Evaluation of Oil-Palm Fungal Disease Infestation with Canopy Hyperspectral Reflectance Data
Abstract
:1. Introduction
2. Materials and Methods
2.1. Test site and ground-truth
2.2. Hyperspectral data
2.3. Spectra pre-processing
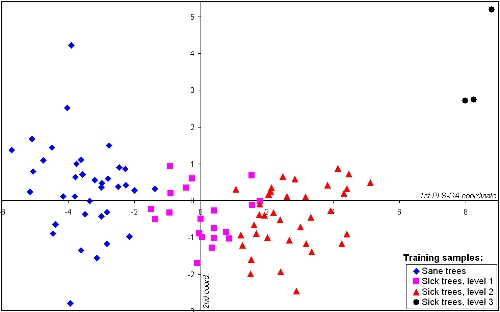
2.4. Partial Least Square Discrimination
- a Partial Least Square Regression (PLSR) [27,28] was calculated between the preprocessed derivative spectra and a disease degree, to transform the spectral data into uncorrelated latent variables that provides an invertible matrix for subsequent factorial discriminant analysis. Compared to data reduction such as classical band selection or vegetation index derivation, it keeps most of the initial wavelength sampling, discarding only spectral domains that provide no information, or information already contained in other domains. Figure 6 shows for instance the contribution of each wavelength to the derived PLSR components, which are significant for all the covered range except between 790 and 880 nm. To perform a simple PLSR, we chose to use it as a predictor of continuous values in the bin 0 to 1 [28,29]. Therefore, to set the disease degree, we have established an arbitrary scale suggested by a trivial linear unmixing based on the two endmembers “healthy” and “critically sick” status. We assigned values of 0 to the healthy trees, 0.4 to the trees in level 1, 0.6 to those in level 2, and 1 to those in level 3 of Ganoderma attack.
- a Linear Discriminant Analysis (LDA) [30] was applied to the first most significant latent variables, enhancing the interclass variability while minimizing the intraclass variability of the sample to build a classification model. The selection of the number of PLSR components is guided by the compromise between minimization of Root Mean Square Error of Cross Validation (RMSECV), gain in determination coefficient (R2) between predicted and reference values, and stability of the model thanks to the fewer number of implied variables [31].
3. Results and Discussion
- - if x < −2, the tree is healthy;
- - if x > 6 or 7, the tree is dramatically sick, almost dead;
- - if −2 < x < 6, the threshold between Level1 and Level2 of disease severity is fuzzier and lays between 1.2 and 1.5.
4. Conclusions
Acknowledgments
References
- Wood, B.J. Opportunities for oil palm R&D in further meeting the challenges of the new dynamics. Planters 2007, 83, 155–177. [Google Scholar]
- Breton, F.; Hasan, Y.; Hariadi; Lubis, Z.; de Franqueville, H. Characterization of parameters for the development of an early screening test for basal stem rot tolerance in oil palm progenies. J. Oil Palm Res 2006, 24–36. [Google Scholar]
- Ganoderma Diseases of Perennial Crops; Flood, J.; Bridge, P.D.; Holderness, M. (Eds.) CABI Publishing: Wallingford, UK, 2000.
- Utomo, C.; Nielpold, F. Development of diagnostic methods for detecting Ganoderma-infected oil palms. J. Phytopathol 2000, 148, 235–247. [Google Scholar]
- Bridge, P.D.; O’Grady, E.B.; Pilotti, C.A. Development of molecular diagnostics for the detection of Ganoderma isolates pathogenic to oil palm. In Ganoderma Diseases of Perennial Crops; CABI Publishing: Wallingford, UK, 2000; pp. 225–234. [Google Scholar]
- Chaerle, L.; van der Straeten, D. Imaging techniques and the early detection of plant stress. Trends Plant Sci 2000, 5, 295–500. [Google Scholar]
- Larsolle, A.; Muhammed, H.H. Measuring crop status using multivariate analysis of hyperspectral field reflectance with application to disease severity and plant density. Prec. Agric 2007, 8, 1385–2256. [Google Scholar]
- Liew, O.W.; Chong, P.C.J.; Li, G.; Asundi, A.K. Signature optical clues: emerging technologies for monitoring plant health. Sensors 2008, 8, 3205–3239. [Google Scholar]
- Muhammed, H.H. Hyperspectral crop reflectance data for characterizing and estimating Fungal Disease Severity in Wheat. Biosyst. Eng 2005, 91, 9–20. [Google Scholar]
- Wang, W.; Zhang, M.; Zhu, J.; Geng, S. Spectral prediction of phytophtora infestans infection on tomatoes using artificial neural netword (ANN). Int. J. Remote Sens 2008, 29, 1693–1706. [Google Scholar]
- Nilsson, H.E. Remote sensing and image analysis in plant pathology. Ann. Rev. Phytopathol 1995, 15, 489–527. [Google Scholar]
- Apan, A.; Held, A.; Phinn, S.; Markley, J. Detecting sugarcane ‘orange rust’ disease using EO-1 hyperion hyperspectral imagery. Int. J. Remote Sens 2004, 25, 489–498. [Google Scholar]
- Goodwin, N.; Coops, C.C.; Stone, C. Assessing plantation canopy condition from airborne imagery using spectral mixture analysis and fractional abundances. Int. J. Appl. Earth Observ. Geoinf 2005, 7, 11–28. [Google Scholar]
- Shafri, H.Z.M.; Hamdam, N. Hyperspectral imagery for mapping disease infection in oil palm plantation using vegetation indices and red edge techniques. Am. J. Appl. Sci 2009, 6, 1031–1035. [Google Scholar]
- Shafri, H.Z.M.; Anuar, M.I. Hyperspectral analysis for detecting disease infection in oil palms. Proceedings of International Conference on Computer and Electrical Engineering, Phuket, Thailand, December 20–22, 2008; pp. 312–316.
- Jorgensen, R.N.; Christensen, L.K.; Bros, R. Spectral reflectance at sub-leaf scale including the spatial distribution discriminating NPK stress characteristics in barley using multiway partial least square regression. Int. J. Remote Sens 2007, 28, 943–962. [Google Scholar]
- Huang, J.F.; Apan, A. Detection of sclerotinia rot disease on celery using hyperspectral data and partial least squares regression. J. Space Sci 2006, 51, 129–142. [Google Scholar]
- Zhang, M.; Liu, X.; O’Neill, M. Spectral discrimination of Phytophtora infestans infection on tomatoes based on principal component and cluster analyses. Int. J. Remote Sens 2002, 23, 1095–1107. [Google Scholar]
- Fairhurst, T.; Caliman, J.P.; Härdter, R.; Witt, C. Oil Palm: Nutrient Disorders and Nutrient Management; Oil Palm Series; PPI/PPIC-IPI: Singapore, 2005; Volume 7. [Google Scholar]
- Savitsky, A.; Golay, M.J.E. Smoothing and differentiation of data by simplified least-squares procedures. Anal. Chem 1964, 36, 1627–1639. [Google Scholar]
- Tsai, F.; Philpot, W. Derivative analysis of hyperspectral data. Remote Sens. Environ 1998, 66, 41–51. [Google Scholar]
- Estep, L.; Carter, G.A. Derivative analysis of AVIRIS data for crop stress detection. Photogram. Eng. Remote Sens 2005, 71, 1417–1421. [Google Scholar]
- Ruffin, C.; King, R.L.; Younani, N.H. A combined derivative spectroscopy and Savitsky-Golay filtering method for the analysis of hyperspectral data. Geosci. Remote Sens 2008, 45, 1–15. [Google Scholar]
- Browne, M.; Mayer, N.; Cutmore, T.R.H. A multiscale polynomial filter for adaptive smoothing. Digital Signal Process 2007, 17, 69–75. [Google Scholar]
- Roger, J.M.; Palagos, B.; Guillaume, S.; Bellon-Maurel, V. Discriminating from highly multivariate data by focal eigen function discriminant analysis; application to NIR spectra. Chemometr. Intell. Lab. Syst 2005, 79, 31–41. [Google Scholar]
- Gorretta, N.; Roger, J.M.; Aubert, M.; Bellon-Maurel, V.; Campan, F.; Roumet, P. Determining vitreousness of durum wheat kernels using near infrared hyperspectral imaging. J. Near Infrared Spectrosc 2006, 14, 231–239. [Google Scholar]
- Martens, H.; Naes, T. Multivariate Calibration; Wiley & Sons: Chichester, UK, 1992. [Google Scholar]
- Luinge, H.J.; vander Maas, J.H.; Visser, T. Partial least squares regression as a multivariate tool for the interpretation of infrared spectra. Chemiometr. Intell. Lab. Syst 1995, 28, 129–138. [Google Scholar]
- Preda, C.; Saporta, G. PLS regression on a stochastic process. Comput. Stat. Data Anal 2005, 48, 149–158. [Google Scholar]
- Lebart, L.; Morineau, A.; Warwick, K. Multivariate Descriptive Statistical Analysis: Correspondence Analysis and Related Techniques for Large Matrices; Wiley & Sons: New York, NY, USA, 1984. [Google Scholar]
- Seasholtz, M.B.; Kowalski, B. The parsimoy principle applied to multivariate calibration. Anal. Chim. Acta 1993, 277, 165–177. [Google Scholar]
- Anderssen, E.; Dyrstad, K.; Westad, F.; Martens, H. Reducing over-optimism in variable selection by cross-model validation. Chemiometr. Intell. Lab. Syst 2006, 84, 69–74. [Google Scholar]
- Bro, R.; Kjeldahl, K.; Smilde, A.K.; Kiers, H.A.L. Cross-validation of components models: A critical look at current methods. Anal. Bioanal. Chem 2008, 390, 1241–1251. [Google Scholar]







| Infection degree | Evolution of stem conditions | Evolution of canopy structure |
|---|---|---|
| Level 1 | Presence of mycelium in the stem bark, or crumbly wood | Yellowing or drying of some leaves. One or two new leaves remain as unopened spears. |
| Level 2 | Presence of fruiting bodies (mushrooms) at the bottom of the stem | Apparition of leaf necrosis. Three to five new leaves remain as unopened spears. Declination of older leaves. |
| Level 3 | Rotten stem | Largely spread leaf necrosis. No new leaf. No new bunch. «Skirt-like» shape of crown due to total leaf declination. |
| Classification result | ||||||
|---|---|---|---|---|---|---|
| Level | 0 | 1 | 2 | 3 | % of good classification | |
| Actual status | 0 | 34 | 2 | 0 | 0 | 94 % |
| 1 | 0 | 16 | 2 | 0 | 89 % | |
| 2 | 0 | 2 | 36 | 0 | 95 % | |
| 3 | 0 | 0 | 0 | 3 | 100 % | |
©2010 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/)
Share and Cite
Lelong, C.C.D.; Roger, J.-M.; Brégand, S.; Dubertret, F.; Lanore, M.; Sitorus, N.A.; Raharjo, D.A.; Caliman, J.-P. Evaluation of Oil-Palm Fungal Disease Infestation with Canopy Hyperspectral Reflectance Data. Sensors 2010, 10, 734-747. https://doi.org/10.3390/s100100734
Lelong CCD, Roger J-M, Brégand S, Dubertret F, Lanore M, Sitorus NA, Raharjo DA, Caliman J-P. Evaluation of Oil-Palm Fungal Disease Infestation with Canopy Hyperspectral Reflectance Data. Sensors. 2010; 10(1):734-747. https://doi.org/10.3390/s100100734
Chicago/Turabian StyleLelong, Camille C. D., Jean-Michel Roger, Simon Brégand, Fabrice Dubertret, Mathieu Lanore, Nurul A. Sitorus, Doni A. Raharjo, and Jean-Pierre Caliman. 2010. "Evaluation of Oil-Palm Fungal Disease Infestation with Canopy Hyperspectral Reflectance Data" Sensors 10, no. 1: 734-747. https://doi.org/10.3390/s100100734
APA StyleLelong, C. C. D., Roger, J.-M., Brégand, S., Dubertret, F., Lanore, M., Sitorus, N. A., Raharjo, D. A., & Caliman, J.-P. (2010). Evaluation of Oil-Palm Fungal Disease Infestation with Canopy Hyperspectral Reflectance Data. Sensors, 10(1), 734-747. https://doi.org/10.3390/s100100734