Insight into the Relationships Between Chemical, Protein and Functional Variables in the PBP/GOBP Family in Moths Based on Machine Learning
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Data and Preproccessing
4.1.1. Data
- Odorant-binding proteins
- Volatile organic compounds
- Binding affinity (Ki)
4.1.2. Preprocessing
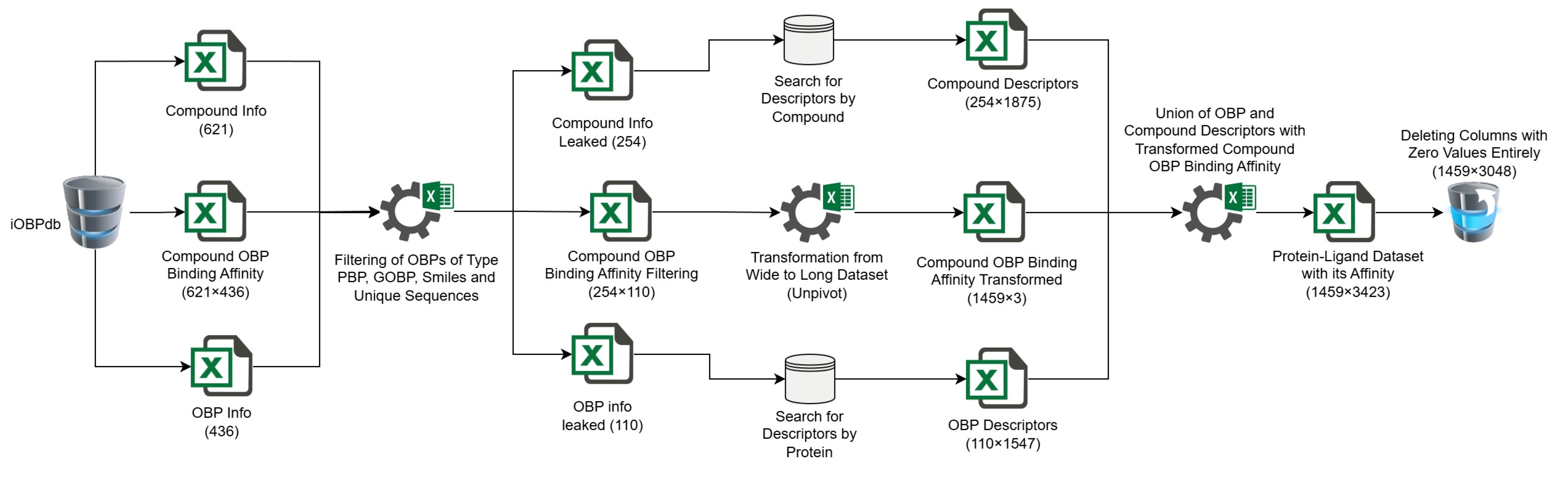
4.1.3. Extraction of Descriptors
4.1.4. Dataset Creation
4.1.5. Machine Learning Models
- Training and testing data.
- Dataset preprocessing
- -
- X is the original value of the feature;
- -
- μ is the mean of the feature in the dataset;
- -
- σ is the standard deviation of the feature.
- -
- Ki represents the value of the inhibition constant in units of molarity (M).
- -
- The factor (or 109 nM) is used to convert Ki to nanomolarity (nM) so that the resulting logarithmic values are on a comparable scale.
4.2. Models’s Implementation
- XGBoost Regressor
- LightGBM Regressor
- Gradient Boosting Regressor
- AdaBoost Regressor
- Random Forest Regressor
- Support Vector Regressor
Hyperparameter Optimization and Cross-Validation
4.3. Models’ Performance Evaluation
4.3.1. Root-Mean-Square Error (RMSE)
- -
- n: total number of observations.
- -
- : real value of the observation i.
- -
- : predicted value for the observation i.
4.3.2. Coefficient of Determination
- -
- : real value of the observation i.
- -
- : predicted value for the observation i.
- -
- : mean of all real values .
- -
- n: total number of observations (256).
4.3.3. Mean Absolute Error
- -
- n: total number of observations.
- -
- : real value of the observation i.
- -
- : predicted value for the observation i.
4.3.4. Confidence Interval
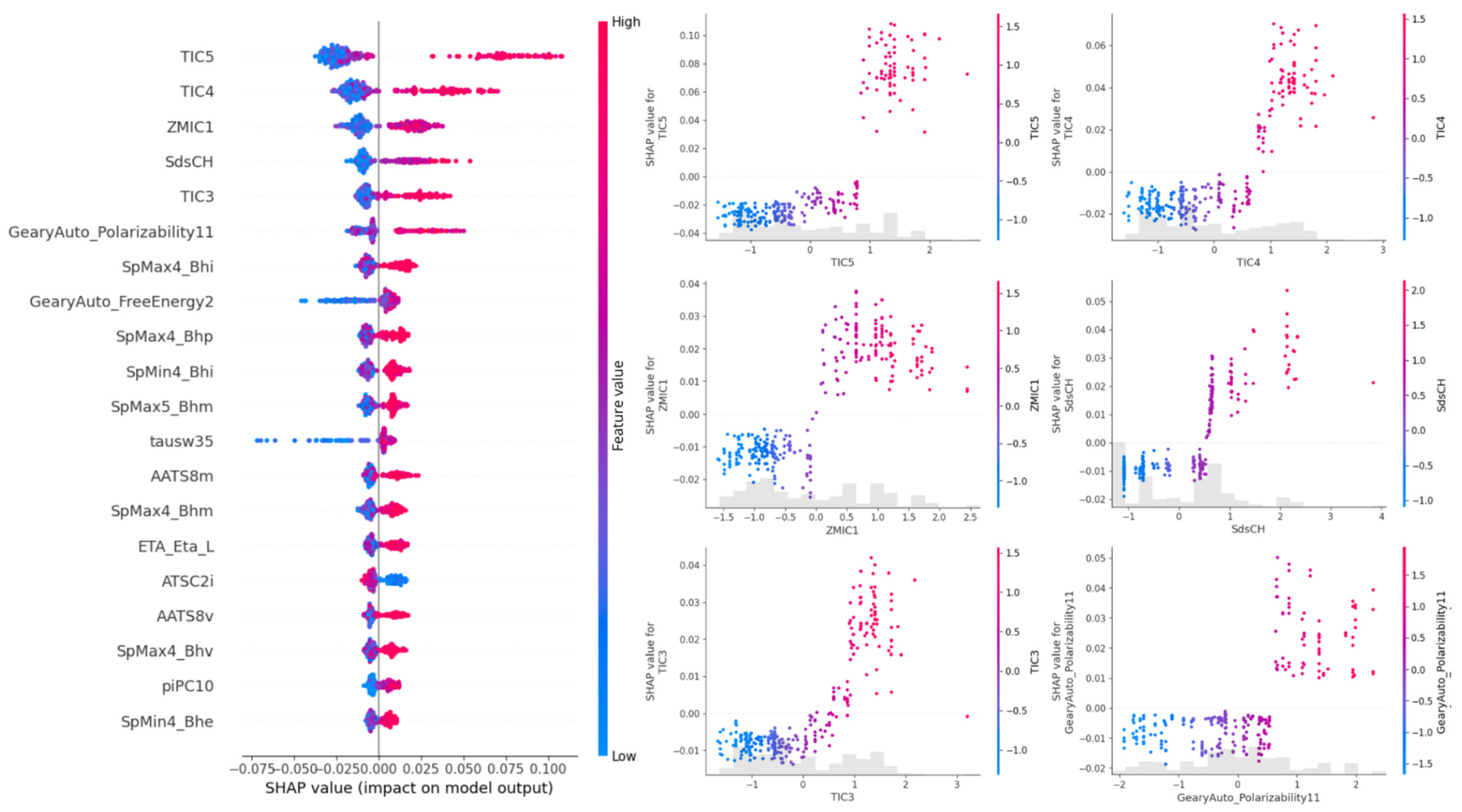
4.4. SHAP Values
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hill, M.P.; Clusella-Trullas, S.; Terblanche, J.S.; Richardson, D.M. Drivers, impacts, mechanisms and adaptation in insect invasions. Biol. Invasions 2016, 18, 883–891. [Google Scholar] [CrossRef]
- Bertelsmeier, C. Globalization and the anthropogenic spread of invasive social insects. Curr. Opin. Insect Sci. 2021, 46, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.H.; Gratton, C. Insects as drivers of ecosystem processes. Curr. Opin. Insect Sci. 2014, 2, 26–32. [Google Scholar] [CrossRef]
- Balaško, M.K.; Bažok, R.; Mikac, K.M.; Lemic, D.; Živković, I.P. Pest Management Challenges and Control Practices in Codling Moth: A Review. Insects 2020, 11, 38. [Google Scholar] [CrossRef]
- Zhang, D.-D.; Löfstedt, C. Moth pheromone receptors: Gene sequences, function, and evolution. Front. Ecol. Evol. 2015, 2, 00105. [Google Scholar] [CrossRef]
- Sullivan, B.T.; Clarke, S.R. Semiochemicals for management of the southern pine beetle (Coleoptera: Curculionidae: Scolytinae): Successes, failures, and obstacles to progress. Can. Entomol. 2020, 153, 36–61. [Google Scholar] [CrossRef]
- Vassiliou, V.A. Effectiveness of Insecticides in Controlling the First and Second Generations of the Lobesia botrana (Lepidoptera: Tortricidae) in Table Grapes. J. Econ. Entomol. 2011, 104, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Mori, N.; Noge, K. Recent advances in chemical ecology: Complex interactions mediated by molecules. Biosci. Biotechnol. Biochem. 2021, 85, 33–41. [Google Scholar] [CrossRef]
- Zhou, J. Odorant-binding proteins in insects. Vitam. Horm. 2010, 83, 241–272. [Google Scholar] [CrossRef] [PubMed]
- Leal, W.S. Odorant Reception in Insects: Roles of Receptors, Binding Proteins, and Degrading Enzymes. Annu. Rev. Entomol. 2013, 58, 373–391. [Google Scholar] [CrossRef]
- Pelosi, P.; Iovinella, I.; Zhu, J.; Wang, G.; Dani, F.R. Beyond chemoreception: Diverse tasks of soluble olfactory proteins in insects. Biol. Rev. 2018, 93, 184–200. [Google Scholar] [CrossRef] [PubMed]
- Rihani, K.; Ferveur, J.-F.; Briand, L. The 40-Year Mystery of Insect Odorant-Binding Proteins. Biomolecules 2021, 11, 509. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sweeney, J.D.; McLean, J.A.; Friskie, L.M. Roles of minor components in pheromone-mediated behavior of western spruce budworm male moths. J. Chem. Ecol. 1990, 16, 1517–1530. [Google Scholar] [CrossRef]
- Vogt, R.G.; Riddiford, L.M. Pheromone binding and inactivation by moth antennae. Nature 1981, 293, 161–163. [Google Scholar] [CrossRef] [PubMed]
- Pelosi, P.; Maida, R. Odorant-binding proteins in insects. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 1995, 111, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Venthur, H.; Mutis, A.; Zhou, J.; Quiroz, A. Ligand binding and homology modelling of insect odorant-binding proteins. Physiol. Entomol. 2014, 39, 183–198. [Google Scholar] [CrossRef]
- Venthur, H.; Zhou, J.-J. Odorant Receptors and Odorant-Binding Proteins as Insect Pest Control Targets: A Comparative Analysis. Front. Physiol. 2018, 9, 1163. [Google Scholar] [CrossRef]
- Ha, T.S.; Smith, D.P. Recent Insights into Insect Olfactory Receptors and Odorant-Binding Proteins. Insects 2022, 13, 926. [Google Scholar] [CrossRef]
- Venthur, H.; Machuca, J.; Godoy, R.; Palma-Millanao, R.; Zhou, J.; Larama, G.; Bardehle, L.; Quiroz, A.; Ceballos, R.; Mutis, A. Structural investigation of selective binding dynamics for the pheromone-binding protein 1 of the grapevine moth, Lobesia botrana. Arch. Insect Biochem. Physiol. 2019, 101, e21557. [Google Scholar] [CrossRef]
- Luo, Y.; Chen, X.; Xu, S.; Li, B.; Luo, K.; Li, G. Functional Role of Odorant-Binding Proteins in Response to Sex Pheromone Component Z8-14:Ac in Grapholita molesta (Busck). Insects 2024, 15, 918. [Google Scholar] [CrossRef]
- Yang, H.-H.; Li, S.-P.; Yin, M.-Z.; Zhu, X.-Y.; Li, J.-B.; Zhang, Y.-N.; Li, X.-M. Functional differentiation of two general odorant-binding proteins to sex pheromones in Spodoptera frugiperda. Pestic. Biochem. Physiol. 2023, 191, 105348. [Google Scholar] [CrossRef] [PubMed]
- Lizana, P.; Mutis, A.; Quiroz, A.; Venthur, H. Insights Into Chemosensory Proteins From Non-Model Insects: Advances and Perspectives in the Context of Pest Management. Front. Physiol. 2022, 13, 924750. [Google Scholar] [CrossRef]
- El-Sayed, A.M.; Ganji, S.; Gross, J.; Giesen, N.; Rid, M.; Lo, P.L.; Kokeny, A.; Unelius, C.R. Climate change risk to pheromone application in pest management. Sci. Nat. 2021, 108, 47. [Google Scholar] [CrossRef]
- Campanacci, V.; Krieger, J.; Bette, S.; Sturgis, J.N.; Lartigue, A.; Cambillau, C.; Breer, H.; Tegoni, M. Revisiting the Specificity of Mamestra brassicaeand Antheraea polyphemus Pheromone-binding Proteins with a Fluorescence Binding Assay. J. Biol. Chem. 2001, 276, 20078–20084. [Google Scholar] [CrossRef] [PubMed]
- Ban, L.; Scaloni, A.; D’Ambrosio, C.; Zhang, L.; Yan, Y.; Pelosi, P. Biochemical characterization and bacterial expression of an odorant-binding protein from Locusta migratoria. Cell. Mol. Life Sci. 2003, 60, 390–400. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Shukla, S.; Nakano-Baker, O.; Godin, D.; MacKenzie, D.; Sarikaya, M. iOBPdb A Database for Experimentally Determined Functional Characterization of Insect Odorant Binding Proteins. Sci. Data 2023, 10, 295. [Google Scholar] [CrossRef]
- Gong, D.-P.; Zhang, H.-J.; Zhao, P.; Xia, Q.-Y.; Xiang, Z.-H. The Odorant Binding Protein Gene Family from the Genome of Silkworm, Bombyx mori. BMC Genom. 2009, 10, 332. [Google Scholar] [CrossRef]
- Vogt, R.G.; Große-Wilde, E.; Zhou, J.-J. The Lepidoptera Odorant Binding Protein gene family: Gene gain and loss within the GOBP/PBP complex of moths and butterflies. Insect Biochem. Mol. Biol. 2015, 62, 142–153. [Google Scholar] [CrossRef] [PubMed]
- Sandler, B.H.; Nikonova, L.; Leal, W.S.; Clardy, J. Sexual attraction in the silkworm moth: Structure of the pheromone-binding-protein–bombykol complex. Chem. Biol. 2000, 7, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Lautenschlager, C.; Leal, W.S.; Clardy, J. Coil-to-helix transition and ligand release of Bombyx mori pheromone-binding protein. Biochem. Biophys. Res. Commun. 2005, 335, 1044–1050. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.-J.; Robertson, G.; He, X.; Dufour, S.; Hooper, A.M.; Pickett, J.A.; Keep, N.H.; Field, L.M. Characterisation of Bombyx mori Odorant-binding Proteins Reveals that a General Odorant-binding Protein Discriminates Between Sex Pheromone Components. J. Mol. Biol. 2009, 389, 529–545. [Google Scholar] [CrossRef] [PubMed]
- Maida, R.; Mameli, M.; Müller, B.; Krieger, J.; Steinbrecht, R.A. The expression pattern of four odorant-binding proteins in male and female silk moths, Bombyx mori. J. Neurocytol. 2005, 34, 149–163. [Google Scholar] [CrossRef] [PubMed]
- Torresen, J. A Review of Future and Ethical Perspectives of Robotics and AI. Front. Robot. AI 2018, 4, 75. [Google Scholar] [CrossRef]
- López-Cortés, X.A.; Matamala, F.; Venegas, B.; Rivera, C. Machine-Learning Applications in Oral Cancer: A Systematic Review. Appl. Sci. 2022, 12, 5715. [Google Scholar] [CrossRef]
- López-Cortés, X.A.; Nachtigall, F.M.; Olate, V.R.; Araya, M.; Oyanedel, S.; Diaz, V.; Jakob, E.; Ríos-Momberg, M.; Santos, L.S. Fast detection of pathogens in salmon farming industry. Aquaculture 2017, 470, 17–24. [Google Scholar] [CrossRef]
- Tapia-Castillo, A.; Carvajal, C.A.; López-Cortés, X.; Vecchiola, A.; Fardella, C.E. Novel metabolomic profile of subjects with non-classic apparent mineralocorticoid excess. Sci. Rep. 2021, 11, 17156. [Google Scholar] [CrossRef]
- Olate-Olave, V.R.; Guzmán, L.; López-Cortés, X.A.; Cornejo, R.; Nachtigall, F.M.; Doorn, M.; Santos, L.S.; Bejarano, A. Comparison of Chilean honeys through MALDI-TOF-MS profiling and evaluation of their antioxidant and antibacterial potential. Ann. Agric. Sci. 2021, 66, 152–161. [Google Scholar] [CrossRef]
- López-Cortés, X.A.; Manríquez-Troncoso, J.M.; Kandalaft-Letelier, J.; Cuadros-Orellana, S. Machine learning and matrix-assisted laser desorption/ionization time-of-flight mass spectra for antimicrobial resistance prediction: A systematic review of recent advancements and future development. J. Chromatogr. A 2024, 1734, 465262. [Google Scholar] [CrossRef] [PubMed]
- López-Cortés, X.A.; Manríquez-Troncoso, J.M.; Hernández-García, R.; Peralta, D. MSDeepAMR: Antimicrobial resistance prediction based on deep neural networks and transfer learning. Front. Microbiol. 2024, 15, 1361795. [Google Scholar] [CrossRef]
- Astudillo, C.A.; López-Cortés, X.A.; Ocque, E.; Manríquez-Troncoso, J.M. Multi-label classification to predict antibiotic resistance from raw clinical MALDI-TOF mass spectrometry data. Sci. Rep. 2024, 14, 31283. [Google Scholar] [CrossRef]
- López-Cortés, X.A.; Manríquez-Troncoso, J.M.; Sepúlveda, A.Y.; Soto, P.S. Integrating Machine Learning with MALDI-TOF Mass Spectrometry for Rapid and Accurate Antimicrobial Resistance Detection in Clinical Pathogens. Int. J. Mol. Sci. 2025, 26, 1140. [Google Scholar] [CrossRef] [PubMed]
- Alzubi, J.; Nayyar, A.; Kumar, A. Machine Learning from Theory to Algorithms: An Overview. J. Phys. Conf. Ser. 2018, 1142, 012012. [Google Scholar] [CrossRef]
- Lima, M.C.F.; Leandro, M.E.D.d.A.; Valero, C.; Coronel, L.C.P.; Bazzo, C.O.G. Automatic Detection and Monitoring of Insect Pests—A Review. Agriculture 2020, 10, 161. [Google Scholar] [CrossRef]
- Balduque-Gil, J.; Lacueva-Pérez, F.J.; Labata-Lezaun, G.; Del-Hoyo-Alonso, R.; Ilarri, S.; Sánchez-Hernández, E.; Martín-Ramos, P.; Barriuso-Vargas, J.J. Big Data and Machine Learning to Improve European Grapevine Moth (Lobesia botrana) Predictions. Plants 2023, 12, 633. [Google Scholar] [CrossRef]
- Almeyda, E.; Paiva, J.; Ipanaque, W. Pest Incidence Prediction in Organic Banana Crops with Machine Learning Techniques. In Proceedings of the 2020 IEEE Engineering International Research Conference (EIRCON), Lima, Peru, 21–23 October 2020; Publishing House: Lima, Peru, 2020; pp. 1–4. [Google Scholar]
- Caballero-Vidal, G.; Bouysset, C.; Grunig, H.; Fiorucci, S.; Montagné, N.; Golebiowski, J.; Jacquin-Joly, E. Machine learning decodes chemical features to identify novel agonists of a moth odorant receptor. Sci. Rep. 2020, 10, 1655. [Google Scholar] [CrossRef]
- Yuvaraj, J.K.; Roberts, R.E.; Sonntag, Y.; Hou, X.-Q.; Grosse-Wilde, E.; Machara, A.; Zhang, D.-D.; Hansson, B.S.; Johanson, U.; Löfstedt, C.; et al. Putative ligand binding sites of two functionally characterized bark beetle odorant receptors. BMC Biol. 2021, 19, 16. [Google Scholar] [CrossRef]
- Sims, C.; Withall, D.M.; Oldham, N.; Stockman, R.; Birkett, M. Computational investigation of aphid odorant receptor structure and binding function. J. Biomol. Struct. Dyn. 2022, 41, 3647–3658. [Google Scholar] [CrossRef]
- Yi, S.-C.; Wu, Y.-H.; Yang, R.-N.; Li, D.-Z.; Abdelnabby, H.; Wang, M.-Q. A Highly Expressed Antennae Odorant-Binding Protein Involved in Recognition of Herbivore-Induced Plant Volatiles in Dastarcus helophoroides. Int. J. Mol. Sci. 2023, 24, 3464. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- ACS Publications. Bombykol—Molecule of the Week Archive. American Chemical Society. Available online: https://www.acs.org/molecule-of-the-week (accessed on 27 November 2024).
- Wikipedia Contributors. Bombykol. Wikipedia. Available online: https://en.wikipedia.org/wiki/Bombykol (accessed on 27 November 2024).
- The Pherobase: Database of Pheromones and Semiochemicals. The World Largest Database of Behavioural Modifying Chemicals. Available online: https://pherobase.com/ (accessed on 12 February 2025).
- Larsson, M.C. Pheromones and Other Semiochemicals for Monitoring Rare and Endangered Species. J. Chem. Ecol. 2016, 42, 853–868. [Google Scholar] [CrossRef]
- Caballero-Vidal, G.; Bouysset, C.; Gévar, J.; Mbouzid, H.; Nara, C.; Delaroche, J.; Golebiowski, J.; Montagné, N.; Fiorucci, S.; Jacquin-Joly, E. Reverse chemical ecology in a moth: Machine learning on odorant receptors identifies new behaviorally active agonists. Cell. Mol. Life Sci. 2021, 78, 6593–6603. [Google Scholar] [CrossRef]
- Kepchia, D.; Xu, P.; Terryn, R.; Castro, A.; Schürer, S.C.; Leal, W.S.; Luetje, C.W. Use of machine learning to identify novel, behaviorally active antagonists of the insect odorant receptor co-receptor (Orco) subunit. Sci. Rep. 2019, 9, 4055. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Kumar, R.; Ranjta, S.; Varadwaj, P.K. SMILES to Smell: Decoding the Structure–Odor Relationship of Chemical Compounds Using the Deep Neural Network Approach. J. Chem. Inf. Model. 2021, 61, 676–688. [Google Scholar] [CrossRef] [PubMed]
- Bo, W.; Yu, Y.; He, R.; Qin, D.; Zheng, X.; Wang, Y.; Ding, B.; Liang, G. Insight into the Structure–Odor Relationship of Molecules: A Computational Study Based on Deep Learning. Foods 2022, 11, 2033. [Google Scholar] [CrossRef]
- Pugalenthi, G.; Tang, K.; Suganthan, P.; Archunan, G.; Sowdhamini, R. A machine learning approach for the identification of odorant binding proteins from sequence-derived properties. BMC Bioinform. 2007, 8, 351. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Zaremska, V.; Lim, S.; Knoll, W.; Pelosi, P. Probe-dependence of competitive fluorescent ligand binding assays to odorant-binding proteins. Anal. Bioanal. Chem. 2020, 412, 547–554. [Google Scholar] [CrossRef]
- The iOBPdb Database of Insect Odorant Binding Proteins. Available online: https://www.iobpdb.com/Home (accessed on 12 February 2025).
- Herrera-Acevedo, C.; Perdomo-Madrigal, C.; Herrera-Acevedo, K.; Coy-Barrera, E.; Scotti, L.; Scotti, M.T. Machine learning models to select potential inhibitors of acetylcholinesterase activity from SistematX: A natural products database. Mol. Divers. 2021, 25, 1553–1568. [Google Scholar] [CrossRef] [PubMed]
- Goldwaser, E.; Laurent, C.; Lagarde, N.; Fabrega, S.; Nay, L.; Villoutreix, B.O.; Jelsch, C.; Nicot, A.B.; Loriot, M.-A.; Miteva, M.A. Machine learning-driven identification of drugs inhibiting cytochrome P450 2C9. PLOS Comput. Biol. 2022, 18, e1009820. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, Y.; Cheng, J.; Zheng, Y.; Zhang, Y. A point cloud-based deep learning strategy for protein–ligand binding affinity prediction. Biomolecules 2021, 11, 1078. [Google Scholar] [CrossRef]
- Öztürk, H.; Özgür, A.; Ozkirimli, E. DeepDTA: Deep drug–target binding affinity prediction. Bioinformatics 2018, 34, i821–i829. [Google Scholar] [CrossRef]
- Thafar, M.A.; Alshahrani, M.; Albaradei, S.; Gojobori, T.; Essack, M.; Gao, X. Affinity2Vec: Drug-target binding affinity prediction through representation learning, graph mining, and machine learning. Sci. Rep. 2022, 12, 4751. [Google Scholar] [CrossRef]
- Zhang, Q.; Wu, J.; Lin, L.; Wang, Y. DeepRLI: A Multi-Objective Framework for Universal Protein–Ligand Interaction Prediction. Biomolecules 2024, 40, 89. [Google Scholar] [CrossRef]
- ALee, S.; Kim, J.; Kim, S. PIGNet2: A versatile deep learning-based protein–ligand interaction prediction model for binding affinity scoring and virtual screening. Molecules 2023, 28, 3456. [Google Scholar] [CrossRef]
- The Pandas Development Team. Pandas: Python Data Analysis Library. Available online: https://pandas.pydata.org/ (accessed on 21 November 2024).
- He, T.; Heidemeyer, M.; Ban, F.; Cherkasov, A.; Ester, M. SimBoost: A read-across approach for predicting drug–target binding affinities using gradient boosting machines. J. Cheminform. 2017, 9, 24. [Google Scholar] [CrossRef]
- Yap, C.W. PaDEL-descriptor: An open source software to calculate molecular descriptors and fingerprints. J. Comput. Chem. 2011, 32, 1466–1474. [Google Scholar] [CrossRef] [PubMed]
- Python Software Foundation. Python Programming Language. Available online: https://www.python.org/ (accessed on 21 November 2024).
- Cao, D.-S.; Xu, Q.-S.; Liang, Y.-Z. propy: A tool to generate various modes of Chou’s PseAAC. Bioinformatics 2013, 29, 960–962. [Google Scholar] [CrossRef]
- Scikit-learn Developers. Scikit-learn: Machine Learning in Python. Available online: https://scikit-learn.org/stable/ (accessed on 21 November 2024).
- Friedman, J.H. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.-Y. LightGBM: A highly efficient gradient boosting decision tree. In Proceedings of the 31st International Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; Alicioglu, G., Sun, B., Eds.; Publishing House: Long Beach, CA, USA, 2017; pp. 3149–3157. [Google Scholar]
- Bergstra, J.; Komer, B.; Eliasmith, C.; Yamins, D.; Cox, D.D. Hyperopt: A Python library for model selection and hyperparameter optimization. Comput. Sci. Discov. 2015, 8, 014008. [Google Scholar] [CrossRef]
- Chicco, D.; Warrens, M.J.; Jurman, G. The coefficient of determination R-squared is more informative than SMAPE, MAE, MAPE, MSE and RMSE in regression analysis evaluation. PeerJ Comput. Sci. 2021, 7, e623. [Google Scholar] [CrossRef]
- James, G.; Witten, D.; Hastie, T.; Tibshirani, R. An Introduction to Statistical Learning with Applications in R; Springer: New York, NY, USA, 2013. [Google Scholar]
- Draper, N.R.; Smith, H. Applied Regression Analysis, 3rd ed.; Wiley: Hoboken, NJ, USA, 1998. [Google Scholar]

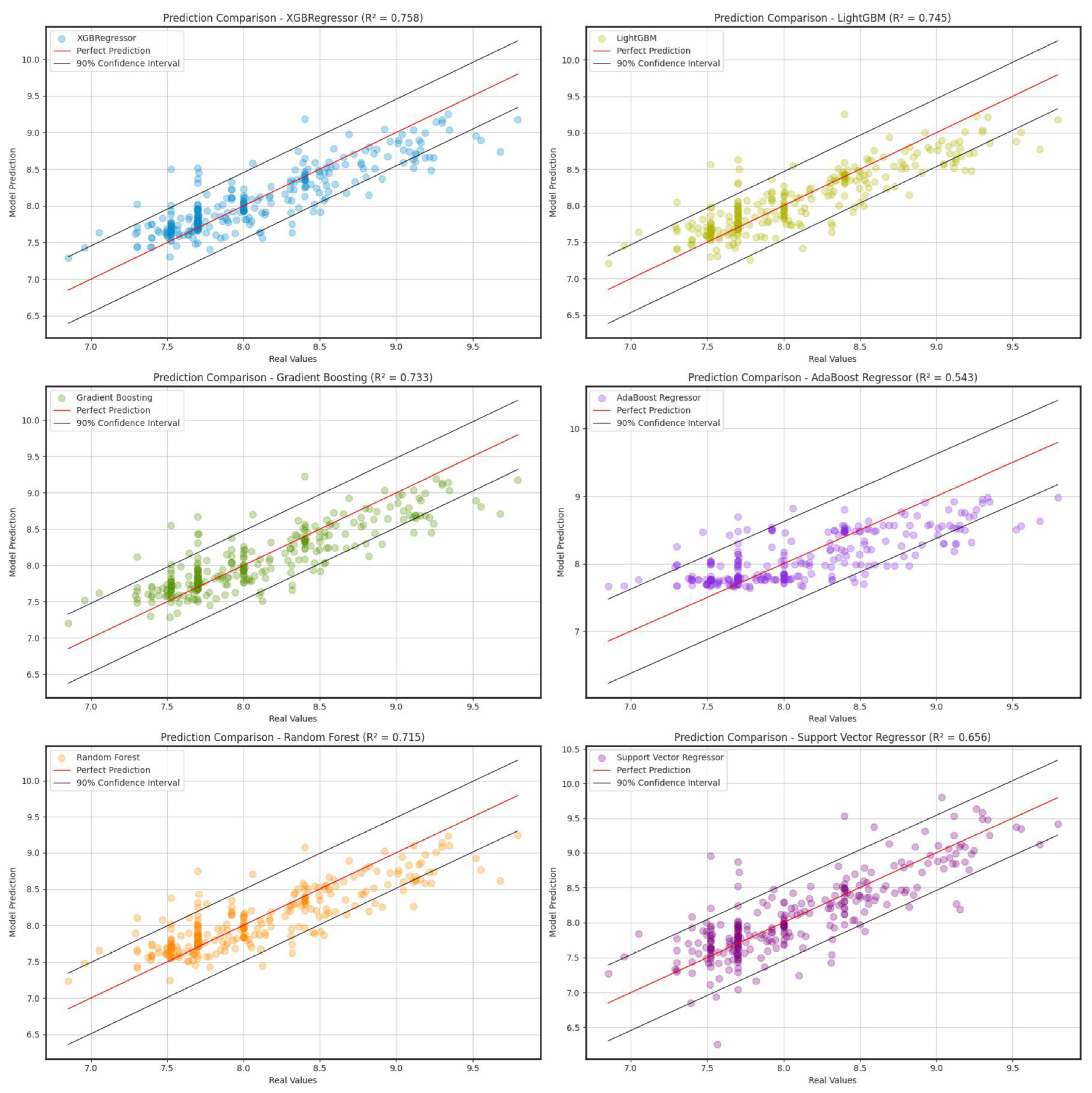
| Models | RMSE | R2 | MAE |
|---|---|---|---|
| XGBoostRegressor | 0.276 | 0.758 | 0.202 |
| LightGBMRegressor | 0.284 | 0.745 | 0.208 |
| GradientBoostingRegressor | 0.290 | 0.733 | 0.216 |
| AdaBoostRegressor | 0.380 | 0.543 | 0.292 |
| RandomForestRegressor | 0.300 | 0.715 | 0.222 |
| SupportVectorRegressor | 0.329 | 0.656 | 0.236 |
| Model | Parameters | Hyperparameter Search | Optimal Value |
|---|---|---|---|
| XGBoostRegressor | n_estimators | [700, 1200] | 800 |
| learning_rate | [0.009, 0.03] | 0.0188610 | |
| max_depth | [10, 15] | 10 | |
| min_child_weight | [5, 10] | 9 | |
| gamma | [0.00, 0.005] | 0.002323 | |
| colsample_bytree | [0.3, 0.6] | 0.392232 | |
| subsample | [0.6, 0.9] | 0.564263 | |
| reg_alpha | [0.5, 1.0] | 0.683823 | |
| reg_lambda | [1.5, 2.0] | 1.711287 | |
| LightGBMRegressor | n_estimators | [700, 1200] | 900 |
| learning_rate | [0.009, 0.03] | 0.0222877 | |
| max_depth | [10, 20] | 13 | |
| num_leaves | [20, 150] | 148 | |
| min_child_weight | [5, 10] | 7 | |
| subsample | [0.5, 1.0] | 0.661123 | |
| colsample_bytree | [0.3, 0.8] | 0.305414 | |
| reg_alpha | [0, 2] | 0.316713 | |
| reg_lambda | [0, 3] | 1.579887 | |
| GradientBoostingRegressor | n_estimators | [100, 500] | 400 |
| learning_rate | [0.009, 0.03] | 0.0265516 | |
| max_depth | [5, 15] | 5 | |
| Subsample | [0.5, 1.0] | 0.687177 | |
| min_samples_split | [2, 10] | 10 | |
| min_samples_leaf | [1, 10] | 1 | |
| max_features | [0.1, 0.5] | 0.260379 | |
| AdaBoostRegressor | n_estimators | [100, 500] | 100 |
| learning_rate | [0.009, 0.03] | 0.00996526 | |
| RandomForestRegressor | n_estimators | [700, 1200] | 1000 |
| max_depth | [3, 20] | 16 | |
| min_samples_split | [2, 20] | 5 | |
| min_samples_leaf | [1, 20] | 1 | |
| SupportVectorRegressor | C | [1000, 5000] | 1000 |
| epsilon | [0.009, 0.03] | 0.029727 | |
| degree | [1, 15] | 13 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Cortés, X.A.; Lara, G.; Fernández, N.; Manríquez-Troncoso, J.M.; Venthur, H. Insight into the Relationships Between Chemical, Protein and Functional Variables in the PBP/GOBP Family in Moths Based on Machine Learning. Int. J. Mol. Sci. 2025, 26, 2302. https://doi.org/10.3390/ijms26052302
López-Cortés XA, Lara G, Fernández N, Manríquez-Troncoso JM, Venthur H. Insight into the Relationships Between Chemical, Protein and Functional Variables in the PBP/GOBP Family in Moths Based on Machine Learning. International Journal of Molecular Sciences. 2025; 26(5):2302. https://doi.org/10.3390/ijms26052302
Chicago/Turabian StyleLópez-Cortés, Xaviera A., Gabriel Lara, Nicolás Fernández, José M. Manríquez-Troncoso, and Herbert Venthur. 2025. "Insight into the Relationships Between Chemical, Protein and Functional Variables in the PBP/GOBP Family in Moths Based on Machine Learning" International Journal of Molecular Sciences 26, no. 5: 2302. https://doi.org/10.3390/ijms26052302
APA StyleLópez-Cortés, X. A., Lara, G., Fernández, N., Manríquez-Troncoso, J. M., & Venthur, H. (2025). Insight into the Relationships Between Chemical, Protein and Functional Variables in the PBP/GOBP Family in Moths Based on Machine Learning. International Journal of Molecular Sciences, 26(5), 2302. https://doi.org/10.3390/ijms26052302






