes explain most of the diversity found in the vertebrate ST8Sia genes. Evol. Biol. 2008, 8, 258. [Google Scholar]
- Jeanneau, C.; Chazalet, V.; Augé, C.; Soumpasis, D.M.; Harduin-Lepers, A.; Delannoy, P.; Imberty, A.; Breton, C. Structure–function analysis of the human sialyltransferase ST3Gal I: Role of n-glycosylation and a novel conserved sialylmotif. J. Biol. Chem. 2004, 279, 13461–13468. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, K.; Kurata, K.; Kojima, N.; Kurosawa, N.; Ohta, S.; Hanai, N.; Tsuji, S.; Nishi, T. Expression cloning of a GM3-specific alpha-2,8-sialyltransferase (GD3 synthase). J. Biol. Chem. 1994, 269, 15950–15956. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, J.; Fukuda, M.N.; Hirabayashi, Y.; Kanamori, A.; Sasaki, K.; Nishi, T.; Fukuda, M. Expression cloning of a human GT3 synthase. GD3 AND GT3 are synthesized by a single enzyme. J. Biol. Chem. 1996, 271, 3684–3691. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.P.; Fujimoto, I.; Rutishauser, U.; Leckband, D.E. Direct evidence that neural cell adhesion molecule (NCAM) polysialylation increases intermembrane repulsion and abrogates adhesion. J. Biol. Chem. 2005, 280, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Seidenfaden, R.; Krauter, A.; Schertzinger, F.; Gerardy-Schahn, R.; Hildebrandt, H. Polysialic acid directs tumor cell growth by con- trolling heterophilic neural cell adhesion molecule interactions. Mol. Cell. Biol. 2003, 23, 5908–5918. [Google Scholar] [CrossRef] [PubMed]
- Eggers, K.; Werneburg, S.; Schertzinger, A.; Abeln, M.; Schiff, M.; Scharenberg, M.A.; Burkhardt, H.; Mühlenhoff, M.; Hildebrandt, H. Psialic acid controls NCAM signals at cell-cell contacts to regulate focal adhesion independent from FGF receptor activity. J. Cell Sci. 2011, 124 Pt 19, 3279–3291. [Google Scholar] [CrossRef] [PubMed]
- Troy, F.A., II. Polysialylation: From bacteria to brains. Glycobiology 1992, 2, 5–23. [Google Scholar] [CrossRef] [PubMed]
- Petit, D.; Teppa, E.; Mir, A.M.; Vicogne, D.; Thisse, C.; Thisse, B.; Filloux, C.; Harduin-Lepers, A. Integrative view of α2,3- sialyltransferases (ST3Gal) molecular and functional evolution in deuterostomes: Significance of lineage-specific losses. Mol. Biol. Evol. 2015, 32, 906–927. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.-P.; Liao, S.-M.; Chen, D.; Huang, R.-B. The Cooperative Effect between Polybasic Region (PBR) and Polysialyltransferase Domain (PSTD) within Tumor-Target Polysialyltranseferase ST8Sia II. Curr. Top. Med. Chem. 2020, 19, 2831–2841. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.M.; Lu, B.; Liu, X.H.; Lu, Z.L.; Liang, S.J.; Chen, D.; Troy Ii, F.A.; Huang, R.B.; Zhou, G.P. Molecular Interactions of the Polysialytransferase Domain (PSTD) in ST8Sia IV with CMP-Sialic Acid and Polysialic Acid Required for Polysialylation of the Neural Cell Adhesion Molecule Proteins: An NMR Study. Int. J. Mol. Sci. 2020, 21, 1590. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.M.; Liu, X.H.; Peng, L.X.; Lu, B.; Huang, R.B.; Zhou, G.P. Molecular Mechanism of Inhibition of Polysialyltransferase Domain (PSTD) by Heparin. Curr. Top. Med. Chem. 2021, 21, 1113–1120. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.; Liu, X.H.; Liao, S.M.; Lu, Z.L.; Chen, D.; Troy, F.A., II; Huang, R.B.; Zhou, G.P. A Possible Modulation Mechanism of Intramolecular and Intermolecular Interactions for NCAM Polysialylation and Cell Migration. Curr. Top. Med. Chem. 2019, 19, 2271–2282. [Google Scholar] [CrossRef]
- Zhou, G.P. The Medicinal Chemistry of Structure-based Inhibitor/Drug Design: Current Progress and Future Prospective. Curr. Top. Med. Chem. 2021, 21, 1097–1098. [Google Scholar] [CrossRef] [PubMed]
- Nakata, D.; Zhang, L.; Troy, F.A. Molecular basis for polysialylation: A novel polybasic polysialyltransferase domain (PSTD) of 32 amino acids unique to the α2,8-polysialyltransferases is essential for polysialylation. Glycoconj. J. 2006, 23, 423–436. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.P.; Chou, K.C. Two Latest Hot Researches in Medicinal Chemistry. Curr. Top. Med. Chem. 2020, 20, 264–265. [Google Scholar] [CrossRef] [PubMed]
- Guan, F.; Wang, X.; He, F. Promotion of Cell Migration by Neural Cell Adhesion Molecule (NCAM) Is Enhanced by PSA in a Polysialyltransferase-Specific Manner. PLoS ONE 2015, 10, e0124237. [Google Scholar] [CrossRef] [PubMed]
- Elkashef, S.M.; Allison, S.J.; Sadiq, M.; Basheer, H.A.; Morais, G.R.; Loadman, P.M.; Pors, K.; Falconer, R.A. Polysialic acid sustains cancer cell survival and migratory capacity in a hypoxic environment. Sci. Rep. 2016, 6, 33026. [Google Scholar] [CrossRef]
- Al-Saraireh, Y.M.; Sutherland, M.; Springett, B.R.; Freiberger, F.; Morais, G.R.; Loadman, P.R.; Errington, R.J.; Smith, P.J.; Fukuda, M.; Gerardy-Schahn, R.; et al. Pharmacological inhibition of polysialyltransferase ST8SiaII modulates tumour cell migration. PLoS ONE 2013, 8, e73366. [Google Scholar] [CrossRef]
- Li, J.; Dai, G.; Cheng, Y.B.; Qi, X.; Geng, M.Y. Polysialylation promotes neural cell adhesion molecule-mediated cell migration in a fibroblast growth factor receptor-dependent manner, but independent of adhesion capability. Glycobiology 2011, 21, 1010–1018. [Google Scholar] [CrossRef]
- Peng, L.X.; Liu, X.H.; Lu, B.; Liao, S.M.; Zhou, F.; Huang, J.M.; Chen, D.; Troy, I.I.F.A.; Zhou, G.P.; Huang, R.B. The inhibition of polysialyltranseferase st8siaiv through heparin binding to polysialyltransferase domain (PSTD). Med. Chem. 2019, 15, 486–495. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.C.; Liu, W.; Maggiora, G.M.; Zhang, C.T. Prediction and Classification of Domain Structural Classes. Proteins Struct. Funct. Bioinform. 1998, 31, 97–103. [Google Scholar] [CrossRef]
- Chou, K.C.; Maggiora, G.M. Domain Structural Class Prediction. Protein Eng. 1998, 11, 523–538. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.-C.; Zhang, C.-T.; Maggiora, G.M. Disposition of amphiphilic helices in heteropolar environments. Proteins Struct. Funct. Genet. 1997, 28, 99–108. [Google Scholar] [CrossRef]
- Jungck, J.R.; Cebeci, M. Wenxiang 3.0: Evolutionary Visualization of α, π, and 3/10 Helices. Evol. Bioinform. 2022, 18, 11769343221101014. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.P.; Huang, R.B. The Graphical Studies of the Major Molecular Interactions for Neural Cell Adhesion Molecule (NCAM) Polysialylation by Incorporating Wenxiang Diagram into NMR Spectroscopy. Int. J. Mol. Sci. 2022, 23, 15128. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.-C. The Significant and Profound Impacts of Chou’s “wenxiang” Diagram. Voice Publ. 2020, 6, 102–103. [Google Scholar] [CrossRef]
- Chou, K.-C.; Lin, W.-Z.; Xiao, X. Wenxiang: A web-server for drawing wenxiang diagrams. Nat. Sci. 2011, 3, 862–865. [Google Scholar] [CrossRef]
- Babikian, V.L.; Kase, C.S.; Pessin, M.S.; Norrving, B.; Gorelick, P.B. Intracerebral hemorrhage in stroke patients anticoagulated with heparin. Stroke 1989, 20, 1500–1503. [Google Scholar] [CrossRef]
- Lu, B.; Liao, S.M.; Liu, X.H.; Liang, S.J.; Huang, J.; Lin, M.; Meng, L.; Wang, Q.Y.; Huang, R.B.; Zhou, G.P. The NMR studies of CMP inhibition of polysialylation. J. Enzyme Inhib. Med. Chem. 2023, 38, 2248411. [Google Scholar] [CrossRef]
- Bellamy, W.; Takase, M.; Yamauchi, K.; Wakabayashi, H.; Kawase, K.; Tomita, M. Identification of the bactericidal domain of lactoferrin. Biochim. Et Biophys. Acta (BBA)-Protein Struct. Mol. Enzymol. 1992, 1121, 130–136. [Google Scholar]
- Gifford, J.L.; Hunter, H.N.; Vogel, H.J. Lactoferricin: A lactoferrin-derived peptide with antimicrobial, antiviral, antitumor and immunological properties. Cell. Mol. Life Sci. 2005, 62, 2588–2598. [Google Scholar] [CrossRef]
- Kang, J.H.; Lee, M.K.; Kim, K.L.; Hahm, K.S. Structure-biological activity relationships of 11-residue highly basic peptide segment of bovine lactoferrin. Int. J. Peptide Protein Res. 1996, 48, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Kühnle, A.; Veelken, R.; Galuska, C.E.; Saftenberger, M.; Verleih, M.; Schuppe, H.C.; Rudlo, S.; Kunz, C.; Galuska, S.P. Polysialic acid interacts with lactoferrin and supports its activity to inhibit the release of neutrophil extracellular traps. Carbohydr. Polym. 2019, 208, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Kühnle, A.; Galuska, C.E.; Zlatina, K.; Galuska, S.P. The Bovine Antimicrobial Peptide Lactoferricin Interacts with Polysialic Acid without Loss of Its Antimicrobial Activity against Escherichia coli. Animals 2020, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Kühnle, A.; Lutteke, T.; Bornho, K.F.; Galuska, S.P. Polysialic acid modulates the binding of external lactoferrin in neutrophil extracellular traps. Biology 2019, 8, 20. [Google Scholar] [CrossRef]
- Ortiz, A.I.; Reglero, A.; Rodriguez-Aparicio, L.B.; Luengo, J.M. In vitro synthesis of colominic acid by membrane-bound sialyltransferase of Escherichia coli K-235. Kinetic properties of this enzyme and inhibition by CMP and other cytidine nucleotides. Eur. J. Biochem. 1989, 178, 741–749. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Piai, A.; Chen, W.; Xia, K.; Chou, J.J. Structure determination protocol for transmembrane domain oligomers. Nat. Protoc. 2019, 14, 2483–2520. [Google Scholar] [CrossRef]
- Schnell, J.R.; Zhou, G.P.; Zweckstetter, M.; Rigby, A.C.; Chou, J.J. Rapid and accurate structure determination of coiled-coil domains using NMR dipolar couplings: Application to cGMP-dependent protein kinase Ialpha. Protein Sci. 2005, 14, 2421–2428. [Google Scholar] [CrossRef]
- Sharma, A.K.; Zhou, G.P.; Kupferman, J.; Surks, H.K.; Christensen, E.N.; Chou, J.J.; Mendelsohn, M.E.; Rigby, A.C. Probing the interaction between the coiled coil leucine zipper of cGMP-dependent protein kinase Ialpha and the C terminus of the myosin binding subunit of the myosin light chain phosphatase. J. Biol. Chem. 2008, 283, 32860–32869. [Google Scholar] [CrossRef]
- Berardi, M.J.; Shih, W.M.; Harrison, S.C.; Chou, J.J. Mitochondrial uncoupling protein 2 structure determined by NMR molecular fragment searching. Nature 2011, 476, 109–113. [Google Scholar] [CrossRef] [PubMed]
- OuYang, B.; Xie, S.; Berardi, M.J.; Zhao, X.; Dev, J.; Yu, W.; Sun, B.; Chou, J.J. Unusual architecture of the p7 channel from hepatitis C virus. Nature 2013, 498, 521–525. [Google Scholar] [CrossRef]
- Oxenoid, K.; Dong, Y.; Cao, C.; Cui, T.; Sancak, Y.; Markhard, A.L.; Grabarek, Z.; Kong, L.; Liu, Z.; Ouyang, B.; et al. Architecture of the mitochondrial calcium uniporter. Nature 2016, 533, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; OuYang, B.; Chou, J.J. Critical Effect of the Detergent:Protein Ratio on the Formation of the Hepatitis C Virus p7 Channel. Biochemistry 2019, 58, 3834–3837. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Fu, T.M.; Cruz, A.C.; Sengupta, P.; Thomas, S.K.; Wang, S.; Siegel, R.M.; Wu, H.; Chou, J.J. Structural Basis and Functional Role of Intramembrane Trimerization of the Fas/CD95 Death Receptor. Mol. Cell 2016, 61, 602–613. [Google Scholar] [CrossRef] [PubMed]
- Joseph, P.R.; Poluri, K.M.; Sepuru, K.M.; Rajarathnam, K. Characterizing protein-glycosaminoglycan interactions using solution NMR spectroscopy. Methods Mol. Biol. 2015, 1229, 325–333. [Google Scholar]
- Bjorndahl, T.C.; Zhou, G.P.; Liu, X.; Perez-Pineiro, R.; Semenchenko, V.; Saleem, F.; Acharya, S.; Bujold, A.; Sobsey, C.A.; Wishart, D.S. Detailed biophysical characterization of the acid-induced PrP(c) to PrP(beta) conversion process. Biochemistry 2011, 50, 1162–1173. [Google Scholar] [CrossRef]
- Gobl, C.; Madl, T.; Simon, B.; Sattler, M. NMR approaches for structural analysis of multidomain proteins and complexes in solution. Prog. Nucl. Magn. Reson. Spectrosc. 2014, 80, 26–63. [Google Scholar] [CrossRef] [PubMed]
- Becker, W.; Bhattiprolu, K.C.; Gubensak, N.; Zangger, K. Investigating Protein-Ligand Interactions by Solution Nuclear Magnetic Resonance Spectroscopy. ChemPhysChem 2018, 19, 895–906. [Google Scholar] [CrossRef] [PubMed]
- Frueh, D.P. Practical aspects of NMR signal assignment in larger and challenging proteins. Prog. Nucl. Magn. Reson. Spectrosc. 2014, 78, 47–75. [Google Scholar] [CrossRef] [PubMed]
- Chalmers, G.R.; Eletsky, A.; Morris, L.C.; Yang, J.Y.; Tian, F.; Woods, R.J.; Moremen, K.W.; Prestegard, J.H. NMR Resonance Assignment Methodology: Characterizing Large Sparsely Labeled Glycoproteins. J. Mol. Biol. 2019, 431, 2369–2382. [Google Scholar] [CrossRef] [PubMed]
- Vaynberg, J.; Qin, J. Weak protein-protein interactions as probed by NMR spectroscopy. Trends Biotechnol. 2006, 24, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, C.; Boelens, R.; Bonvin, A.M.J.J. HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003, 125, 1731–1737. [Google Scholar] [CrossRef] [PubMed]
- Van Zundert, G.C.P.; Rodrigues, J.P.G.L.M.; Trellet, M.; Schmitz, C.; Kastritis, P.L.; Karaca, E.; Melquiond, A.S.J.; van Dijk, M.; de Vries, S.J.; Bonvin, A.M.J.J. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef] [PubMed]
- González-Ruiz, D.; Gohlke, H. Steering Protein-Ligand Docking with Quantitative NMR Chemical Shift Perturbations. J. Chem. Inf. Model. 2009, 49, 2260–2271. [Google Scholar] [CrossRef] [PubMed]
- Laveglia, V.; Giachetti, A.; Cerofolini, L.; Haubrich, K.; Fragai, M.; Ciulli, A.; Rosato, A. Automated Determination of Nuclear Magnetic Resonance Chemical Shift Perturbations in Ligand Screening Experiments: The PICASSO Web Server. J. Chem. Inf. Model. 2021, 61, 5726–5733. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Unger, S.W.; Filipp, F.V.; Sattler, M.; Szalma, S. Automated evaluation of chemical shift perturbation spectra: New approaches to quantitative analysis of receptor-ligand interaction NMR spectra. J. Biomol. NMR 2004, 29, 491–504. [Google Scholar] [CrossRef] [PubMed]
- Fino, R.; Byrne, R.; Softley, C.A.; Sattler, M.; Schneider, G.; Popowicz, G.M. Introducing the CSP Analyzer: A novel Machine Learning-based application for automated analysis of two-dimensional NMR spectra in NMR fragment-based screening. Comput. Struct. Biotechnol. J. 2020, 18, 603–611. [Google Scholar] [CrossRef]
- Williamson, M.P. Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Magn. Reson. Spectrosc. 2013, 73, 1–16. [Google Scholar] [CrossRef]
- Williamson, R.A.; Carr, M.D.; Frenkiel, T.A.; Feeney, J.; Freedman, R.B. Mapping the binding site for matrix metalloproteinase on the N-terminal domain of the tissue inhibitor of metalloproteinases-2 by NMR chemical shift perturbation. Biochemistry 1997, 36, 13882–13889. [Google Scholar] [CrossRef]
- Yadav, R.; Yoo, D.G.; Kahlenberg, J.M.; Bridges, S.L., Jr.; Oni, O.; Huang, H.; Stecenko, A.; Rada, B. Systemic levels of anti-PAD4 autoantibodies correlate with airway obstruction in cystic fibrosis. J. Cyst. Fibros. 2019, 18, 636–645. [Google Scholar] [CrossRef]
- Brinkmann, V.; Reichard, U.; Goosmann, C.; Fauler, B.; Uhlemann, Y.; Weiss, D.S.; Weinrauch, Y.; Zychlinsky, A. Neutrophil extracellular traps kill bacteria. Science 2004, 303, 1532–1535. [Google Scholar] [CrossRef] [PubMed]
- Yoo, D.G.; Winn, M.; Pang, L.; Moskowitz, S.M.; Malech, H.L.; Leto, T.L.; Rada, B. Release of cystic fibrosis airway inflammatory markers from Pseudomonas aeruginosa–stimulated human neutrophils involves NADPH oxidase-dependent extracellular DNA trap formation. J. Immunol. 2014, 192, 4728–4738. [Google Scholar] [CrossRef] [PubMed]
- Yoo, D.G.; Floyd, M.; Winn, M.; Moskowitz, S.M.; Rada, B. NET formation induced by Pseudomonas aeruginosa cystic brosis isolates measured as release of myeloperoxidase-DNA and neutrophil elastase-DNA complexes. Immunol. Lett. 2014, 160, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Manzenreiter, R.; Kienberger, F.; Marcos, V.; Schilcher, K.; Krautgartner, W.D.; Obermayer, A.; Huml, M.; Stoiber, W.; Hector, A.; Griese, M.; et al. Ultrastructural characterization of cystic fibrosis sputum using atomic force and scanning electron microscopy. J. Cyst. Fibros. 2012, 11, 84–92. [Google Scholar] [CrossRef]
- Papayannopoulos, V.; Staab, D.; Zychlinsky, A. Neutrophil elastase enhances sputum solubilization in cystic brosis patients receiving DNase therapy. PLoS ONE 2011, 6, e28526. [Google Scholar] [CrossRef]
- Li, P.; Li, M.; Lindberg, M.R.; Kennett, M.J.; Xiong, N.; Wang, Y. PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J. Exp. Med. 2010, 207, 1853–1862. [Google Scholar] [CrossRef]
- Wang, Y.; Li, M.; Stadler, S.; Correll, S.; Li, P.; Wang, D.; Hayama, R.; Leonelli, L.; Han, H.; Grigoryev, S.A.; et al. Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. J. Cell Biol. 2009, 184, 205–213. [Google Scholar] [CrossRef]
- Cheng, H.; Chen, X.; Huan, L. cDNA cloning and sequence analysis of human lactoferrin. Chin. J. Biotechnol. 2001, 17, 385–387. [Google Scholar]
- Le Provost, F.; Nocart, M.; Guerin, G.; Martin, P. Characterization of the goat lactoferrin cDNA: Assignment of the relevant locus to bovine U12 synteny group. Biochem. Biophys. Res. Commun. 1994, 203, 1324–1332. [Google Scholar] [CrossRef]
- Dwivedi, N.; Radic, M. Citrullination of autoantigens implicates NETosis in the induction of autoimmunity. Ann. Rheum. Dis. 2014, 73, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Pang, L.; Hayes, C.P.; Buac, K.; Yoo, D.G.; Rada, B. Pseudogout-associated in ammatory calcium pyrophosphate dihydrate microcrystals induce formation of neutrophil extracellular traps. J. Immunol. 2013, 190, 6488–6500. [Google Scholar] [CrossRef] [PubMed]
- Lewis, H.D.; Liddle, J.; Coote, J.E.; Atkinson, S.J.; Barker, M.D.; Bax, B.D.; Bicker, K.L.; Bingham, R.P.; Campbell, M.; Chen, Y.H.; et al. InhibitionofPAD4 activity is sufficient to disrupt mouse and human NET formation. Nat. Chem. Biol. 2015, 11, 189–191. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Ren, Y.; Lu, Q.; Wang, K.; Wu Wang, Y.; Zhang, Y.; Cui, X.; Yang, Z.; Chen, Z. Lactoferrin: A glycoprotein that plays an active role in human health. Front. Nutr. 2023, 9, 1018336. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, J.; Kanwar, R.; Kanwar, J. Lactoferrin and cancer in dierent cancer models. Front. Biosci. 2011, 3, 1080. [Google Scholar] [CrossRef] [PubMed]
- Elzoghby, A.O.; Abdelmoneem, M.A.; Hassanin, I.A.; Abd Elwakil, M.M.; Elnaggar, M.A.; Mokhtar, S.; Fang, J.Y.; Elkhodairy, K.A. Lactoferrin, a multi-functional glycoprotein: Active therapeutic, drug nanocarrier & targeting ligand. Biomaterials 2020, 263, 120355. [Google Scholar] [CrossRef]
- Morgenthau, A.; Pogoutse, A.; Adamiak, P.; Moraes, T.; Schryvers, A. Bacterial receptors for host transferrin and lactoferrin: Molecular mechanisms and role in host-microbe interactions. Future Microbiol. 2013, 8, 1575–1585. [Google Scholar] [CrossRef]
- Kell, D.; Heyden, E.; Pretorius, E. The biology of lactoferrin, an iron-binding protein that can help defend against viruses and bacteria. Front. Immunol. 2020, 11, 1221. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Meletharayil, G.; Kapoor, R.; Abbaspourrad, A. Bioactives in bovine milk: Chemistry, technology, and applications. Nutr. Rev. 2021, 79 (Suppl. 2), 48–69. [Google Scholar] [CrossRef]
- Shende, P.; Khanolkar, B. Human breast milk-based nutritherapy: A blueprint for pediatric healthcare. J. Food Drug Anal. 2021, 29, 203–213. [Google Scholar] [CrossRef]
- Dierick, M.; Vanrompay, D.; Devriendt, B.; Cox, E. Lactoferrin, a versatile natural antimicrobial glycoprotein that modulates the host’s innate immunity. Biochem. Cell Biol. 2021, 99, 61–65. [Google Scholar] [CrossRef] [PubMed]


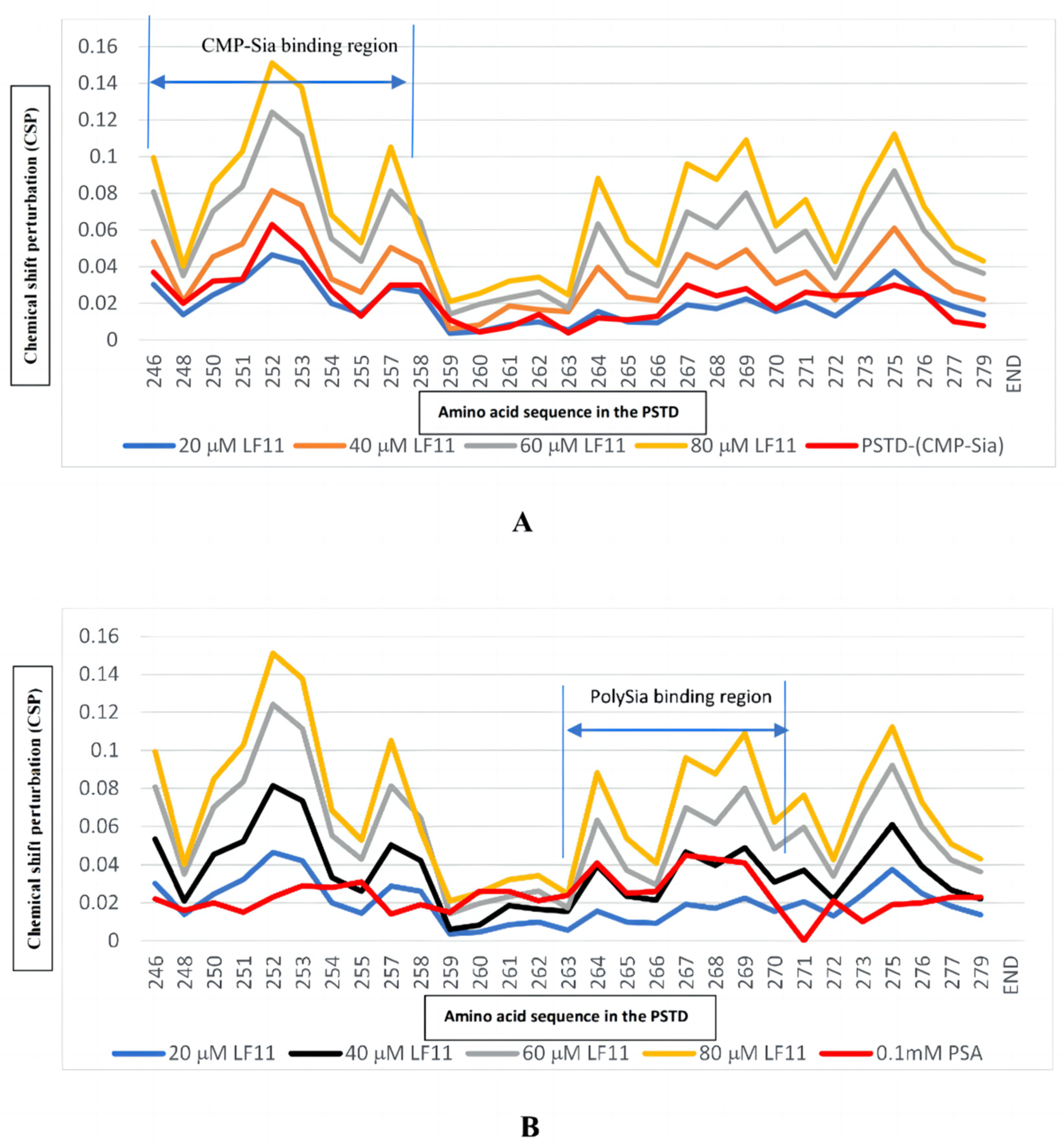

| LFcinB11 Concentration Interacting with the PSTD | Residues in the PSTD That Do Not Change in terms of Chemical Shift | Residues in the PSTD That Changed in relation to Chemical Shift |
|---|---|---|
| 20 μM | 17 residues (K248, A254, Y255, L258, R259, V260, I261, H262, A263, V264, R265, Y267, W268, L269, K272, K276, and S279) | 8 residues (K246, K250, V251, R252, T253, S257, V273, and I275) |
| 40 μM | 6 residues (R259, V260, I261, H262, A263, and K272) | 19 residues (K246, K248, K250, V251, R252, T253, A254, Y255, S257, L258, V264, R265, Y267, W268, L269, V273, I275, R277, and S279) |
| 60 μM | 3 residues (R259, V260, and A263) | 22 residues |
| 80 μM | 0 residues | 25 residues |
| Ligands Binding to the PSTD | The Maximum CSPs in CMP-Sia Binding Region (K246-L258) | The Maximum CSPs in polySia Binding Region (A263-R271) |
|---|---|---|
| CMP-Sia | 0.063 | 0.030 |
| polySia | 0.031 | 0.045 |
| Heparin LMWH (80 μM) | 0.087 | 0.072 |
| CMP (1 mM) | 0.087 | 0.046 |
| LFcinB11 (20 μM) | 0.047 | 0.038 |
| LFcinB11 (40 μM) | 0.081 | 0.061 |
| LFcinB11 (60 μM) | 0.124 | 0.092 |
| LFcinB11 (80 μM) | 0.151 | 0.109 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, B.; Liao, S.-M.; Liang, S.-J.; Peng, L.-X.; Li, J.-X.; Liu, X.-H.; Huang, R.-B.; Zhou, G.-P. The Bifunctional Effects of Lactoferrin (LFcinB11) in Inhibiting Neural Cell Adhesive Molecule (NCAM) Polysialylation and the Release of Neutrophil Extracellular Traps (NETs). Int. J. Mol. Sci. 2024, 25, 4641. https://doi.org/10.3390/ijms25094641
Lu B, Liao S-M, Liang S-J, Peng L-X, Li J-X, Liu X-H, Huang R-B, Zhou G-P. The Bifunctional Effects of Lactoferrin (LFcinB11) in Inhibiting Neural Cell Adhesive Molecule (NCAM) Polysialylation and the Release of Neutrophil Extracellular Traps (NETs). International Journal of Molecular Sciences. 2024; 25(9):4641. https://doi.org/10.3390/ijms25094641
Chicago/Turabian StyleLu, Bo, Si-Ming Liao, Shi-Jie Liang, Li-Xin Peng, Jian-Xiu Li, Xue-Hui Liu, Ri-Bo Huang, and Guo-Ping Zhou. 2024. "The Bifunctional Effects of Lactoferrin (LFcinB11) in Inhibiting Neural Cell Adhesive Molecule (NCAM) Polysialylation and the Release of Neutrophil Extracellular Traps (NETs)" International Journal of Molecular Sciences 25, no. 9: 4641. https://doi.org/10.3390/ijms25094641






