Single-Step Purification of Monomeric l-Selectin via Aptamer Affinity Chromatography
Abstract
:1. Introduction
2. Materials and Methods
2.1. Protein Expression
2.2. Preparation of the Aptamer Column
2.3. Purification of l-Selectin LE-His
2.4. Endoglycosidase Digestion
2.5. SDS-PAGE Staining and Western Blotting
2.6. Selectin Quantification via ELISA and SPR
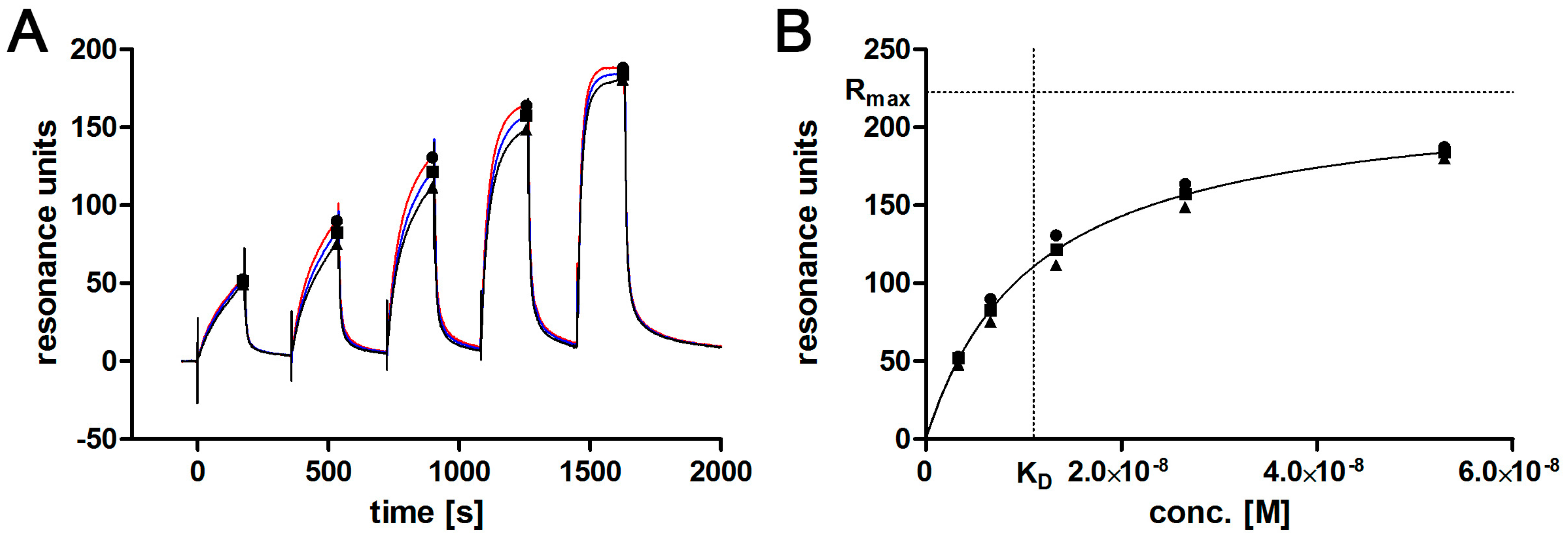
2.7. Affinity Determination via SPR
3. Results and Discussion
3.1. Protein Purification by Ni-NTA and AAC
3.2. Aptamer Affinity to l-Selectin
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Porath, J.; Carlsson, J.; Olsson, I.; Belfrage, G. Metal chelate affinity chromatography, a new approach to protein fractionation. Nature 1975, 258, 598–599. [Google Scholar] [CrossRef] [PubMed]
- Hage, D.S.; Matsuda, R. Affinity chromatography: A historical perspective. Method. Mol. Biol. 2015, 1286, 1–19. [Google Scholar] [CrossRef]
- Zhao, Q.; Wu, M.; Chris, L.X.; Li, X.-F. Applications of aptamer affinity chromatography. TrAC Trends Anal. Chem. 2012, 41, 46–57. [Google Scholar] [CrossRef]
- Keefe, A.D.; Pai, S.; Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010, 9, 537–550. [Google Scholar] [CrossRef] [PubMed]
- Acquah, C.; Danquah, M.K.; Yon, J.L.; Sidhu, A.; Ongkudon, C.M. A review on immobilised aptamers for high throughput biomolecular detection and screening. Anal. Chim. Acta 2015, 888, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Hianik, T.; Ostatna, V.; Zajacova, Z.; Stoikova, E.; Evtugyn, G. Detection of aptamer-protein interactions using QCM and electrochemical indicator methods. Bioorg. Med. Chem. Lett. 2005, 15, 291–295. [Google Scholar] [CrossRef] [PubMed]
- Ley, K.; Laudanna, C.; Cybulsky, M.I.; Nourshargh, S. Getting to the site of inflammation: The leukocyte adhesion cascade updated. Nat. Rev. Immunol. 2007, 7, 678–689. [Google Scholar] [CrossRef] [PubMed]
- Muller, W.A. Leukocyte-endothelial cell interactions in the inflammatory response. Lab. Investig. 2002, 82, 521–533. [Google Scholar] [CrossRef]
- Sperandio, M.; Pickard, J.; Unnikrishnan, S.; Acton, S.T.; Ley, K. Analysis of leukocyte rolling in vivo and in vitro. Method. Enzymol. 2006, 416, 346–371. [Google Scholar]
- Ley, K. The role of selectins in inflammation and disease. Trends Mol. Med. 2003, 9, 263–268. [Google Scholar] [CrossRef]
- Hicke, B.J.; Watson, S.R.; Koenig, A.; Lynott, C.K.; Bargatze, R.F.; Chang, Y.F.; Ringquist, S.; Moon-McDermott, L.; Jennings, S.; Fitzwater, T.; et al. DNA aptamers block l-selectin function in vivo. Inhibition of human lymphocyte trafficking in scid mice. J. Clin. Investig. 1996, 98, 2688–2692. [Google Scholar] [CrossRef] [PubMed]
- Romig, T.S.; Bell, C.; Drolet, D.W. Aptamer affinity chromatography: Combinatorial chemistry applied to protein purification. J. Chromatogr. B Biomed. Sci. Appl. 1999, 731, 275–284. [Google Scholar] [CrossRef]
- Riese, S.B.; Buscher, K.; Enders, S.; Kuehne, C.; Tauber, R.; Dernedde, J. Structural requirements of mono-and multivalent l-selectin blocking aptamers for enhanced receptor inhibition in vitro and in vivo. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 901–908. [Google Scholar] [CrossRef] [PubMed]
- Wedepohl, S.; Kaup, M.; Riese, S.B.; Berger, M.; Dernedde, J.; Tauber, R.; Blanchard, V. N-glycan analysis of recombinant l-selectin reveals sulfated GalNac and GalNac-GalNac motifs. J. Proteome Res. 2010, 9, 3403–3411. [Google Scholar] [CrossRef] [PubMed]
- Christensen, L.L. Theoretical analysis of protein concentration determination using biosensor technology under conditions of partial mass transport limitation. Anal. Biochem. 1997, 249, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Richalet-Secordel, P.M.; Rauffer-Bruyere, N.; Christensen, L.L.; Ofenloch-Haehnle, B.; Seidel, C.; van Regenmortel, M.H. Concentration measurement of unpurified proteins using biosensor technology under conditions of partial mass transport limitation. Anal. Biochem. 1997, 249, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Sigmundsson, K.; Másson, G.; Rice, R.; Beauchemin, N.; Öbrink, B. Determination of active concentrations and association and dissociation rate constants of interacting biomolecules: An analytical solution to the theory for kinetic and mass transport limitations in biosensor technology and its experimental verification. Biochemistry 2002, 41, 8263–8276. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, R.; Katsamba, P.S.; Nordin, H.; Pol, E.; Myszka, D.G. Analyzing a kinetic titration series using affinity biosensors. Anal. Biochem. 2006, 349, 136–147. [Google Scholar] [CrossRef] [PubMed]


| Aptamer | Sequence | Reference |
|---|---|---|
| LD201 | 5′-CAAGGTAACC AGTACAAGGT GCTAAACGTA ATGGCTTCG-3′ | Hicke et al. 1996 |
| LD201mod | 5′-GCGGTAACC AGTACAAGGT GCTAAACGTA ATGGCGC-3′ | Romig et al. 1999 |
| LD201mΔ1 | 5′-GC · · · · C AGTACAAGGT GCTAAACGTA ATGGC-3′ | Riese et al. 2016 |
| Affinity Matrix | l-sel LE-His (µg) (a) | Yield (%) (b) |
|---|---|---|
| Ni-NTA | 167 ± 21 | 17 |
| LD201mΔ1 | 608 ± 23 | 62 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuehne, C.; Wedepohl, S.; Dernedde, J. Single-Step Purification of Monomeric l-Selectin via Aptamer Affinity Chromatography. Sensors 2017, 17, 226. https://doi.org/10.3390/s17020226
Kuehne C, Wedepohl S, Dernedde J. Single-Step Purification of Monomeric l-Selectin via Aptamer Affinity Chromatography. Sensors. 2017; 17(2):226. https://doi.org/10.3390/s17020226
Chicago/Turabian StyleKuehne, Christian, Stefanie Wedepohl, and Jens Dernedde. 2017. "Single-Step Purification of Monomeric l-Selectin via Aptamer Affinity Chromatography" Sensors 17, no. 2: 226. https://doi.org/10.3390/s17020226
APA StyleKuehne, C., Wedepohl, S., & Dernedde, J. (2017). Single-Step Purification of Monomeric l-Selectin via Aptamer Affinity Chromatography. Sensors, 17(2), 226. https://doi.org/10.3390/s17020226






