Docking of monomers
The most common reaction mechanism of phenol substrates oxidation with heme peroxidases that appears to be universally accepted today is depicted in the following scheme:
where E is a resting (native) form of a peroxidase, S stands for the substrate and S
• for the corresponding radical. The resting ferric enzyme reacts rapidly with peroxide to form compound I, i.e. an oxy-ferryl (Fe=O) derivative, in which one electron has been withdrawn from the heme group to form a porphyrin π-cation radical. This intermediate is reduced in two sequential one-electron oxidation steps through compound II formation.
Compound I and II have bound an oxygen atom to a heme of iron forming Fe=O. The intermolecular orientation of substrate and (porphyrin)Fe=O complex must be favorable for an electron and a proton transfer. The experimental and modeling studies of ARP associated reactions showed that a proton transfer is determined by a proper hydrogen bonding network inside the active center [
11]. The absence of hydrogen bonding between oxygen of ferryl group and hydroxyl group of phenol derivative produces “unproductive” complex that determines the decrease of the reaction rate almost thousand times [
11].
The dockings showed that phenol derivatives
b1,
na1 and
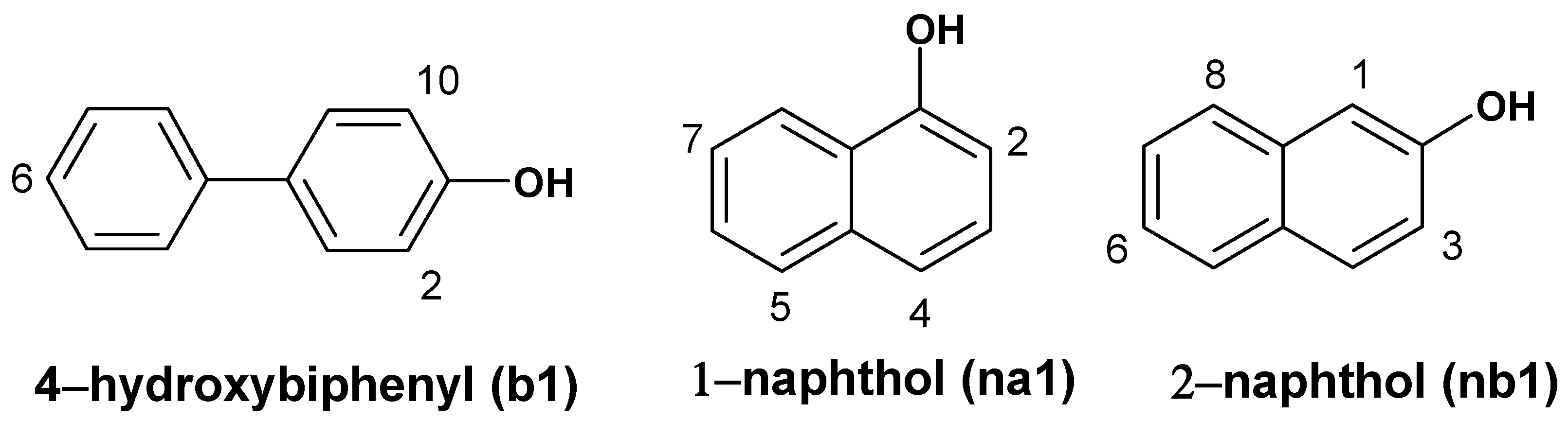
nb1 docked in the active center with highest affinity, or the lowest docking energy, in the whole ARP structure. All monomers placed OH groups inside the active center near Fe=O and formed strong hydrogen bonds with the oxygen atom of Fe=O fragment (
Figure 2). The lengths of hydrogen bonds are 2.1 Å, 1.8 Å and 1.6 Å respectively of 4
-hydroxybiphenyl, 1-naphthol and 2-naphthol.
Figure 2.
The dockings of 1-naphthol (yellow), 2-naphthol (green) and 4-hydroxy-biphenyl (cyan) in the active center of ARP. Hydrogen bonds (length depicted in Å) are formed between OH and Fe=O groups.
Figure 2.
The dockings of 1-naphthol (yellow), 2-naphthol (green) and 4-hydroxy-biphenyl (cyan) in the active center of ARP. Hydrogen bonds (length depicted in Å) are formed between OH and Fe=O groups.
The docking energies of monomers were -7.0 kcal/mol, -6.4 kcal/mol and -6.4 kcal/mol (
Table 2). The docking results show that phenol substances form a “productive” complex. For that reason they should be considered as active substrates. Experimental measurements confirm the high activity of these substances. The constants of 1-naphthol and 2-naphthol oxidation were 1.4·10
7 M
-1·s
-1 and 5.4·10
7 M
-1·s
-1 and approached the rate of diffusion limited for enzyme catalysis [
5] .
Docking of oligomers
In this work the hypothesis was made that unproductive bonding of oligomers may determinate peroxidase inhibition during phenol derivatives oxidation. To confirm or reject this hypothesis the docking of oligomer structures in the active center of ARP has been performed. The calculations show that all oligomer structures dock in the active center of ARP with the highest affinity. The complexes of oligomers are even stronger since the docking energy is less in comparison to monomer substances (
Table 2).
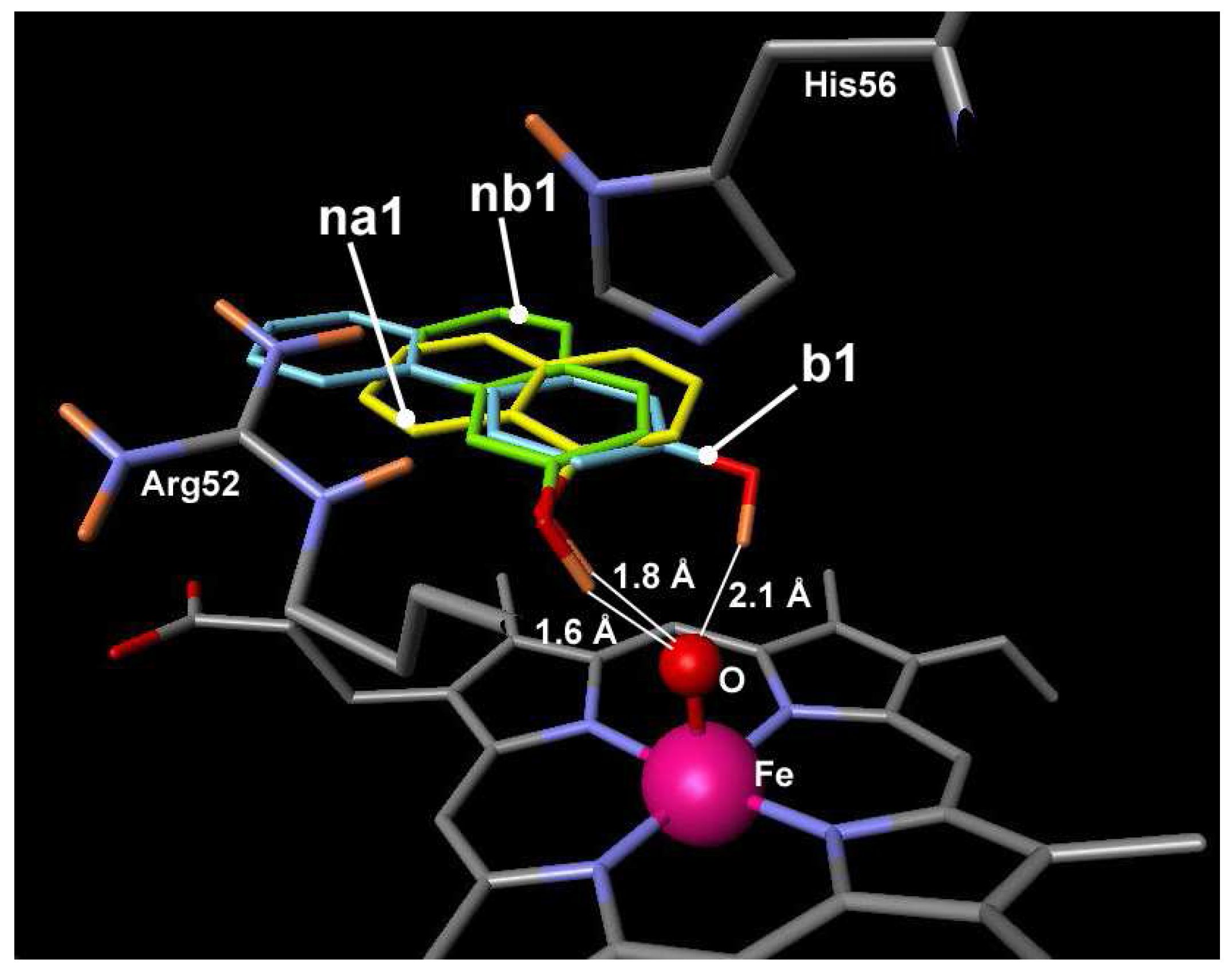
The dockings revealed some amino acids that are capable to make hydrogen bonds with hydroxyl group of the oligomers. All oligomers are capable to make hydrogen bonds with the amino acids that are common for 4-hydroxybiphenyl, 1-naphthols and 2-naphthols compounds: Arg52, Pro154, Asn158, Glu190, Gly191 (
blue in
Figure 3). All interacting amino acids make the heme pocket environment or are the part of the active site itself. Among these amino acids only Arg52 is hydrophilic, while other amino acids are hydrophobic. Hydrogen bonding is made through oxygen atom of C=O group in peptide fragment of the amino acids. Naphthols have more amino acids capable to participate in H-bonding and almost all of these amino acids are hydrophobic too: Ile153, Pro91, Leu192, Leu228. Naphthols make hydrogen bonding with oxygen atom from Fe=O and propionate residue of heme, too.
Figure 3.
Amino acids of ARP forming hydrogen bonding with oligomers. Amino acids in blue are common for 1- naphthol, 2-naphthol and 4-hydroxybiphenyl oligomers. Amino acids in magenta are characteristic to naphthols oligomers.
Figure 3.
Amino acids of ARP forming hydrogen bonding with oligomers. Amino acids in blue are common for 1- naphthol, 2-naphthol and 4-hydroxybiphenyl oligomers. Amino acids in magenta are characteristic to naphthols oligomers.
Table 2.
The docking energy of phenol derivatives and oligomers.
Table 2.
The docking energy of phenol derivatives and oligomers.
| 1-naphthol derivative | Docking energy, kcal/mol | 2-naphthol derivative | Docking energy, kcal/mol | 4-hydroxy-biphenyl derivative | Docking energy, kcal/mol |
|---|
|
|
|
|
|
|
| na1 | -6.4 | nb2 | -6.4 | b1 | -7.0 |
|
|
|
|
|
|
| na2a | -8.4 | nb2a | -7.6 | b2a | -9.0 |
| na2b | -5.6 | nb2b | -6.8 | b2b | -10.5 |
| na2c | -5.4 | nb2c | -9.1 | | |
| na2d | -9.3 | nb2d | -8.5 | | |
| na2e | -9.9 | nb2e | -7.1 | | |
| na2f | -8.6 | nb2f | -8.8 | | |
| na2g | -8.9 | nb2g | -10.0 | | |
| na2h | -9.2 | nb2h | -9.1 | | |
| na2i | -8.7 | nb2i | -9.1 | | |
| na2j | -8.6 | nb2j | -7.1 | | |
| na2k | -8.9 | | | | |
| na2l | -8.3 | | | | |
| na2m | -8.4 | | | | |
|
|
|
|
|
|
| na3a | -5.0 | nb3a | -11.6 | b3a | -11.8 |
| na3b | -10.4 | nb3b | -8.6 | b3b | -11.6 |
| na3c | -8.3 | nb3c | -9.3 | b3c | -12.4 |
| na3d | -11.5 | nb3d | -8.8 | | |
| na3e | -10.3 | | | | |
| na3f | -9.5 | | | | |
|
|
|
|
|
|
| | | | | b4a | -10.5 |
| | | | | b4b | -12.2 |
|
|
|
|
|
|
| na5a | -7.6 | nb5a | -11.4 | | |
| na5b | -13.3 | nb5b | -13.9 | | |
| na5c | -8.0 | nb5c | -8.1 | | |
| na5d | -7.3 | nb5d | -11.4 | | |
| na5e | -9.8 | | | | |
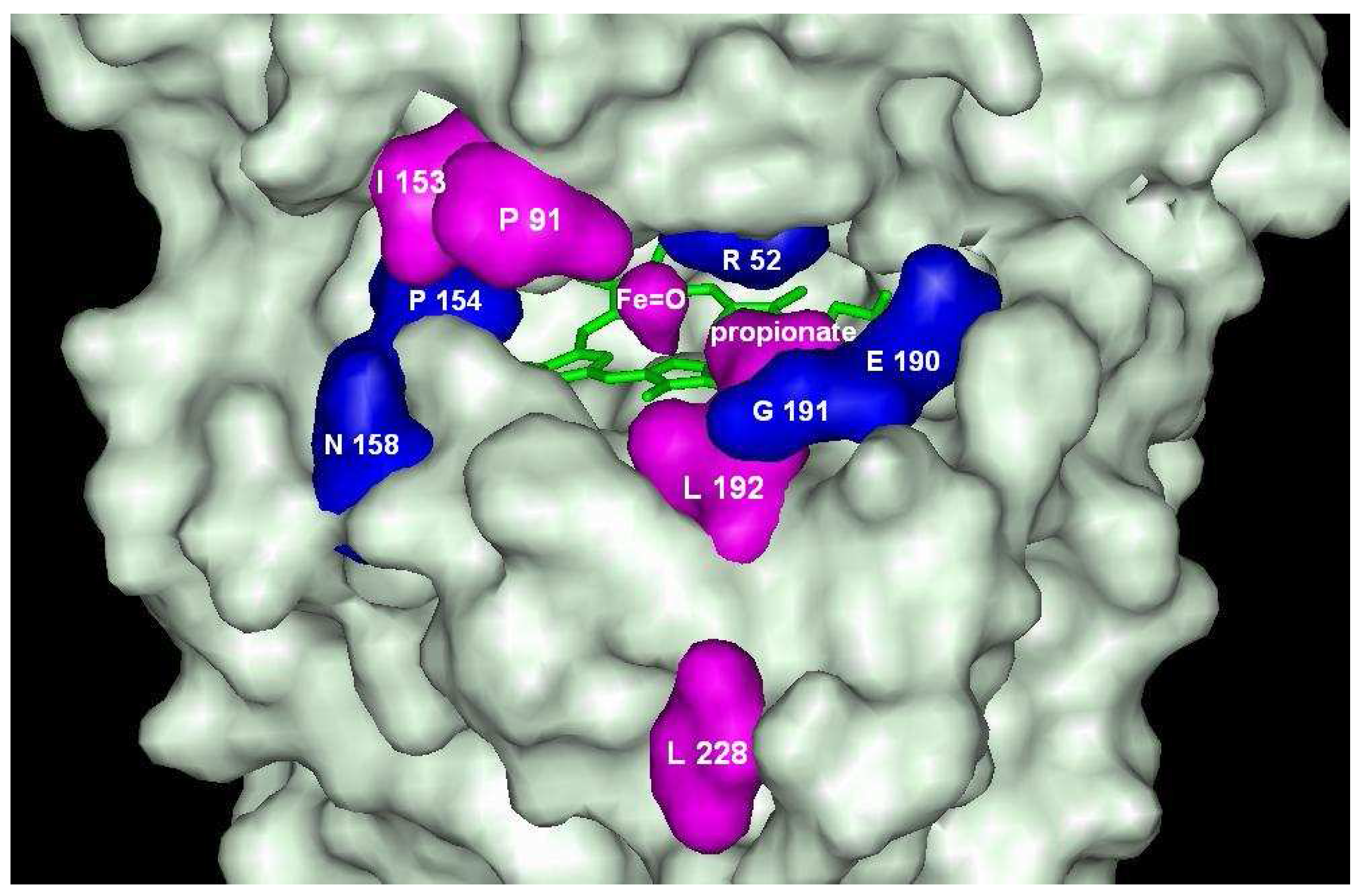
The oligomers of 1-naphthol are docked at the entrance of the active center, but are not able to make hydrogen bond with Fe=O group of ARP. The complex formation energies vary between –5.5 kcal/mol and –9.9 kcal/mol for ten C-dimers and three O-dimers of naphthol derivatives. The lowest energies are found for dimers having bonds through position 7 (
Figure 1),
i.e. for
na2d,
na2e,
na2g and
na2h. The highest energy is calculated for compounds
na2b and
na2c having bonding in position 5. Despite distinct differences in affinities, these compounds occupy almost the same position at the entrance of the heme pocket (
Figure 4). Dockings show that the difference in docking energies are determined by docking peculiarities of the naphthol fragment outside the heme pocket. All dimers have no ability to make hydrogen bonds with Fe=O fragment with exception of derivative
na2e and
na2i. These derivatives may form a complex in which OH groups is located at larger than 2.5 Å distance from Fe=O. In this case hydrogen bond should be very weak. The lowest docking energies for trimers were found for
na3b (-10.4 kcal/mol),
na3d (-11.5 kcal/mol) and
na3e (-10.3 kcal/mol) compounds, which have bonding at the position 7. The remaining trimers made bonding mostly at position 4. The lowest docking energy –13.3 kcal/mol was indicated for pentamer
na5b.
The studied oligomeric O-derivatives of 1-naphthols have only one OH group. Therefore, these compound have limited abilities to form hydrogen bond inside the active center of ARP. However oligomeric O-derivatives have similar and lower docking energies than other oligomeric C-derivatives, which have two and more OH groups. The current finding reveals that hydrogen bonding is not the main factor responsible for binding of oligomers and the hydrophobic forces play much more important role.
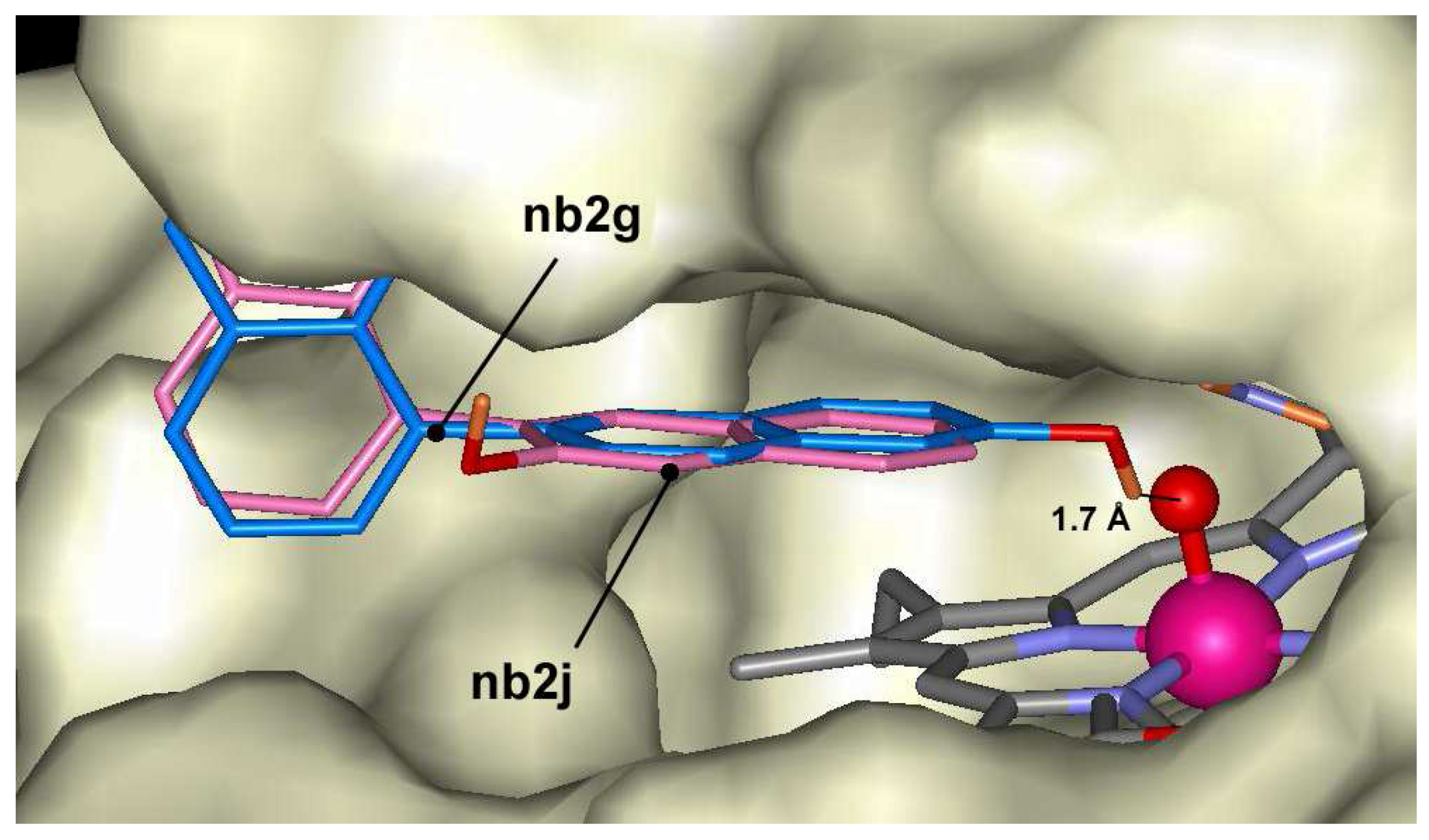
2-naphthol dimers may form 10 dimers through C-C bonding, and the docking of these dimers was calculated in ARP structure. The lowest docking energy was found for compound
nb2g (-10.0 kcal/mol) and some other dimers having bonding at positions 6 and 8 like compounds
nb2i,
nb2h or
nb2c. The highest energies were calculated for compounds
nb2b,
nb2e and
nb2j having bonding at position 3. All mentioned dimers occupy the same position in the heme pocket (
Figure 5). The possible explanation of energy differences might be related to the fact that
nb2b,
nb2e and
nb2j compounds do not form hydrogen bond with Fe=O group like as
nb2g,
nb2h or
nb2c. The last three compounds are exception among oligomers and may form hydrogen bonds with Fe=O group at distances 1.7 Å, 2.0 Å and 1.9 Å, respectively. Trimers of 2-naphthol have the lowest docking energies between –8.6 kcal/mol and –11.6 kcal/mol. Like as dimers the lowest docking energy was calculated for
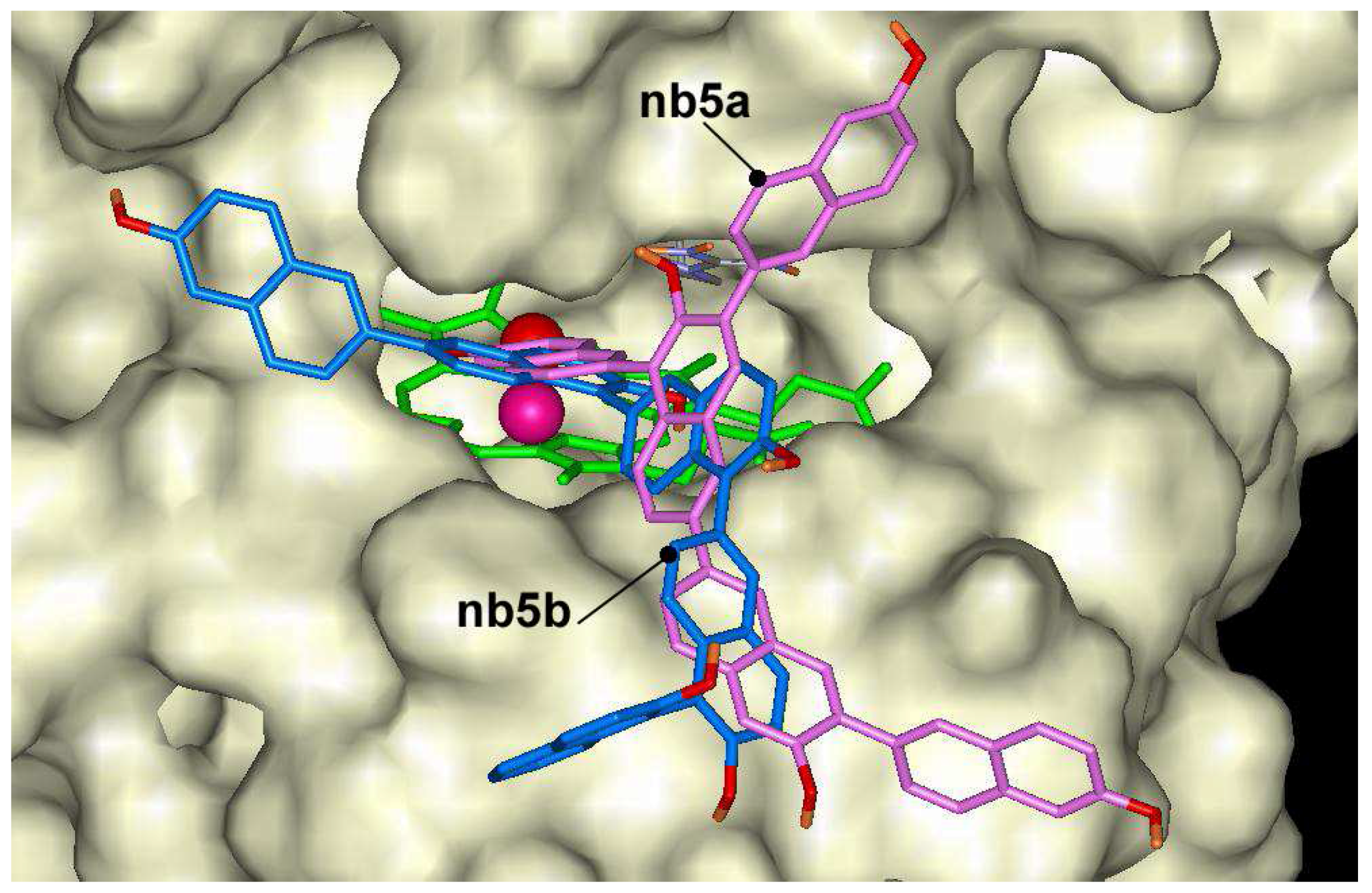
nb3a producing oligomers at 6 th position. The pentamers showed lower docking energies. The lowest docking energy was calculated for pentamer
nb5b (–13.9 kcal/mol) that is formed with bonds at position 6 and 1 (
Figure 6). In general, the lowest affinities of 2-naphthol oligomers were found for ones having bonding through position 3.
Figure 4.
Dockings of na2b (magenta) and na2d (dark orange) in the active center of ARP.
Figure 4.
Dockings of na2b (magenta) and na2d (dark orange) in the active center of ARP.
Figure 5.
Dockings of nb2g (blue) and nb2j (magenta) in the active center of ARP.
Figure 5.
Dockings of nb2g (blue) and nb2j (magenta) in the active center of ARP.
Figure 6.
Dockings of nb5a (magenta) and nb5b (blue) in the active center of ARP.
Figure 6.
Dockings of nb5a (magenta) and nb5b (blue) in the active center of ARP.
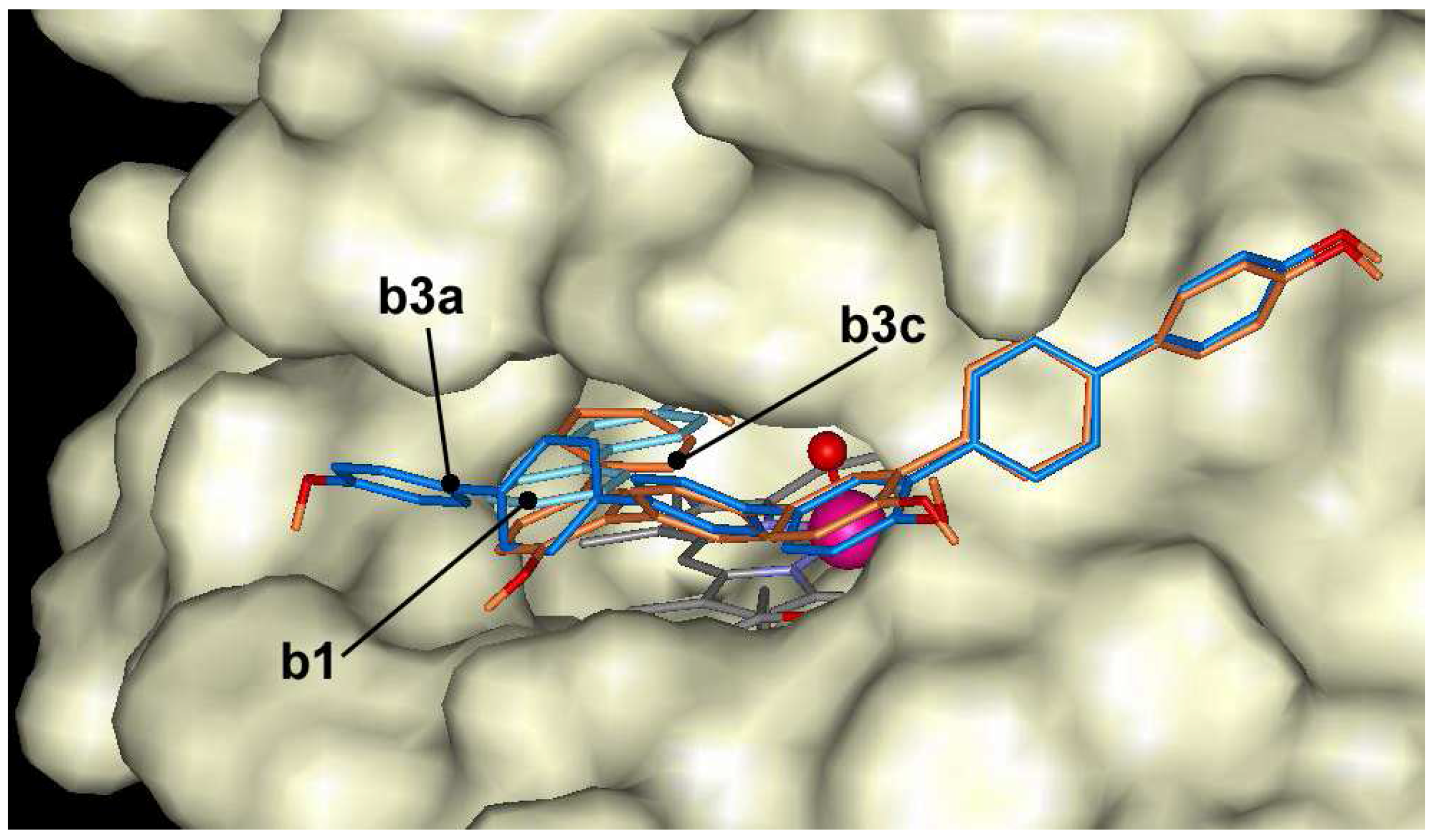
The oligomers of 4-hydroxybiphenyl dock at the entrance of the active center, but are not able to make hydrogen bond with Fe=O group of ARP like as naphthol derivatives. The docking energies vary in the range of –7.0 and –12.4 kcal/mol. 4-hydroxybiphenyl derivatives with linear-shape like
b2a,
b3a and
b4a dock across the entrance to the heme pocket blocking the entrance and can not form hydrogen bond with Fe=O group (
Figure 7). Compounds with crooked shape like
b2b,
b3b and
b4b dock across the entrance to the heme pocket with some hydroxyls in the heme pocket. However, these hydroxyls are located too far from Fe=O group to form hydrogen bond. The compounds with crooked shape have about 1 kcal/mol higher affinity to the active center of ARP than linear compounds.
Figure 7.
The docking of 4-hydroxybiphenyl (b1 - cyan) and trimer (b3a - blue, b3c – dark orange) at the entrance of the active center of ARP.
Figure 7.
The docking of 4-hydroxybiphenyl (b1 - cyan) and trimer (b3a - blue, b3c – dark orange) at the entrance of the active center of ARP.
1-naphthol and 2-naphthol derivatives are more compact compounds and have lower flexibility degree than 4-hydroxybiphenyl and its derivatives, because naphthol derivatives do not have one extra flexible bond. However, 4-hydroxybiphenyl tetramers have too many freely rotating bonds and the docking results is distorted due to high flexibility. The decreasing affinity of pentamers and tetramers can be related to increasing multi-center binding of oligomers. Multi-center binding limits the possibility to exploit all favorable docking places in ARP, especially to compounds with a linear shape.
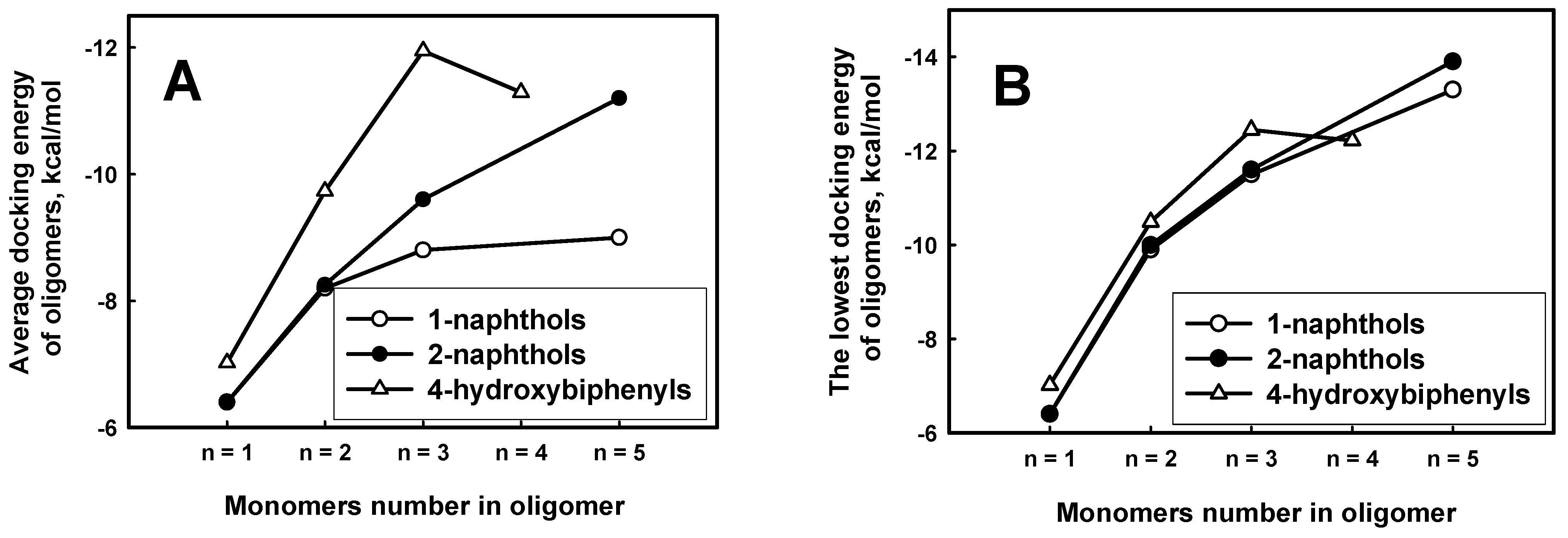
The calculated docking energies of phenol derivatives revealed the obvious tendency that the larger molecular weight of oligomer had the large affinity to ARP (
Figure 8 A,B). This dependence was demonstrated for all derivatives of naphthols. There is also tendency that the average docking energy of explored 4-hydroxybiphenyl oligomers drops faster along with growing oligomer length. It means that 4-hydroxybiphenyl oligomers binds stronger than 1-naphthols and 2-naphthols. According to the average docking energies the oligomers can be placed in decreasing order of binding to ARP: 4-hydroxybiphenyl, 1-naphthols and 2-naphthols. That order is comparable to the decreasing monomer length: the longer monomer length in oligomers chain, the higher affinity to ARP. However, the dependence of lowest docking energies on oligomer lengths is similar to 4-hydroxybiphenyl, 1-naphthols and 2-naphthols (
Figure 8 B) compounds than it is expressed in
Figure 8A.
Figure 8.
The average (A) and the lowest (B) docking energies of 1-hydroxybiphenyl, 1-naphthol and 2-naphthol oligomers through carbon for each length of oligomer.
Figure 8.
The average (A) and the lowest (B) docking energies of 1-hydroxybiphenyl, 1-naphthol and 2-naphthol oligomers through carbon for each length of oligomer.
The docking results reveal that all explored oligomers have the highest binding affinity in the active center of ARP. It means that oligomer might act as substrates of ARP. Even more, the calculations show that they form a stronger complexes in comparison to monomers. The calculations, however, revealed that the most of these compounds did not form hydrogen bonding network inside the active center of ARP. Three 2-naphthol dimers can make weak hydrogen bonds in the active center of ARP, but oligomers of this compound with elongated chain like as trimers, tetramers or pentamers do not form hydrogen bonds at all.
The docking results of oligomers reveal that oligomers do not fulfill the demands for productive binding, which is crucial for substrate oxidation by ARP [
11]. Even the dimer stage for 1-naphthol and 4-hydroxybiphenyl and the trimer stage for 2-naphthol forms unproductive complex. The absence of the proper hydrogen bonding network does not make favorable structure for proton transfer in order an oxidation reaction could appear. Therefore, oligomers of 4-hydroxybiphenyl, 1-naphthol and 2-naphthol serve as concurrent inhibitors, which block the entrance to the active site of ARP and prevent the binding of monomer. The higher affinity of oligomer derivatives guarantees the stable but unproductive oligomer ARP complex formation. That could lead further to the formation of the aggregates of ARP with oligomers or the aggregates of pure oligomers.