Effects of UV-B Radiation Exposure on Transgenerational Plasticity in Grain Morphology and Proanthocyanidin Content in Yuanyang Red Rice
Abstract
:1. Introduction
2. Results
2.1. Effects of UV-B Radiation Exposure on Rice Seeds and Proanthocyanidin Content
2.2. Effect of UV-B Radiation on Transgenerational Plasticity in Rice
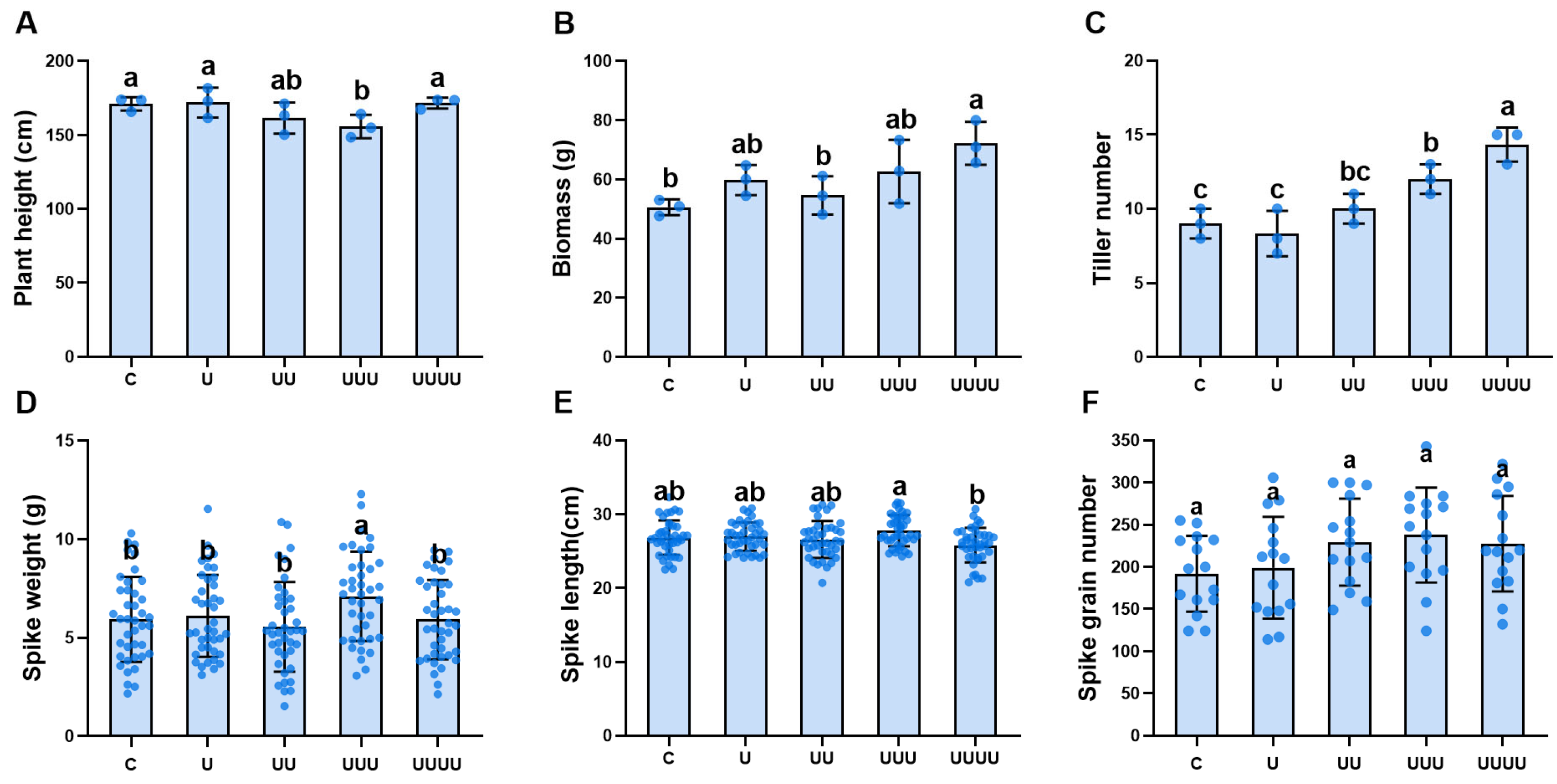
2.2.1. Transgenerational Plasticity and Rice Growth
2.2.2. Transgenerational Plasticity in Rice Grain Morphology
2.2.3. Transgenerational Plasticity in MDA/Proanthocyanidin Contents
2.2.4. Transgenerational Plasticity in Proanthocyanidin Synthase
2.3. Effects of UV-B Stress Memory on Rice Offspring
2.3.1. Memory Effect on Offspring Growth
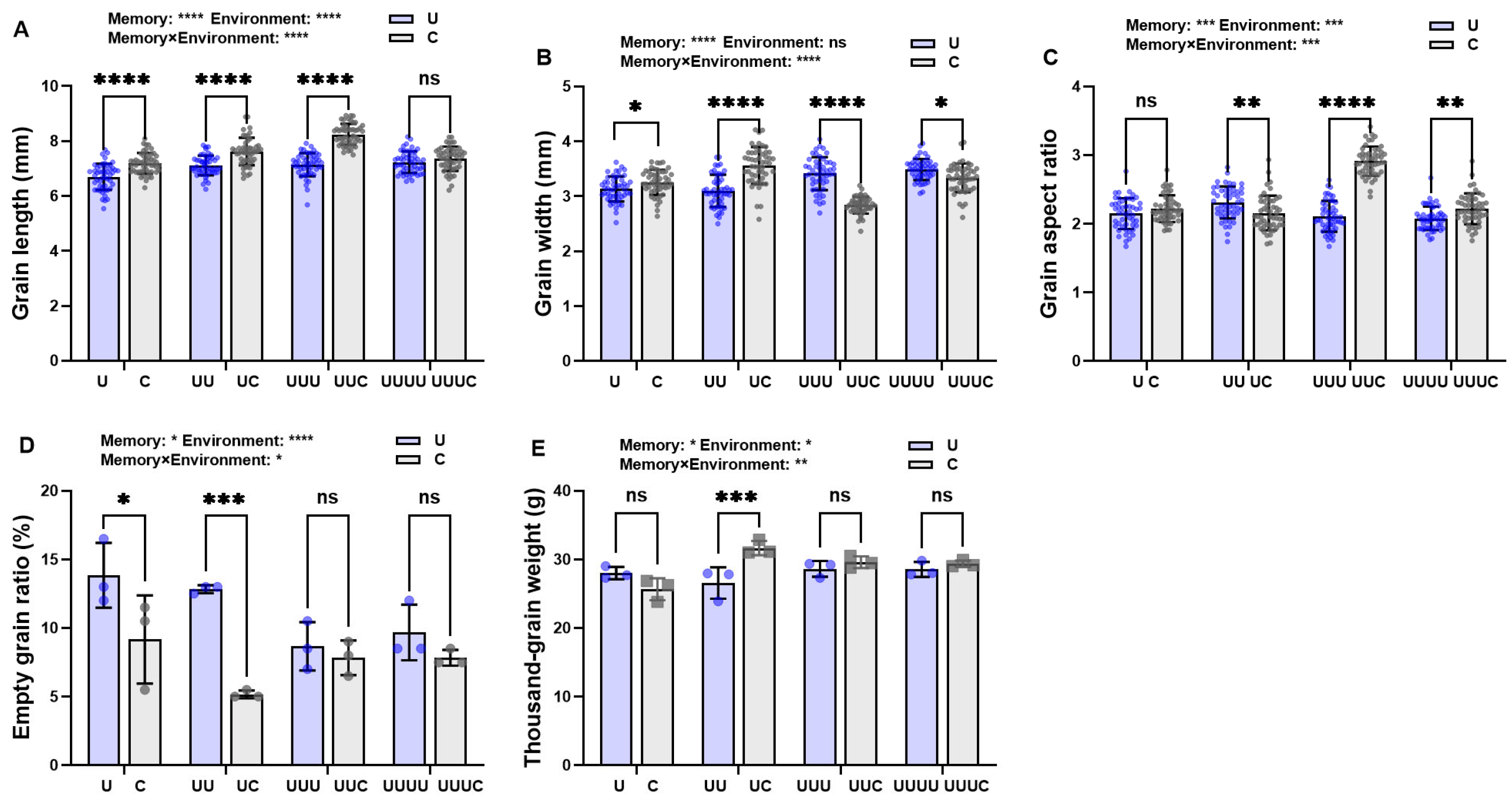
2.3.2. Memory Effect on Seed Morphology in Offspring
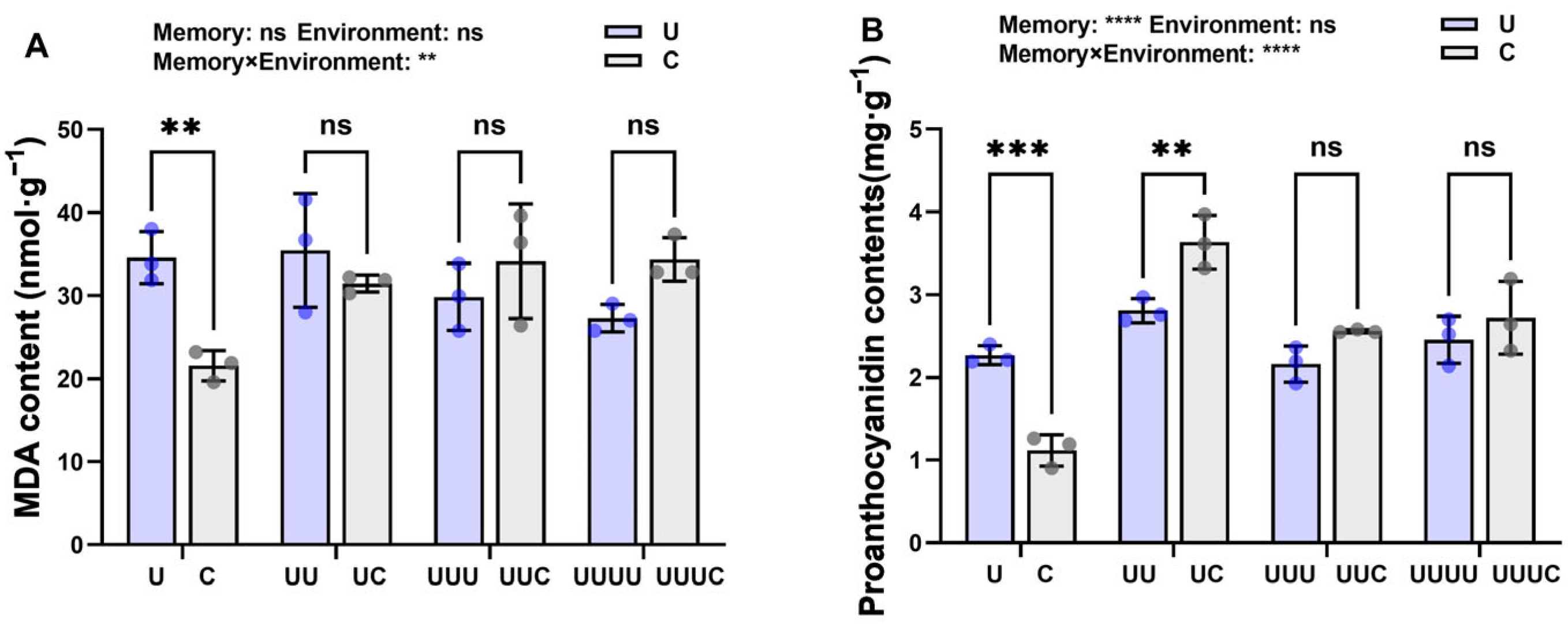
2.3.3. Memory Effect on Offspring MDA and Proanthocyanidin Content
2.3.4. Memory Effect on Enzyme Activity Regulation of Proanthocyanidin Content
2.4. Transcriptome Analysis
2.4.1. Quality Assessment of Sequencing Data
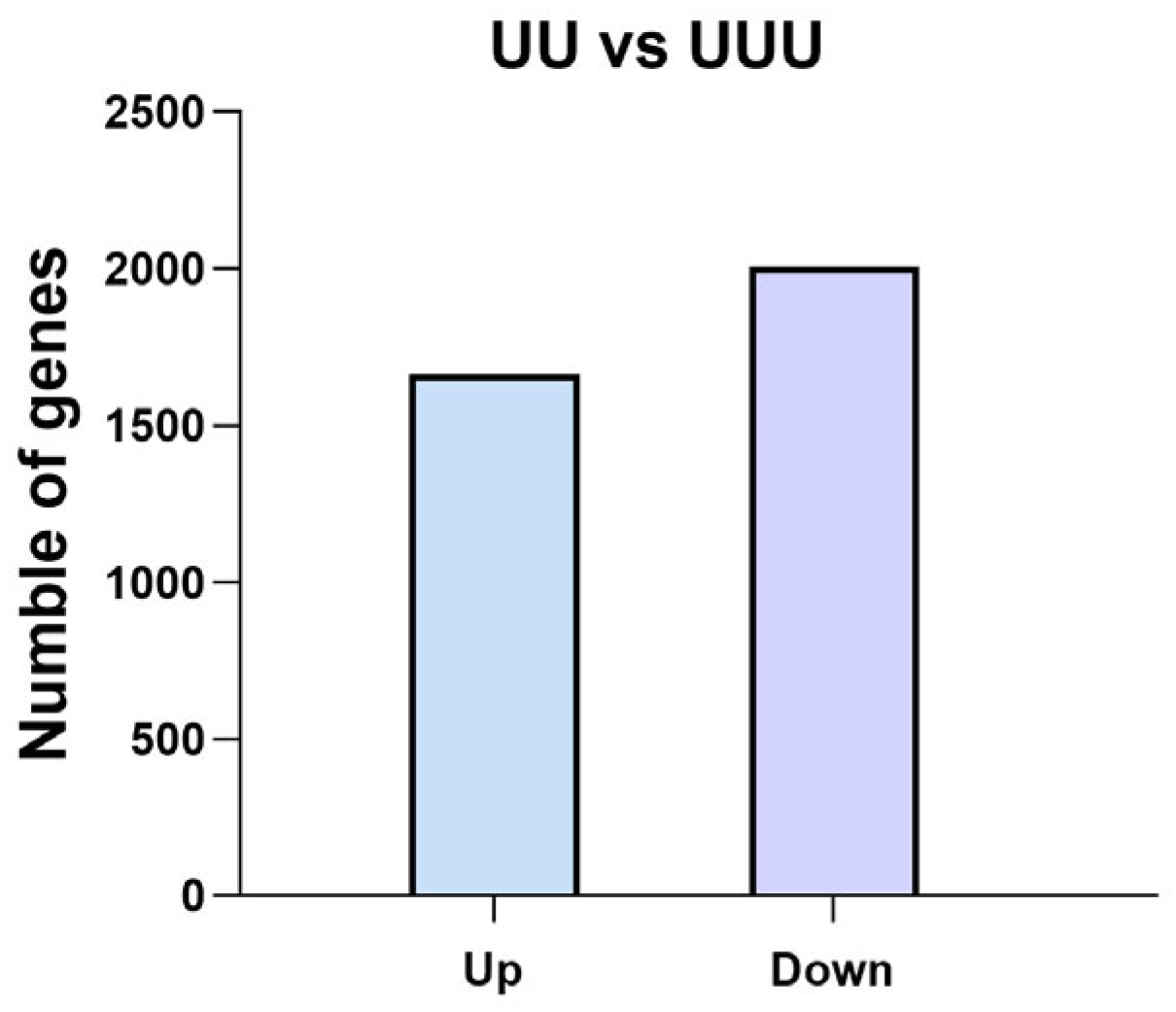
2.4.2. Number of Differentially Expressed Genes
2.4.3. GO Enrichment Analysis
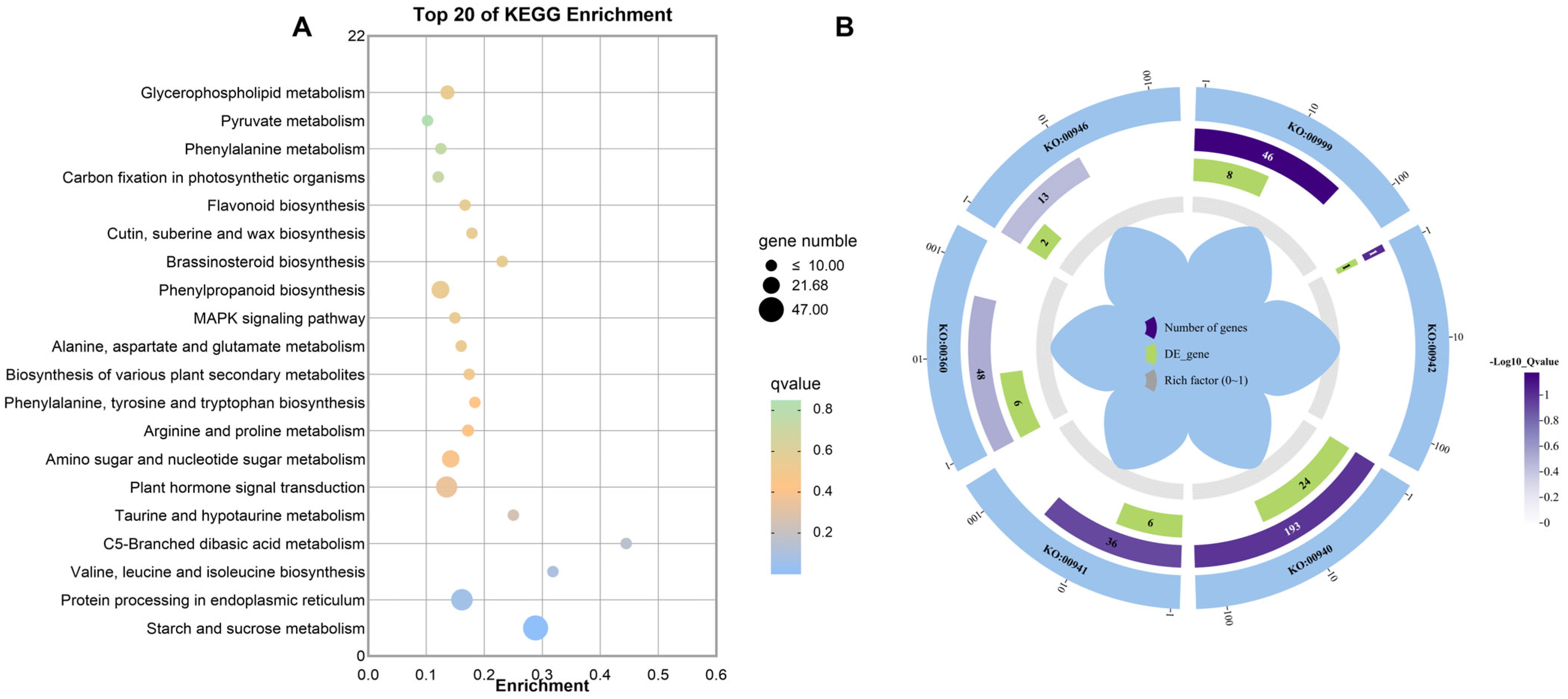
2.4.4. KEGG Enrichment Analysis
2.4.5. Screening of Target Genes
3. Discussion
3.1. Effects of Different Intensities of UV-B Radiation on Rice
3.2. Transgenerational Plasticity in Rice in Response to UV-B Radiation
3.2.1. Transgenerational Plasticity Response Characterization
3.2.2. Mechanism of Transgenerational Changes in Proanthocyanidin Content
3.3. Stress Memory Effect on Transgenerational Plasticity
4. Materials and Methods
4.1. Overview of Test Site
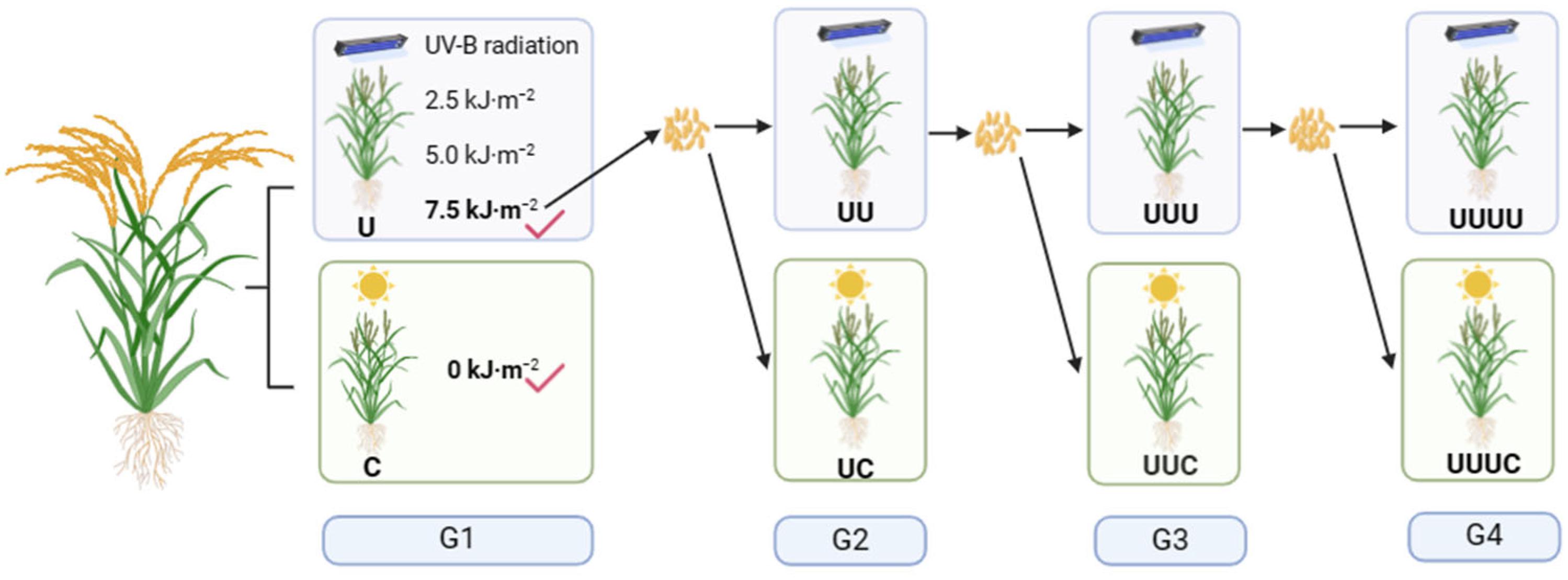
4.2. UV-B Radiation Treatment
4.3. Plant Material and Treatment
4.4. Indicator Measurements
4.4.1. Growth Traits
4.4.2. Panicle and Grain Morphological Traits
4.4.3. Physiological Traits
4.5. Transcriptome Sequencing
4.6. Plasticity Response Index
4.7. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Sample | Clean Data (bp) | AF_Q20 (%) | AF_Q30 (%) | AF_N (%) | AF_GC (%) |
|---|---|---|---|---|---|
| UU1 | 7,215,007,346 | 98.85% | 95.48% | 0.00% | 51.76% |
| UU2 | 7,386,387,904 | 98.82% | 95.33% | 0.00% | 50.99% |
| UU3 | 6,857,180,340 | 99.00% | 95.94% | 0.00% | 51.16% |
| UUU1 | 7,621,922,903 | 98.90% | 95.72% | 0.00% | 53.06% |
| UUU2 | 7,813,042,637 | 98.94% | 95.86% | 0.00% | 51.42% |
| UUU3 | 7,536,985,572 | 99.02% | 96.16% | 0.00% | 51.40% |
| Sample | Total | Unmapped | Unique Mapped | Multiple Mapped | Total Mapped |
|---|---|---|---|---|---|
| UU1 | 48,933,326 | 2,874,464 (5.87%) | 44,715,560 (91.38%) | 1,343,302 (2.75%) | 46,058,862 (94.13%) |
| UU2 | 47,575,544 | 1,840,318 (3.87%) | 41,360,594 (86.94%) | 4,374,632 (9.20%) | 45,735,226 (96.13%) |
| UU3 | 50,172,210 | 2,042,037 (4.07%) | 42,408,808 (84.53%) | 5,721,365 (11.40%) | 48,130,173 (95.93%) |
| UUU1 | 48,233,244 | 2,366,783 (4.91%) | 43,710,529 (90.62%) | 2,155,932 (4.47%) | 45,866,461 (95.09%) |
| UUU2 | 51,403,966 | 2,769,362 (5.39%) | 47,184,683 (91.79%) | 1,449,921 (2.82%) | 48,634,604 (94.61%) |
| UUU3 | 52,697,298 | 3,191,709 (6.06%) | 47,900,776 (90.90%) | 1,604,813 (3.05%) | 49,505,589 (93.94%) |
References
- Purohit, P.; Borgford-Parnell, N.; Klimont, Z.; Höglund-Isaksson, L. Achieving Paris climate goals calls for increasing ambition of the Kigali Amendment. Nat. Clim. Chang. 2022, 12, 339–342. [Google Scholar] [CrossRef]
- Bernhard, G.H.; Bais, A.F.; Aucamp, P.J.; Klekociuk, A.R.; Liley, J.B.; McKenzie, R.L. Stratospheric ozone, UV radiation, and climate interactions. Photochem. Photobiol. Sci. 2023, 22, 937–989. [Google Scholar] [CrossRef] [PubMed]
- Beyaz, R. Impact of gamma irradiation pretreatment on the growth of common vetch (Vicia sativa L.) seedlings grown under salt and drought stress. Int. J. Radiat. Biol. 2019, 96, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Robson, T.M.; Klem, K.; Urban, O.; Jansen, M.A. Re-interpreting plant morphological responses to UV-B radiation. Plant Cell Environ. 2015, 38, 856–866. [Google Scholar] [CrossRef] [PubMed]
- Julkunen-Tiitto, R.; Nenadis, N.; Neugart, S.; Robson, M.; Agati, G.; Vepsäläinen, J.; Zipoli, G.; Nybakken, L.; Winkler, B.; Jansen, M.A.K. Assessing the response of plant flavonoids to UV radiation: An overview of appropriate techniques. Phytochem. Rev. 2014, 14, 273–297. [Google Scholar] [CrossRef]
- Puy, J.; Bello, F.d.; Dvořáková, H.; Medina, N.G.; Latzel, V.; Carmona, C.P. Competition-induced transgenerational plasticity influences competitive interactions and leaf decomposition of offspring. New Phytol. 2021, 229, 3497–3507. [Google Scholar] [CrossRef] [PubMed]
- Turcotte, M.M.; Levine, J.M. Phenotypic Plasticity and Species Coexistence. Trends Ecol. Evol. 2016, 31, 803–813. [Google Scholar] [CrossRef]
- Kooke, R.; Johannes, F.; Wardenaar, R.; Becker, F.; Etcheverry, M.; Colot, V.; Vreugdenhil, D.; Keurentjes, J.J.B. Epigenetic Basis of Morphological Variation and Phenotypic Plasticity inArabidopsis thaliana. Plant Cell 2015, 27, 337–348. [Google Scholar] [CrossRef]
- Bell, A.M.; Hellmann, J.K. An Integrative Framework for Understanding the Mechanisms and Multigenerational Consequences of Transgenerational Plasticity. Annu. Rev. Ecol. Evol. Syst. 2019, 50, 97–118. [Google Scholar] [CrossRef]
- Liu, J.; Feng, L.; Li, J.; He, Z. Genetic and epigenetic control of plant heat responses. Front. Plant Sci. 2015, 6, 133426. [Google Scholar] [CrossRef]
- Ding, Y.; Fromm, M.; Avramova, Z. Multiple exposures to drought ‘train’ transcriptional responses in Arabidopsis. Nat. Commun. 2012, 3, 740. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Zhao, Y.; Liu, F.; Shi, M.; Guan, Y.; Zhang, T.; Zhao, F.; Qiao, Q.; Geng, Y. Transcriptional memory of gene expression across generations participates in transgenerational plasticity of field pennycress in response to cadmium stress. Front. Plant Sci. 2022, 13, 953794. [Google Scholar] [CrossRef]
- Li, S.; Xia, Q.; Wang, F.; Yu, X.; Ma, J.; Kou, H.; Lin, X.; Gao, X.; Liu, B. Laser Irradiation-Induced DNA Methylation Changes Are Heritable and Accompanied with Transpositional Activation of mPing in Rice. Front. Plant Sci. 2017, 8, 363. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, M.; Souri, M.K. Growth and mineral content of coriander (Coriandrum sativum L.) plants under mild salinity with different salts. Acta Physiol. Plant. 2018, 40, 194. [Google Scholar] [CrossRef]
- Souri, M.K.; Hatamian, M. Aminochelates in plant nutrition: A review. J. Plant Nutr. 2018, 42, 67–78. [Google Scholar] [CrossRef]
- Zhang, X.; Li, C.; Tie, D.; Quan, J.; Yue, M.; Liu, X. Epigenetic memory and growth responses of the clonal plant Glechoma longituba to parental recurrent UV-B stress. Funct. Plant Biol. 2021, 48, 827–838. [Google Scholar] [CrossRef]
- Yan, Y.; Stoddard, F.L.; Neugart, S.; Oravec, M.; Urban, O.; Sadras, V.O.; Aphalo, P.J. The transgenerational effects of solar short-UV radiation differed in two accessions of Vicia faba L. from contrasting UV environments. J. Plant Physiol. 2020, 248, 153145. [Google Scholar] [CrossRef]
- Donelson, J.M.; Salinas, S.; Munday, P.L.; Shama, L.N.S. Transgenerational plasticity and climate change experiments: Where do we go from here? Glob. Chang. Biol. 2018, 24, 13–34. [Google Scholar] [CrossRef]
- Ramírez, D.A.; Rolando, J.L.; Yactayo, W.; Monneveux, P.; Mares, V.; Quiroz, R. Improving potato drought tolerance through the induction of long-term water stress memory. Plant Sci. 2015, 238, 26–32. [Google Scholar] [CrossRef]
- Auler, P.A.; Souza, G.M.; da Silva Engela, M.R.G.; do Amaral, M.N.; Rossatto, T.; da Silva, M.G.Z.; Furlan, C.M.; Maserti, B.; Braga, E.J.B. Stress memory of physiological, biochemical and metabolomic responses in two different rice genotypes under drought stress: The scale matters. Plant Sci. 2021, 311, 110994. [Google Scholar] [CrossRef]
- Lv, C.; Hu, Z.; Wei, J.; Wang, Y. Transgenerational effects of elevated CO2 on rice photosynthesis and grain yield. Plant Mol. Biol. 2022, 110, 413–424. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; Li, K.; Sun, Y.; Chen, B.; Pan, Z.; Wang, Z.; Pang, B.; He, S.; Miao, Y.; Du, X. Physiological and transcriptional analyses reveal formation of memory under recurring drought stresses in seedlings of cotton (Gossypium hirsutum). Plant Sci. 2024, 338, 111920. [Google Scholar] [CrossRef] [PubMed]
- Gourlay, G.; Hawkins, B.J.; Albert, A.; Schnitzler, J.P.; Peter Constabel, C. Condensed tannins as antioxidants that protect poplar against oxidative stress from drought and UV-B. Plant Cell Environ. 2021, 45, 362–377. [Google Scholar] [CrossRef] [PubMed]
- Henry-Kirk, R.A.; Plunkett, B.; Hall, M.; McGhie, T.; Allan, A.C.; Wargent, J.J.; Espley, R.V. Solar UV light regulates flavonoid metabolism in apple (Malus × domestica). Plant Cell Environ. 2018, 41, 675–688. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Geng, F.; Chang, R.; Ruan, M.; Bian, Y.; Xu, L.; Feng, F.; Chen, Q. Comprehensive Evaluation of the Effect of Ultraviolet Stress on Functional Phytochemicals of Hulless Barley (Qingke) Grass in Different Growth Times at Vegetative Stage. ACS Omega 2020, 5, 31810–31820. [Google Scholar] [CrossRef] [PubMed]
- Marfil, C.; Ibanez, V.; Alonso, R.; Varela, A.; Bottini, R.; Masuelli, R.; Fontana, A.; Berli, F. Changes in grapevine DNA methylation and polyphenols content induced by solar ultraviolet-B radiation, water deficit and abscisic acid spray treatments. Plant Physiol. Biochem. 2019, 135, 287–294. [Google Scholar] [CrossRef]
- Weifeng, Y.; Penglin, Z.; Shaojun, L.; Yajun, G.; Guiquan, Z.; Shaokui, W. Research progress of grain shape genetics in rice. J. South China Agric. Univ. 2019, 40, 203–210. [Google Scholar]
- Marathi, B.; Guleria, S.K.; Mohapatra, T.; Parsad, R.; Muthialu, N.; Kurungara, V.K.; Atwal, S.S.; Prabhu, K.V.; Singh, N.; Singh, A.K. QTL analysis of novel genomic regions associated with yield and yield related traits in new plant type based recombinant inbred lines of rice (Oryza sativa L.). BMC Plant Biol. 2012, 12, 137. [Google Scholar] [CrossRef] [PubMed]
- Xiulian, M.; Yanwei, L.; Chuandong, L.; Yan, G.; Zhiwei, L.; Wenhui, Z. Effects of Enhancing UV-B Radiation and Elevating Carbon Dioxide on Yield and Quality of Rice (Oryza sativa L.). Crops 2015, 164, 138–142. [Google Scholar]
- Suchar, V.A.; Robberecht, R. Integration and scaling of UV-B radiation effects on plants: From DNA to leaf. Ecol. Evol. 2015, 5, 2544–2555. [Google Scholar] [CrossRef]
- Linyu, L.; Qunce, H.; Shuyin, Z.; Shuaipeng, Z. Effects of low-energy N+ ion beam irradiation on photosynthetic and transpiration in rice under enhanced UV-B radiation. Trans. CSAE 2013, 29, 136–144. [Google Scholar]
- Beyaz, R.; Sancak, C.; Yildiz, Ç.; Kuşvuran, Ş.; Yildiz, M. Physiological responses of the M1 sainfoin (Onobrychis viciifolia Scop) plants to gamma radiation. Appl. Radiat. Isot. 2016, 118, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Sheng, J.; Li, Z.; He, Y.; Zu, Y.; Li, Y. Enhanced UV-B Radiation Induced the Proanthocyanidins Accumulation in Red Rice Grain of Traditional Rice Cultivars and Increased Antioxidant Capacity in Aging Mice. Int. J. Mol. Sci. 2023, 24, 3397. [Google Scholar] [CrossRef] [PubMed]
- Haixia, W.; Peng, C.; Hanzhu, L.; Wenzhe, L. Research Advances about the Defense Mechanisms for Plants to Enhanced Ultraviolet-B Radiation. Acta Bot. Boreal. Occident. Sin. 2007, 27, 1491–1497. [Google Scholar]
- Xiupin, W.; Xiang, L.; Zuran, L.; Chengting, L.; Yongmei, H.; Yuan, L. Research progress on the characteristics and mechanisms of transgenerational plasticity of plants in response to enhanced UV-B radiation. Plant Physiol. J. 2022, 58, 797–805. [Google Scholar] [CrossRef]
- Quan, J.; Latzel, V.; Tie, D.; Zhang, Y.; Munzbergova, Z.; Chai, Y.; Liu, X.; Yue, M. Ultraviolet B Radiation Triggers DNA Methylation Change and Affects Foraging Behavior of the Clonal Plant Glechoma longituba. Front. Plant Sci. 2021, 12, 633982. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wolfe, K.; Cobb, G.P. Exposure to Copper Oxide Nanoparticles and Arsenic Causes Intergenerational Effects on Rice (Oryza sativa japonica Koshihikari) Seed Germination and Seedling Growth. Environ. Toxicol. Chem. 2019, 38, 1978–1987. [Google Scholar] [CrossRef]
- Kumar, S.; Seem, K.; Mohapatra, T. Biochemical and Epigenetic Modulations under Drought: Remembering the Stress Tolerance Mechanism in Rice. Life 2023, 13, 1156. [Google Scholar] [CrossRef] [PubMed]
- Yin, R.; Arongaus, A.B.; Binkert, M.; Ulm, R. Two Distinct Domains of the UVR8 Photoreceptor Interact with COP1 to Initiate UV-B Signaling in Arabidopsis. Plant Cell 2015, 27, 202–213. [Google Scholar] [CrossRef]
- Lu, N.; Jun, J.H.; Liu, C.; Dixon, R.A. The flexibility of proanthocyanidin biosynthesis in plants. Plant Physiol. 2022, 190, 202–205. [Google Scholar] [CrossRef]
- Luo, X.; Zhao, H.; Yao, P.; Li, Q.; Huang, Y.; Li, C.; Chen, H.; Wu, Q. An R2R3-MYB Transcription Factor FtMYB15 Involved in the Synthesis of Anthocyanin and Proanthocyanidins from Tartary Buckwheat. J. Plant Growth Regul. 2017, 37, 76–84. [Google Scholar] [CrossRef]
- Thomas, D.T.T.; Challabathula, D.; Puthur, J.T. UV-B priming of Oryza sativa var. Kanchana seedlings augments its antioxidative potential and gene expression of stress-response proteins under various abiotic stresses. 3 Biotech 2019, 9, 375. [Google Scholar] [CrossRef] [PubMed]
- Rius, S.P.; Emiliani, J.; Casati, P. P1 Epigenetic Regulation in Leaves of High Altitude Maize Landraces: Effect of UV-B Radiation. Front. Plant Sci. 2016, 7, 523. [Google Scholar] [CrossRef] [PubMed]
- Su, N.; Lu, Y.; Wu, Q.; Liu, Y.; Xia, Y.; Xia, K.; Cui, J. UV-B-induced anthocyanin accumulation in hypocotyls of radish sprouts continues in the dark after irradiation. J. Sci. Food Agric. 2016, 96, 886–892. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Quan, J.; Wang, X.; Zhang, Z.; Liu, X.; Zhang, R.; Yue, M. Predictability of parental ultraviolet-B environment shapes the growth strategies of clonal Glechoma longituba. Front. Plant Sci. 2022, 13, 949752. [Google Scholar] [CrossRef] [PubMed]
- Latzel, V.; Rendina Gonzalez, A.P.; Rosenthal, J. Epigenetic Memory as a Basis for Intelligent Behavior in Clonal Plants. Front. Plant Sci. 2016, 7, 1354. [Google Scholar] [CrossRef] [PubMed]
- Srikant, T.; Drost, H.G. How Stress Facilitates Phenotypic Innovation through Epigenetic Diversity. Front. Plant Sci. 2020, 11, 606800. [Google Scholar] [CrossRef]
- Wang, S. Phenotypic Plasticity and Exotic Plant Invasiveness: Effects of Estimators, Species, Traits and Environments; Shenyang Agricultural University: Shenyang, China, 2022; pp. 23–28. [Google Scholar]






| Treatment | Spike Length/cm | Spike Weight/g | Grain Length/mm | Grain Width/mm | Thousand-Grain Weight/g | Empty Grain Ratio/% | Proanthocyanidin Content /mg·g−1 | PPIG1 |
|---|---|---|---|---|---|---|---|---|
| 0 kJ·m−2 | 26.09 ± 0.32 a | 5.59 ± 0.26 a | 7.50 ± 0.48 a | 3.21 ± 0.23 b | 28.75 ± 0.13 a | 8.38 ± 0.66 c | 1.12 ± 0.19 c | |
| 2.5 kJ·m−2 | 25.87 ± 0.39 a | 4.70 ± 0.23 b | 7.07 ± 0.35 b | 3.44 ± 0.25 a | 26.33 ± 0.24 b | 10.13 ± 0.24 b | 1.84 ± 0.26 b | 1.59 |
| 5 kJ·m−2 | 25.91 ± 0.30 a | 5.06 ± 0.25 ab | 6.96 ± 0.42 b | 3.12 ± 0.25 c | 26.03 ± 0.26 b | 11.50 ± 0.58 b | 2.63 ± 0.38 a | 1.09 |
| 7.5 kJ·m−2 | 26.44 ± 0.37 a | 5.00 ± 0.19 ab | 6.39 ± 0.39 c | 2.95 ± 0.23 d | 24.68 ± 015 c | 17.13 ± 0.47 a | 2.27 ± 0.41 ab | 2.06 |
| GO Term | Description of GO Term | Genes | Up | Down |
|---|---|---|---|---|
| GO:0051553 | flavone biosynthetic process | 4 | 2 | 2 |
| GO:1900000 | regulation of anthocyanin catabolic process | 2 | 0 | 2 |
| GO:2000762 | regulation of phenylpropanoid metabolic process | 14 | 4 | 10 |
| GO:0010224 | response to UV-B | 9 | 4 | 5 |
| GO:0009314 | response to radiation | 87 | 42 | 45 |
| GO:0031537 | regulation of anthocyanin metabolic process | 5 | 1 | 4 |
| GO:0009812 | flavonoid metabolic process | 17 | 6 | 11 |
| GO:0009698 | phenylpropanoid metabolic process | 12 | 1 | 11 |
| GO:0009813 | flavonoid biosynthetic process | 9 | 4 | 5 |
| GO:0046283 | anthocyanin-containing compound metabolic process | 7 | 3 | 4 |
| GO:0009699 | phenylpropanoid biosynthetic process | 10 | 2 | 8 |
| GO:0009962 | regulation of flavonoid biosynthetic process | 3 | 1 | 2 |
| GO:0009718 | anthocyanin-containing compound biosynthetic process | 1 | 1 | 0 |
| GO:0033729 | anthocyanidin reductase activity | 1 | 0 | 1 |
| GO:0009094 | L-phenylalanine biosynthetic process | 2 | 1 | 1 |
| GO:0009411 | response to UV | 14 | 7 | 7 |
| GO:0046284 | anthocyanin-containing compound catabolic process | 2 | 0 | 2 |
| KO Term | Description of GO Term | Differential Genes | Up | Down |
|---|---|---|---|---|
| KO:00999 | Biosynthesis of various plant secondary metabolites | 8 | 4 | 4 |
| KO:00942 | Anthocyanin biosynthesis | 1 | 1 | 0 |
| KO:00940 | Phenylpropanoid biosynthesis | 24 | 7 | 17 |
| KO:00941 | Flavonoid biosynthesis | 6 | 2 | 4 |
| KO:00360 | Phenylalanine metabolism | 6 | 1 | 5 |
| KO:00946 | Degradation of flavonoids | 2 | 1 | 1 |
| Gene ID | Gene Name | Changes | Description |
|---|---|---|---|
| Os02g0467000 | OsC4H | down | putative cinnamate 4-hydroxylase |
| Os10g0578950 | OsACS2 | down | 4-coumarate-CoA ligase-like 2 |
| Os08g0448000 | Os4CL5 | down | probable 4-coumarate-CoA ligase 5 |
| Os08g0441500 | OsCCR | down | cinnamoyl-CoA reductase 1 |
| Os06g0623200 | OsCCR28 | down | cinnamoyl-CoA reductase 2 |
| Os03g0112900 | OsF5HL2 | down | cytochrome P450 84A1 |
| Os02g0467600 | C4H1 | down | cytochrome P450 CYP73A100 |
| Os02g0767300 | FLS | up | flavonol synthase/flavanone 3-hydroxylase |
| Os05g0578500 | OsCCR23 | down | cinnamoyl-CoA reductase-like SNL6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Wang, X.; Zu, Y.; He, Y.; Li, Z.; Li, Y. Effects of UV-B Radiation Exposure on Transgenerational Plasticity in Grain Morphology and Proanthocyanidin Content in Yuanyang Red Rice. Int. J. Mol. Sci. 2024, 25, 4766. https://doi.org/10.3390/ijms25094766
Zhang L, Wang X, Zu Y, He Y, Li Z, Li Y. Effects of UV-B Radiation Exposure on Transgenerational Plasticity in Grain Morphology and Proanthocyanidin Content in Yuanyang Red Rice. International Journal of Molecular Sciences. 2024; 25(9):4766. https://doi.org/10.3390/ijms25094766
Chicago/Turabian StyleZhang, Lin, Xiupin Wang, Yanqun Zu, Yongmei He, Zuran Li, and Yuan Li. 2024. "Effects of UV-B Radiation Exposure on Transgenerational Plasticity in Grain Morphology and Proanthocyanidin Content in Yuanyang Red Rice" International Journal of Molecular Sciences 25, no. 9: 4766. https://doi.org/10.3390/ijms25094766





