NMR-based Structural Studies of the Glycosylated MUC1 Tandem Repeat Peptide
Abstract
:Introduction
Materials and Methods
Results and Discussion

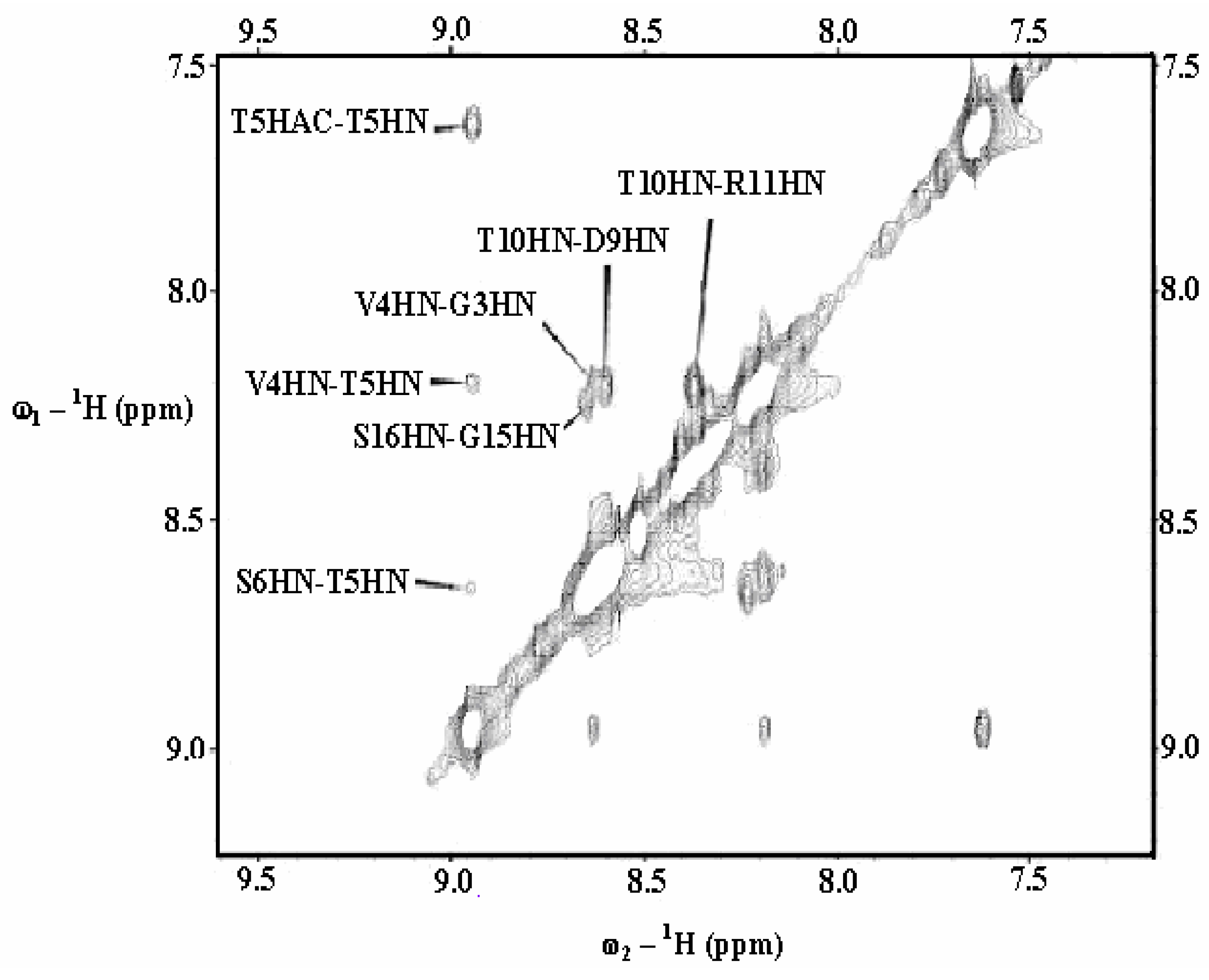
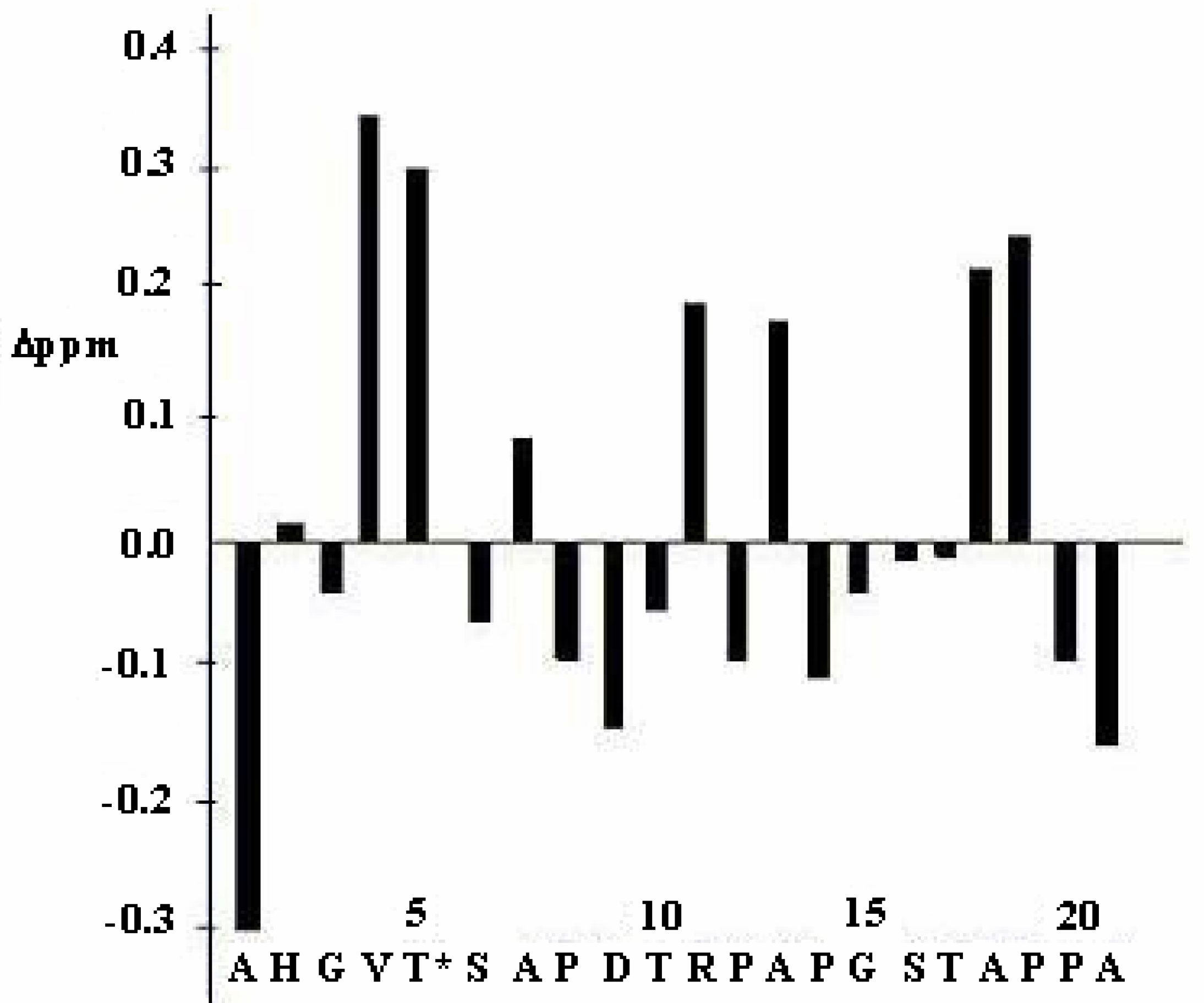
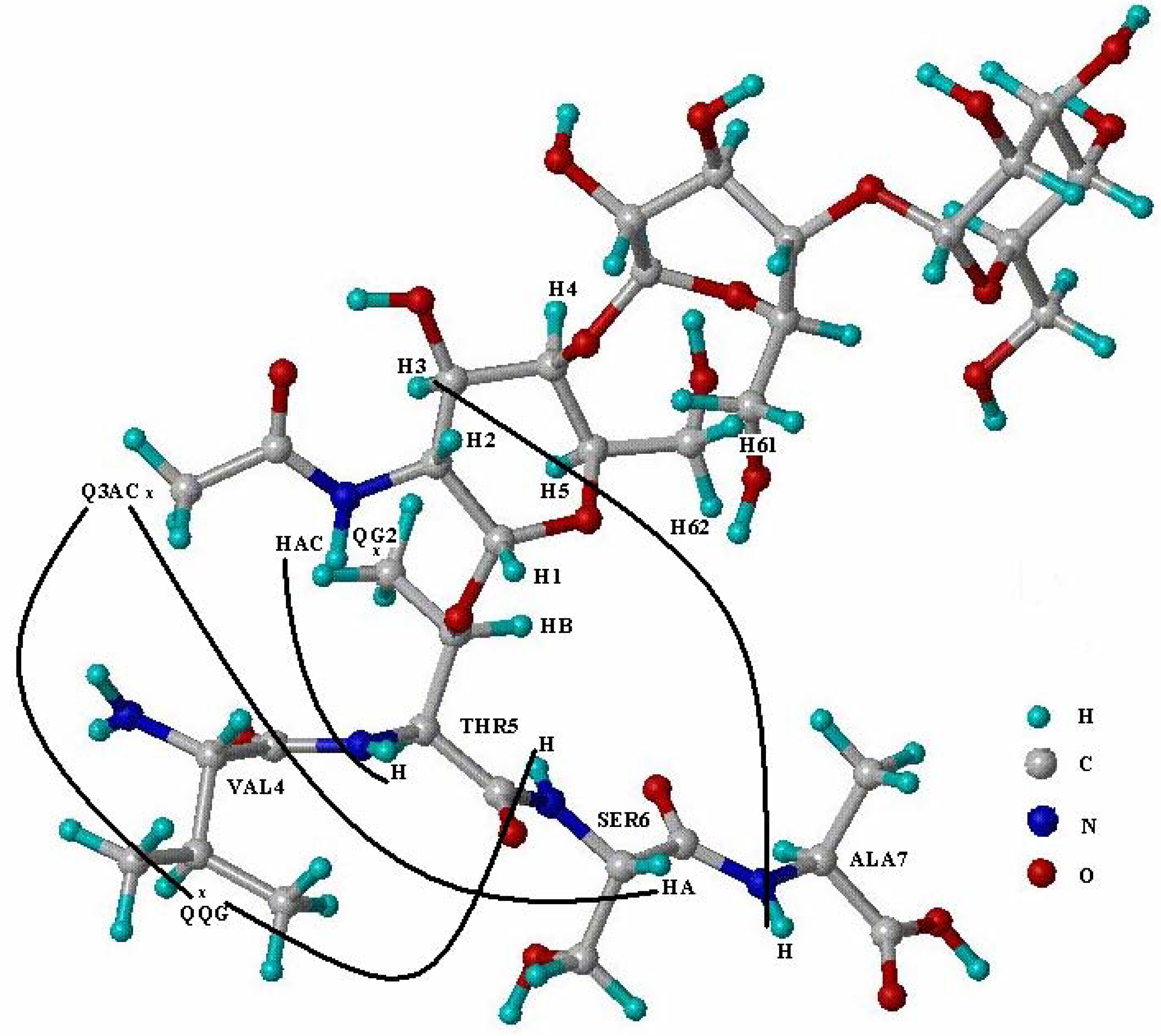
| Gly3 | Val4 | Thr5 | Ser6 | Ala7 | ||||||
| φ | Ψ | φ | ψ | φ | ψ | φ | ψ | φ | ψ | |
| Cluster1 | 171 | –157 | –56 | 113 | –109 | 121 | –142 | 131 | –94 | 164 |
| Cluster2 | –152 | –136 | –83 | 75 | –117 | 117 | –128 | 134 | –103 | 139 |
| Cluster3 | –87 | –112 | –149 | 164 | –110 | 127 | –81 | 175 | –113 | 142 |
Acknowledgements
References
- Lan, M. S.; Batra, S. K.; Qi, W. N.; Metzgar, R. S.; Hollingsworth, M. A. Cloning and sequencing of a human pancreatic tumor-mucin cDNA. J. Biol. Chem. 1990, 265, 15294–15299. [Google Scholar] [PubMed]
- McDermott, K. M.; Crocker, P. R.; Harris, A.; Burdick, M. D.; Hinoda, Y.; Hayashi, T.; Imai, K.; Hollingsworth, M. A. Overexpression of MUC1 reconfigures the binding properties of tumor cells. Int. J. Cancer 2001, 94, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Wandall, H. H.; Hassan, M.; Mirgorodskaya, F.; Kristensen, A. K.; Roepstorff, P.; Bennett, E. P.; Nielsen, P. A.; Hollingsworth, M. A.; Burchell, J.; Taylor Papadimitriou, J.; Clausen, H. Specificities of three members of the human UDP-N-acetyl-alpha-D-galactosamine: Polypeptide N-acetylgalactosaminyltransferase family, GalNAc-T1, -T2, and -T3. J. Biol. Chem. 1997, 272, 23503–23514. [Google Scholar] [CrossRef] [PubMed]
- Stadie, T.; Chai, W.; Lawson, A. M.; Byfield, P.; Hanisch, F. G. Studies on the order and site-specificity of GalNAc-transfer to MUC1 tandem repeats by UDP-GalNAc: polypeptide N-acetylgalactosaminyltransferase from milk or mammary carcinoma cells. J. Biol. Chem. 1995, 274, 27867–27874. [Google Scholar]
- Nishimori, I.; Johnson, N. R.; Sanderson, S. D.; Perini, R.; Mountjoy, K.; Cernag, R. L.; Gross, M. L.; Hollingsworth, M. A. Influence of acceptor substrate primary amino acid sequence on the activity of human UDP-N-acetylgalactosamine: polypeptide N-acetylgalactosaminyltransferase. Studies with the MUC1 tandem repeat. J. Biol. Chem. 1994, 269, 16123–16130. [Google Scholar] [PubMed]
- Bennett, E. P.; Hassan, H.; Mandel, U.; Hollingsworth, M. A.; Akisawa, N.; Ikematsu, Y.; Merkx, G.; van Kessel, A. G.; Olofsson, S.; Clausen, H. Cloning and characterization of a close homologue of human UDP-GalNAc: polypeptide N-acetylgalactosaminyl-transferase-T3, designated GalNAc-T6. Evidence for genetic but not functional redundancy. J. Biol. Chem. 1998, 274, 25362–25370. [Google Scholar] [CrossRef]
- Hanisch, F. G.; Muller, S.; Hassan, H.; Clausen, H.; Zachara, N.; Gooley, A. A.; Paulsen, H.; Alving, K.; Peter-Katalinic, J. Dynamic epigenetic regulation of initial O-glycosylation by UDP-N-Acetylgalactosamine: Peptide N-acetylgalactosaminyl- transferases. Site-specific glycosylation of MUC1 repeat peptide influences the substrate qualities at adjacent or distant Ser/Thr positions. J. Biol. Chem. 1999, 274, 9946–9954. [Google Scholar] [CrossRef] [PubMed]
- Bennett, E. P.; Hassan, H.; Hollingsworth, M. A.; Clausen, H. A novel human UDP-N-acetyl-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase, GalNAc-T7, with specificity for partial GalNAc- glycosylated acceptor substrates. FEBS Lett. 1999, 460, 226–230. [Google Scholar] [CrossRef] [PubMed]
- Hassan, H.; Reis, C.; Bennett, E. P.; Mirgorodskaya, E.; Roepstorff, P.; Hollingsworth, M. A.; Burchell, J.; Taylor-Papadimitriou, J.; Clausen, H. The lectin domain of UDP-N-acetyl-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase-T4 directs its glycopeptide specificities. J. Biol. Chem. 2000, 275, 38197–38205. [Google Scholar] [CrossRef] [PubMed]
- Hanisch, F. G.; Reis, C. A.; Clausen, H.; Paulsen, H. Evidence for glycosylation- dependent activities of polypeptide N-acetylgalactosaminyltransferases rGalNAc-T2 and -T4 on mucin glycopeptides. Glycobiology 2001, 11, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Kirnarsky, L.; Prakash, O.; Vogen, S. M.; Nomoto, M.; Hollingsworth, M. A.; Sherman, S. Structural effects of O-glycosylation on a 15-residue peptide from the mucin (MUC1) core protein. Biochemistry 2000, 39, 12076–12082. [Google Scholar] [CrossRef] [PubMed]
- Wüthrich, K. NMR of proteins and nucleic acids. John Wiley and Sons: New York, 1986. [Google Scholar]
- Goddard, T. D.; Kneller, D. G. SPARKY 3. University of California: San Francisco.
- Güntert, P.; Braun, W.; Wüthrich, K. Efficient computation of three-dimensional protein structures in solution from nuclear magnetic resonance data using the program DIANA and the supporting programs CALIBA, HABAS and GLOMSA. J. Mol. Biol. 1991, 217, 517–530. [Google Scholar] [CrossRef] [PubMed]
- Shats, O.; Sherman, S. The FiSiNOE-3 Program for Determination of Protein and Peptide Conformations from NMR Data. Third Electronic Computational Chemistry Conference (ECCC-3). 1996. http://bims.unmc.edu/ECCC3/fisinoe3.htm.
- Güntert, P.; Wüthrich, K. Automated Stereo Specific 1H NMR Assignments and their Impact on the Precision of Protein Structure Determinations. J. Am. Chem. Soc. 1989, 111, 3997–4004. [Google Scholar] [CrossRef]
- Tripos Associates. St. Louis. version 6.6.
- Weiner, S. J.; Kollman, P. A.; Case, D. A.; Singh, U. C.; Ghio, C.; Alagona, G.; Profeta, S., Jr.; Weiner, P. A New Forcefield for Molecular Mechanical Simulation of Nucleic Acids and Proteins. J. Am. Chem. Soc. 1984, 106, 765–784. [Google Scholar] [CrossRef]
- Woods, R. J.; Edge, C. J.; Wormald, M. R.; Dwek, R. A. A generalized parameter set for molecular dynamics simulations of glycoproteins and oligosaccharides. Application to the structure and dynamics of a disaccharide related to oligomannose. In Complex Carbohydrates in Drug Research; Bock, K., Clausen, H., Krogsagaard-Larsen, P., Kofod, H., Eds.; Alfred Benzon Symposium; 1993; Volume 36, pp. 15–26. [Google Scholar]
- Güentert, P.; Mumenthaler, C.; Wüthrich, K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 1997, 273, 283–298. [Google Scholar] [CrossRef] [PubMed]
- Rubinstein, A.; Shats, O.; Sherman, S. Role of the Local Interactions in Intrinsic Conformational and Structural Propensitites of Amino Acid Residues in Proteins. Seventh Electronic Computational Chemistry Conference (ECCC-7). 2001. http://bims.unmc.edu/ECCC7.
- Ramachandran, G. N.; Ramakrishnan, C.; Sasisekharan, V. Stereochemistry of polypeptide chain conformations. J. Mol. Biol. 1963, 7, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D. S.; Sykes, B. D.; Richards, F. M. The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry 1992, 31, 1647–1651. [Google Scholar] [CrossRef] [PubMed]
© 2004 by MDPI (http://www.mdpi.net). Reproduction for noncommercial purposes permitted.
Share and Cite
Suryanarayanan, G.; Keifer, P.A.; Wang, G.; Kinarsky, L.; Hollingsworth, M.A.; Sherman, S. NMR-based Structural Studies of the Glycosylated MUC1 Tandem Repeat Peptide. Int. J. Mol. Sci. 2004, 5, 84-92. https://doi.org/10.3390/i5030084
Suryanarayanan G, Keifer PA, Wang G, Kinarsky L, Hollingsworth MA, Sherman S. NMR-based Structural Studies of the Glycosylated MUC1 Tandem Repeat Peptide. International Journal of Molecular Sciences. 2004; 5(3):84-92. https://doi.org/10.3390/i5030084
Chicago/Turabian StyleSuryanarayanan, G., P. A. Keifer, G. Wang, L. Kinarsky, M. A. Hollingsworth, and S. Sherman. 2004. "NMR-based Structural Studies of the Glycosylated MUC1 Tandem Repeat Peptide" International Journal of Molecular Sciences 5, no. 3: 84-92. https://doi.org/10.3390/i5030084
APA StyleSuryanarayanan, G., Keifer, P. A., Wang, G., Kinarsky, L., Hollingsworth, M. A., & Sherman, S. (2004). NMR-based Structural Studies of the Glycosylated MUC1 Tandem Repeat Peptide. International Journal of Molecular Sciences, 5(3), 84-92. https://doi.org/10.3390/i5030084




