Detection of Matrix Metalloproteinase Activity by Bioluminescence via Intein-Mediated Biotinylation of Luciferase
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. RLuc8-pep-GyrA Plasmid Construction
2.3. Preparation for RLuc8-m7-Bio and Rluc8-pep-C via Intein-Mediated Ligation
2.4. Sample Preparation
2.5. Determination of Biotinylation and Unbiotinylation of Proteins Using Biolayer Interferometry
2.6. MMP Assays on the Microplate
3. Results and Discussion
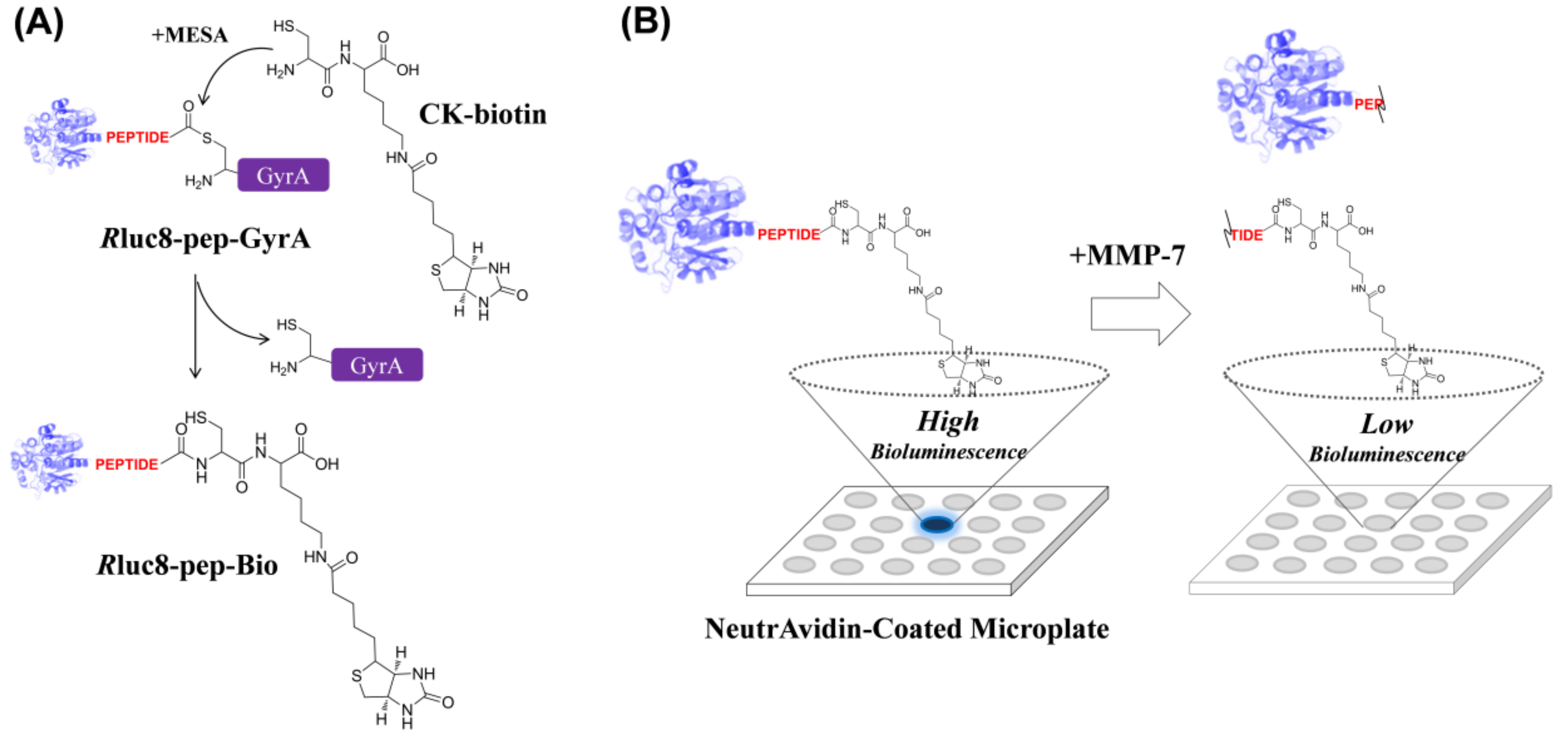
3.1. Intein-Mediated Site-Specific Biotinylation of Luciferase
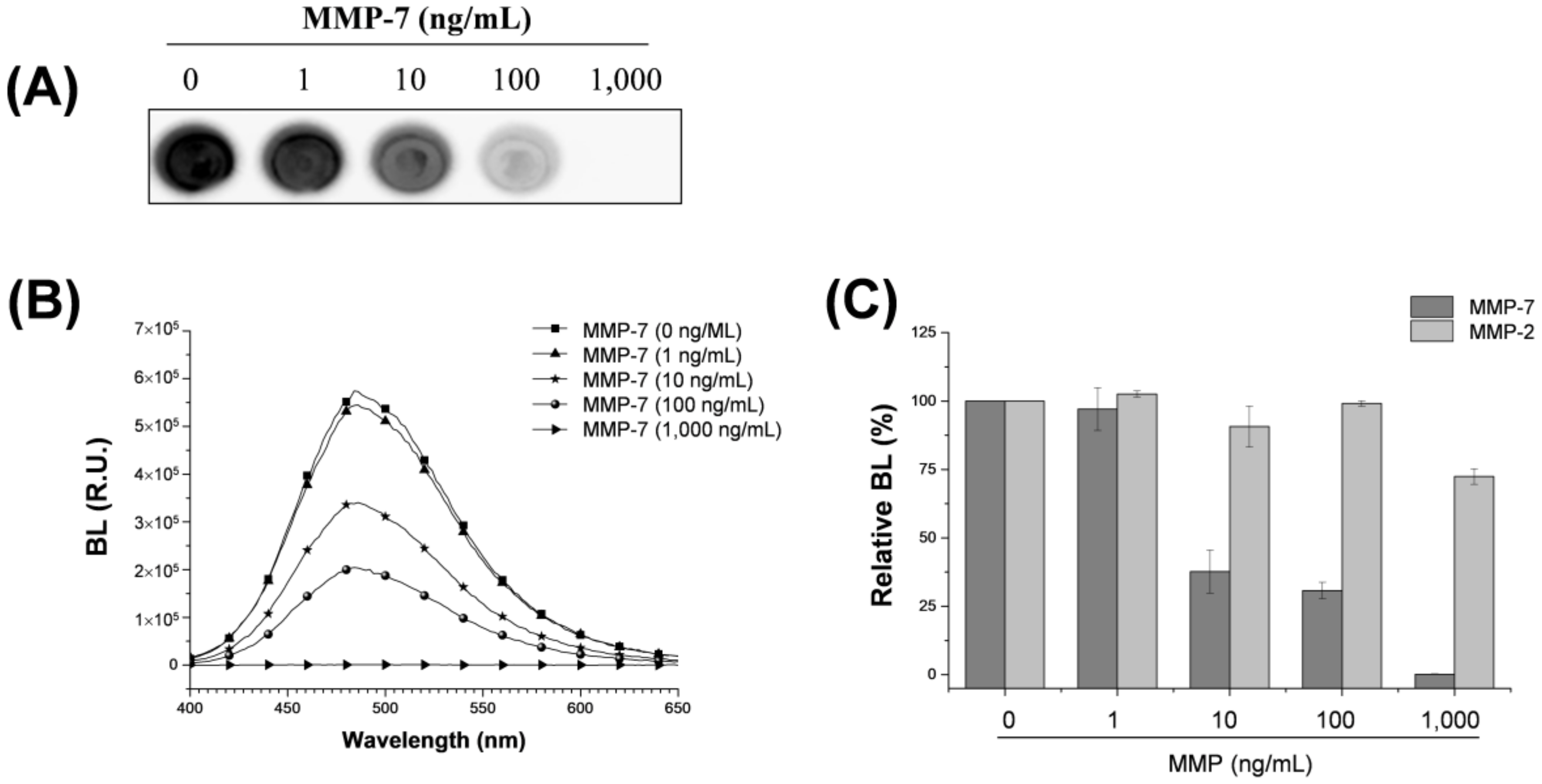
3.2. Assaying MMP-7 Activity
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Jeftinija, S. The story of cell secretion: Events leading to the discovery of the ’porosome’—The universal secretory machinery in cells. J. Cell. Mol. Med. 2006, 10, 273–279. [Google Scholar] [PubMed]
- Page-McCaw, A.; Ewald, A.J.; Werb, Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat. Rev. Mol. Cell Biol. 2007, 8, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Gialeli, C.; Theocharis, A.D.; Karamanos, N.K. Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. FEBS J. 2011, 278, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Snoek-van Beurden, P.A.M.; Von den Hoff, J.W. Zymographic techniques for the analysis of matrix metalloproteinases and their inhibitors. Biotechniques 2005, 38, 73–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, G.B.; Kim, K.H.; Park, Y.H.; Ko, S.; Kim, Y.P. Colorimetric assay of matrix metalloproteinase activity based on metal-induced self-assembly of carboxy gold nanoparticles. Biosens. Bioelectron. 2013, 41, 833–839. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.Q.; Park, Y.M.; Yang, D.; Yoo, T.H.; Yoon, H.C. Development of a matrix metalloproteinase-2 (MMP-2) biosensing system by integrating an enzyme-mediated color development reaction into a common electronics components setup. Biochip J. 2016, 10, 198–207. [Google Scholar] [CrossRef]
- Fernandez, A.; Vendrell, M. Smart fluorescent probes for imaging macrophage activity. Chem. Soc. Rev. 2016, 45, 1182–1196. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, Z.H.; Lin, J.Q.; Lu, J.L.; Liu, B.F.; Zeng, S.Q.; Luo, Q.M. Detection of MMP activity in living cells by a genetically encoded surface-displayed fret sensor. Biochim. Biophys. Acta Mol. Cell Res. 2007, 1773, 400–407. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Kim, Y.P. Fluorescent and bioluminescent nanoprobes for in vitro and in vivo detection of matrix metalloproteinase activity. BMB Rep. 2015, 48, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Lee, S.M.; Kim, G.B.; Kim, Y.P. Gold nanoparticle-based fluorescence quenching via metal coordination for assaying protease activity. Gold Bull. 2012, 45, 213–219. [Google Scholar] [CrossRef]
- Kim, G.B.; Kim, Y.P. Analysis of protease activity using quantum dots and resonance energy transfer. Theranostics 2012, 2, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.P.; Oh, Y.H.; Oh, E.; Ko, S.; Han, M.K.; Kim, H.S. Energy transfer-based multiplexed assay of proteases by using gold nanoparticle and quantum dot conjugates on a surface. Anal. Chem. 2008, 80, 4634–4641. [Google Scholar] [CrossRef] [PubMed]
- Frangioni, J.V. In Vivo near-infrared fluorescence imaging. Curr. Opin. Chem. Biol. 2003, 7, 626–634. [Google Scholar] [CrossRef] [PubMed]
- Owens, E.A.; Henary, M.; El Fakhri, G.; Choi, H.S. Tissue-specific near-infrared fluorescence imaging. Acc. Chem. Res. 2016, 49, 1731–1740. [Google Scholar] [CrossRef] [PubMed]
- Paley, M.A.; Prescher, J.A. Bioluminescence: A versatile technique for imaging cellular and molecular features. MedChemComm 2014, 5, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.O.; Lim, B.; Kim, Y.-P. Bioluminescence resonance energy transfer nanoprobes for imaging. IEEE J. Sel. Top. Quantum Electron. 2014, 20, 1–10. [Google Scholar]
- Kim, Y.P.; Daniel, W.L.; Xia, Z.; Xie, H.; Mirkin, C.A.; Rao, J. Bioluminescent nanosensors for protease detection based upon gold nanoparticle-luciferase conjugates. Chem. Commun. 2010, 46, 76–78. [Google Scholar] [CrossRef] [PubMed]
- Xia, Z.Y.; Xing, Y.; So, M.K.; Koh, A.L.; Sinclair, R.; Rao, J.H. Multiplex detection of protease activity with quantum dot nanosensors prepared by intein-mediated specific bioconjugation. Anal. Chem. 2008, 80, 8649–8655. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.Q.; Zhang, Y.; Xiao, F.; Xia, Z.Y.; Rao, J.H. Quantum dot/bioluminescence resonance energy transfer based highly sensitive detection of proteases. Angew. Chem. Int. Ed. 2007, 46, 4346–4349. [Google Scholar] [CrossRef] [PubMed]
- Dawson, P.E.; Muir, T.W.; Clark-Lewis, I.; Kent, S.B. Synthesis of proteins by native chemical ligation. Science 1994, 266, 776–779. [Google Scholar] [CrossRef] [PubMed]
- Muir, T.W. Semisynthesis of proteins by expressed protein ligation. Annu. Rev. Biochem. 2003, 72, 249–289. [Google Scholar] [CrossRef] [PubMed]
- Machova, Z.; Beck-Sickinger, A.G. Expressed protein ligation for protein semisynthesis and engineering. Methods Mol. Biol. 2005, 298, 105–130. [Google Scholar] [PubMed]
- Schwarzer, D.; Cole, P.A. Protein semisynthesis and expressed protein ligation: Chasing a protein’s tail. Curr. Opin. Chem. Biol. 2005, 9, 561–569. [Google Scholar] [CrossRef] [PubMed]
- Topilina, N.I.; Mills, K.V. Recent advances in in vivo applications of intein-mediated protein splicing. Mob. DNA 2014, 5, 5. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.H.; Park, G.C.; Lee, S.M.; Lee, J.H.; Lim, B.; Hwang, S.M.; Kim, J.H.; Park, H.; Joo, J.; Kim, Y.P. Surface-tunable bioluminescence resonance energy transfer via geometry-controlled zno nanorod coordination. Small 2015, 11, 3469–3475. [Google Scholar] [CrossRef] [PubMed]
- Loening, A.M.; Fenn, T.D.; Wu, A.M.; Gambhir, S.S. Consensus guided mutagenesis of Renilla luciferase yields enhanced stability and light output. Protein Eng. Des. Sel. 2006, 19, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Klabunde, T.; Sharma, S.; Telenti, A.; Jacobs, W.R., Jr.; Sacchettini, J.C. Crystal structure of gyra intein from Mycobacterium xenopi reveals structural basis of protein splicing. Nat. Struct. Biol. 1998, 5, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Turk, B.E.; Huang, L.L.; Piro, E.T.; Cantley, L.C. Determination of protease cleavage site motifs using mixture-based oriented peptide libraries. Nat. Biotechnol. 2001, 19, 661–667. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, Y.; Grunebach, F.; Schmidt, S.M.; Heine, A.; Hantschel, M.; Stevanovic, S.; Rammensee, H.G.; Brossart, P. Matrilysin (MMP-7) is a novel broadly expressed tumor antigen recognized by antigen-specific T cells. Clin. Cancer Res. 2008, 14, 5503–5511. [Google Scholar] [CrossRef] [PubMed]
- Edman, K.; Furber, M.; Hemsley, P.; Johansson, C.; Pairaudeau, G.; Petersen, J.; Stocks, M.; Tervo, A.; Ward, A.; Wells, E.; et al. The discovery of MMP7 inhibitors exploiting a novel selectivity trigger. ChemMedChem 2011, 6, 769–773. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.P.; Lee, B.S.; Kim, E.; Choi, I.S.; Moon, D.W.; Lee, T.G.; Kim, H.S. Activity-based assay of matrix metalloproteinase on nonbiofouling surfaces using time-of-flight secondary ion mass spectrometry. Anal. Chem. 2008, 80, 5094–5102. [Google Scholar] [CrossRef] [PubMed]
- Nuttall, R.K.; Sampieri, C.L.; Pennington, C.J.; Gill, S.E.; Schultz, G.A.; Edwards, D.R. Expression analysis of the entire MMP and TIMP gene families during mouse tissue development. FEBS Lett. 2004, 563, 129–134. [Google Scholar] [CrossRef]
- Pei, D.Q. Identification and characterization of the fifth membrane-type matrix metalloproteinase MT5-MMP. J. Biol. Chem. 1999, 274, 8925–8932. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, D.L.; Kim, H.; Kim, D.; Lee, J.O.; Gye, M.C.; Kim, Y.-P. Detection of Matrix Metalloproteinase Activity by Bioluminescence via Intein-Mediated Biotinylation of Luciferase. Sensors 2018, 18, 875. https://doi.org/10.3390/s18030875
Nguyen DL, Kim H, Kim D, Lee JO, Gye MC, Kim Y-P. Detection of Matrix Metalloproteinase Activity by Bioluminescence via Intein-Mediated Biotinylation of Luciferase. Sensors. 2018; 18(3):875. https://doi.org/10.3390/s18030875
Chicago/Turabian StyleNguyen, Duc Long, Hanim Kim, Dasom Kim, Jin Oh Lee, Myung Chan Gye, and Young-Pil Kim. 2018. "Detection of Matrix Metalloproteinase Activity by Bioluminescence via Intein-Mediated Biotinylation of Luciferase" Sensors 18, no. 3: 875. https://doi.org/10.3390/s18030875
APA StyleNguyen, D. L., Kim, H., Kim, D., Lee, J. O., Gye, M. C., & Kim, Y. -P. (2018). Detection of Matrix Metalloproteinase Activity by Bioluminescence via Intein-Mediated Biotinylation of Luciferase. Sensors, 18(3), 875. https://doi.org/10.3390/s18030875





