Rapid Measurement of Soybean Seed Viability Using Kernel-Based Multispectral Image Analysis
Abstract
:1. Introduction
2. Methods
2.1. Sample Preparation
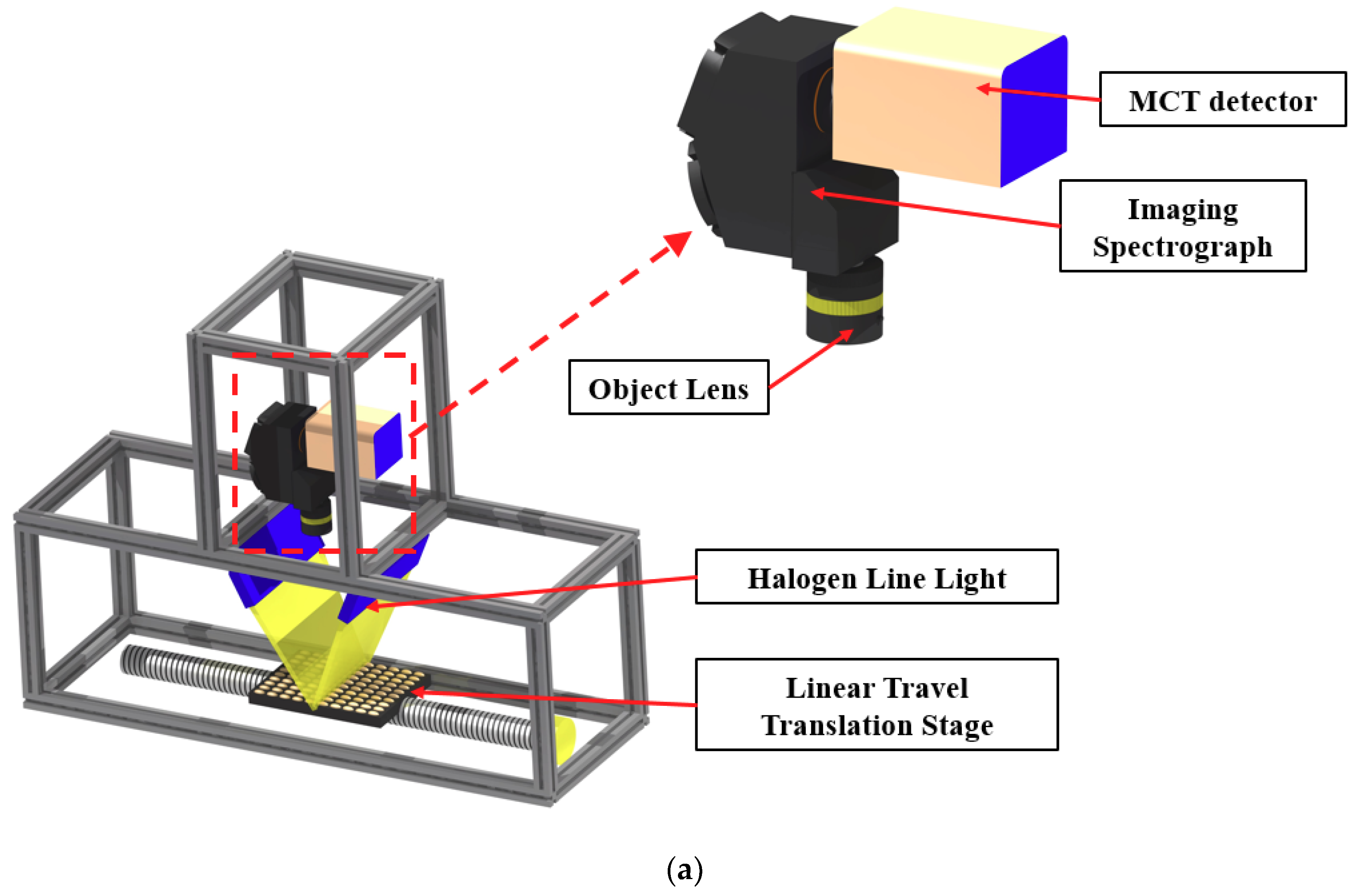
2.2. SWIR Hyperspectral Imaging System
2.3. Image Acquisition and Correction
2.4. Data Extraction and Preprocessing
2.5. Partial Least-Squares Discriminant Analysis (PLS-DA)
2.6. Variable Importance in Projection (VIP)
2.7. Image Processing
2.8. Germination Test
3. Results and Discussion
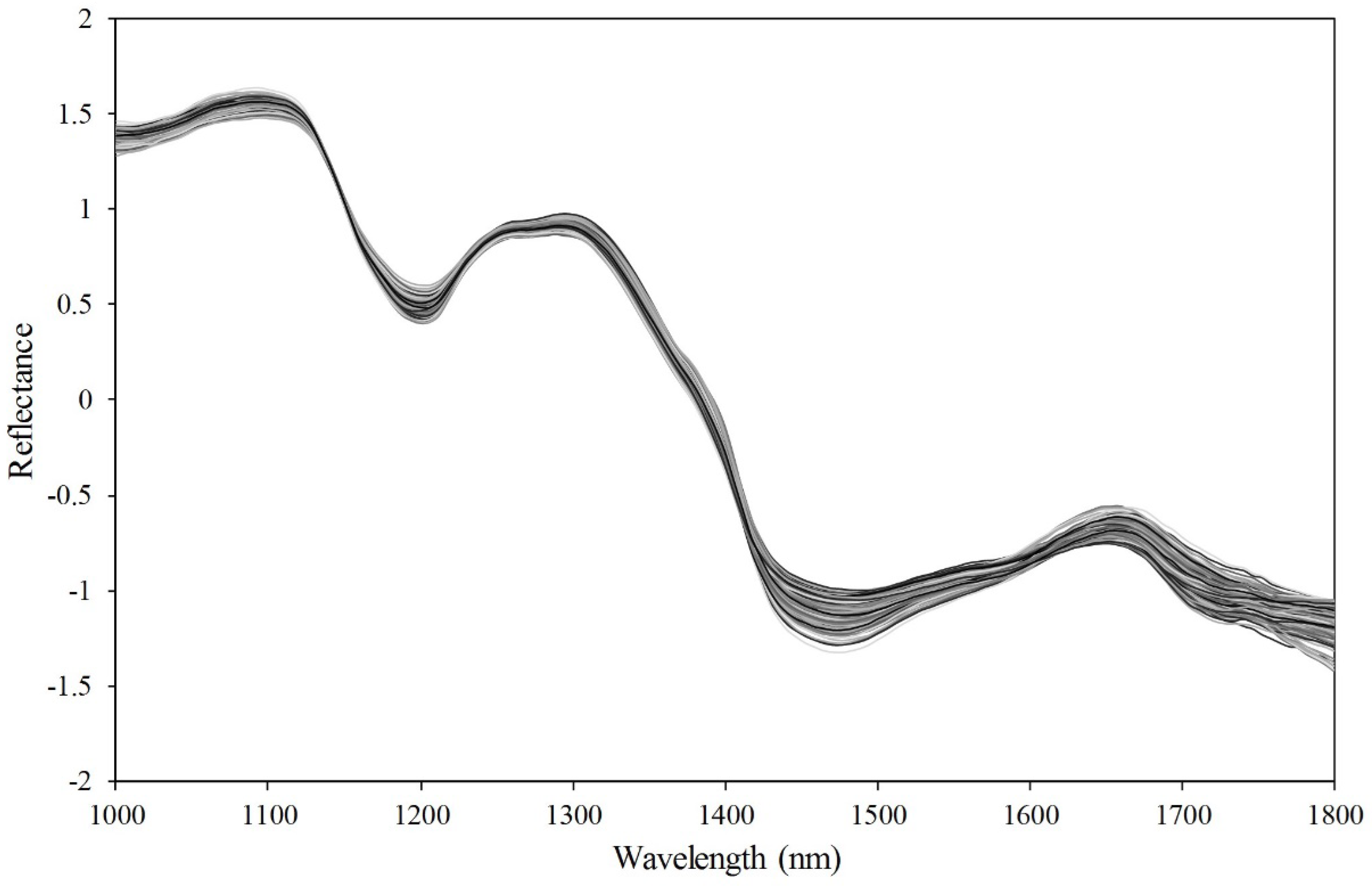
3.1. Spectral Characteristics of Soybean Seeds
3.2. PLS-DA Classification Using Entire Wavelengths
3.3. PLS-DA Classification Using VIP Selected Variables
3.4. Kernel-Based Classification of Viable and Nonviable Soybean Seeds
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Miller-Garvin, J.; Naeve, S.L. United States Soybean Quality—Annual Report; The U.S. Soybean Export Council: St. Paul, MN, USA, 2017; p. 17.
- FAOSTAT. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 9 May 2018).
- Brown-Lima, C.; Cooney, M.; Cleary, D. An Overview of the Brazil-China Soybean Trade and Its Strategic Implications for Conservation; United States Agency for International Development: Washington DC, USA, 2010.
- Lim, J.G.; Kim, G.Y.; Mo, C.Y.; Oh, K.M.; Kim, G.S.; Yoo, H.C.; Ham, H.H.; Kim, Y.T.; Kim, S.M.; Kim, M.S. Rapid and Nondestructive Discrimination of Fusarium Asiaticum and Fusarium Graminearum in Hulled Barley (Hordeum vulgare L.) Using Near-Infrared Spectroscopy. J. Biosyst. Eng. 2017, 42, 301–302. [Google Scholar] [CrossRef]
- Seed Vigor Testing Handbook; AOSA: Ithaca, NY, USA, 2009.
- Lee, H.; Huy, T.Q.; Park, E.; Bae, H.-J.; Baek, I.; Kim, M.S.; Mo, C.; Cho, B.-K. Machine Vision Technique for Rapid Measurement of Soybean Seed Vigor. J. Biosyst. Eng. 2017, 42, 227–233. [Google Scholar] [CrossRef]
- Mo, C.; Lim, J.; Kwon, S.W.; Lim, D.K.; Kim, M.S.; Kim, G.; Kang, J.; Kwon, K.-D.; Cho, B.-K. Hyperspectral Imaging and Partial Least Square Discriminant Analysis for Geographical Origin Discrimination of White Rice. J. Biosyst. Eng. 2017, 42, 293–300. [Google Scholar] [CrossRef]
- Ravikanth, L.; Singh, C.B.; Jayas, D.S.; White, N.D.G. Classification of contaminants from wheat using near-infrared hyperspectral imaging. Biosyst. Eng. 2015, 135, 73–86. [Google Scholar] [CrossRef]
- Wang, D.; Dowell, F.E.; Ram, M.S.; Schapaugh, W.T. Classification of fungal-damaged soybean seeds using near-infrared spectroscopy. Int. J. Food Prop. 2004, 7, 75–82. [Google Scholar] [CrossRef]
- Mahesh, S.; Jayas, D.S.; Paliwal, J.; White, N.D.G. Identification of wheat classes at different moisture levels using near-infrared hyperspectral images of bulk samples. Sens. Instrum. Food Qual. Saf. 2011, 5, 1–9. [Google Scholar] [CrossRef]
- Huang, M.; Tang, J.; Yang, B.; Zhu, Q. Classification of maize seeds of different years based on hyperspectral imaging and model updating. Comput. Electron. Agric. 2016, 122, 139–145. [Google Scholar] [CrossRef]
- Williams, P.J.; Kucheryavskiy, S. Classification of maize kernels using NIR hyperspectral imaging. Food Chem. 2016, 209, 131–138. [Google Scholar] [CrossRef]
- Ambrose, A.; Kandpal, L.M.; Kim, M.S.; Lee, W.-H.; Cho, B.-K. High speed measurement of corn seed viability using hyperspectral imaging. Infrared Phys. Technol. 2016, 75, 173–179. [Google Scholar] [CrossRef]
- Kandpal, L.M.; Lohumi, S.; Kim, M.S.; Kang, J.-S.; Cho, B.-K. Near-infrared hyperspectral imaging system coupled with multivariate methods to predict viability and vigor in muskmelon seeds. Sens. Actuators B Chem. 2016, 229, 534–544. [Google Scholar] [CrossRef]
- Weyer, L.; Workman, J., Jr. Practical Guide to Interpretive Near-Infrared Spectroscopy; CRC Press: Boca Raton, FL, USA, 2007; ISBN 1420018310. [Google Scholar]
- Rinnan, Å.; van den Berg, F.; Engelsen, S.B. Review of the most common pre-processing techniques for near-infrared spectra. TrAC Trends Anal. Chem. 2009, 28, 1201–1222. [Google Scholar] [CrossRef]
- Andersen, C.M.; Bro, R. Variable selection in regression-a tutorial. J. Chemom. 2010, 24, 728–737. [Google Scholar] [CrossRef]
- Marcos Filho, J. Seed vigor testing: An overview of the past, present and future perspective. Sci. Agric. 2015, 72, 363–374. [Google Scholar] [CrossRef]
- Basavarajappa, B.S.; Shekar Shetty, H.; Prakash, H.S. Membrane deterioration and other biochemical-changes, associated with accelerated aging of maize seeds. Seed Sci. Technol. 1991, 19, 279–286. [Google Scholar]
- Rastegar, Z.; Sedghi, M.; Khomari, S. Effects of Accelerated Aging on Soybean Seed Germination Indexes at Laboratory Conditions. Not. Sci. Biol. 2011, 3, 126. [Google Scholar] [CrossRef]
- Wu, D.; He, Y.; Feng, S.-J.; Bao, Y.-D. Application of infrared spectra technique based on LS-support vector machines to the non-destructive measurement of fat content in milk powder. J. Infrared Millim. Waves 2008, 3, 5. [Google Scholar] [CrossRef]
- Lee, J.H.; Choung, M.-G. Nondestructive determination of herbicide-resistant genetically modified soybean seeds using near-infrared reflectance spectroscopy. Food Chem. 2011, 126, 368–373. [Google Scholar] [CrossRef]
- Serranti, S.; Cesare, D.; Marini, F.; Bonifazi, G. Classification of oat and groat kernels using NIR hyperspectral imaging. Talanta 2013, 103, 276–284. [Google Scholar] [CrossRef]
- Chen, H.-Z.; Song, Q.-Q.; Tang, G.-Q.; Xu, L.-L. An optimization strategy for waveband selection in FT-NIR quantitative analysis of corn protein. J. Cereal Sci. 2014, 60, 595–601. [Google Scholar] [CrossRef]
- He, H.-J.; Sun, D.-W. Selection of Informative Spectral Wavelength for Evaluating and Visualising Enterobacteriaceae Contamination of Salmon Flesh. Food Anal. Methods 2015, 8, 2427–2436. [Google Scholar] [CrossRef]
- Riccioli, C.; Pérez-Marín, D.; Garrido-Varo, A. Identifying animal species in NIR hyperspectral images of processed animal proteins (PAPs): Comparison of multivariate techniques. Chemom. Intell. Lab. Syst. 2018, 172, 139–149. [Google Scholar] [CrossRef]
- Siedliska, A.; Baranowski, P.; Mazurek, W. Classification models of bruise and cultivar detection on the basis of hyperspectral imaging data. Comput. Electron. Agric. 2014, 106, 66–74. [Google Scholar] [CrossRef]
- Tigabu, M.; Odén, P.C. Discrimination of viable and empty seeds of Pinus patula Schiede & Deppe with near-infrared spectroscopy. New For. 2003, 25, 163–176. [Google Scholar] [CrossRef]
- Sato, T. New Estimation Method for Fatty Acid Composition in Oil Using Near Infrared Spectroscopy. Biosci. Biotechnol. Biochem. 2002, 66, 2543–2548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGoverin, C.M.; Engelbrecht, P.; Geladi, P.; Manley, M. Characterisation of non-viable whole barley, wheat and sorghum grains using near-infrared hyperspectral data and chemometrics. Anal. Bioanal. Chem. 2011, 401, 2283–2289. [Google Scholar] [CrossRef] [PubMed]
- Jha, S.N. (Ed.) Nondestructive Evaluation of Food Quality; Springer: Berlin/Heidelberg, Germany, 2010; ISBN 978-3-642-43543-0. [Google Scholar]
- Jerry Workman, J. (Ed.) The Handbook of Organic Compounds, Three-Volume Set, 1st ed.; Elsevier: Amsterdam, Netherlands, 2000; ISBN 9780080533650. [Google Scholar]
- Wang, D.; Ram, M.S.; Dowell, F.E. Classification of damaged soybean seeds using near-infrared spectroscopy. Trans. ASAE 2002, 45, 1943–1948. [Google Scholar] [CrossRef]
- Priestley, D.A.; Leopold, A.C. Absence of Lipid Oxidation during Accelerated Aging of Soybean Seeds. Plant Physiol. 1979, 63, 726–729. [Google Scholar] [CrossRef] [Green Version]








| Preprocessing | Latent Variables | RMSECV |
|---|---|---|
| Raw | 14 | 0.312 |
| SNV | 12 | 0.304 |
| Max | 14 | 0.310 |
| Mean | 12 | 0.322 |
| Range | 14 | 0.303 |
| Smoothing | 17 | 0.312 |
| Calibration (n = 149,884) | Raw | SNV | Max | Mean | Range | Smoothing |
|---|---|---|---|---|---|---|
| Viable | 91.0 | 91.4 | 91.2 | 90.7 | 91.4 | 91.0 |
| Non-viable | 92.8 | 92.7 | 92.8 | 93.1 | 92.7 | 92.8 |
| Total | 91.9 | 92.1 | 92.0 | 91.9 | 92.1 | 91.9 |
| Validation (n = 50,336) | ||||||
| Viable | 89.0 | 89.4 | 89.1 | 88.6 | 89.4 | 89.0 |
| Non-viable | 94.6 | 94.8 | 94.9 | 95.1 | 94.7 | 94.6 |
| Total | 91.8 | 92.1 | 92.0 | 91.8 | 92.1 | 91.8 |
| Calibration (n = 149,884) | Raw | SNV | Max | Mean | Range | Smoothing |
|---|---|---|---|---|---|---|
| Viable | 82.8 | 87.1 | 83.2 | 84.9 | 80.2 | 85.9 |
| Non-viable | 84.7 | 88.8 | 86.5 | 89.2 | 79.5 | 88.1 |
| Total | 83.7 | 88.0 | 84.9 | 87.1 | 79.9 | 87.0 |
| Validation (n = 50,336) | ||||||
| Viable | 80.8 | 84.1 | 80.0 | 81.0 | 76.7 | 85.6 |
| Non-viable | 82.7 | 91.1 | 88.3 | 91.0 | 81.8 | 87.3 |
| Total | 81.8 | 87.6 | 84.1 | 86.1 | 79.3 | 84.5 |
| PLS-DA with Full Wavelengths | Optimum Detection Rate (%) | AUC | Calibration (n = 300) | Validation (n = 100) | ||
|---|---|---|---|---|---|---|
| Viable Accuracy (%) | Non-Viable Accuracy (%) | Viable Accuracy (%) | Non-Viable Accuracy (%) | |||
| Raw | 71.3 | 0.9999 | 100 | 99.3 | 96.0 | 100 |
| SNV | 63.4 | 0.9999 | 100 | 99.3 | 98.0 | 100 |
| Max | 56.3 | 0.9999 | 100 | 99.3 | 98.0 | 100 |
| Mean | 49.4 | 0.9998 | 100 | 99.3 | 98.0 | 100 |
| Range | 63.0 | 0.9999 | 100 | 99.3 | 98.0 | 100 |
| Smoothing | 70.9 | 0.9999 | 100 | 99.3 | 98.0 | 100 |
| PLS-DA with VIP | ||||||
| Raw | 52.0 | 0.9947 | 95.3 | 97.3 | 96.0 | 96.0 |
| SNV | 33.9 | 0.9992 | 100 | 98.0 | 96.0 | 98.0 |
| Max | 56.0 | 0.9941 | 94.7 | 97.3 | 96.0 | 98.0 |
| Mean | 43.1 | 0.9959 | 95.3 | 98.0 | 96.0 | 98.0 |
| Range | 49.7 | 0.9717 | 94.0 | 90.0 | 96.0 | 94.0 |
| Smoothing | 55.7 | 0.9996 | 99.3 | 98.7 | 98.0 | 100 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baek, I.; Kusumaningrum, D.; Kandpal, L.M.; Lohumi, S.; Mo, C.; Kim, M.S.; Cho, B.-K. Rapid Measurement of Soybean Seed Viability Using Kernel-Based Multispectral Image Analysis. Sensors 2019, 19, 271. https://doi.org/10.3390/s19020271
Baek I, Kusumaningrum D, Kandpal LM, Lohumi S, Mo C, Kim MS, Cho B-K. Rapid Measurement of Soybean Seed Viability Using Kernel-Based Multispectral Image Analysis. Sensors. 2019; 19(2):271. https://doi.org/10.3390/s19020271
Chicago/Turabian StyleBaek, Insuck, Dewi Kusumaningrum, Lalit Mohan Kandpal, Santosh Lohumi, Changyeun Mo, Moon S. Kim, and Byoung-Kwan Cho. 2019. "Rapid Measurement of Soybean Seed Viability Using Kernel-Based Multispectral Image Analysis" Sensors 19, no. 2: 271. https://doi.org/10.3390/s19020271
APA StyleBaek, I., Kusumaningrum, D., Kandpal, L. M., Lohumi, S., Mo, C., Kim, M. S., & Cho, B. -K. (2019). Rapid Measurement of Soybean Seed Viability Using Kernel-Based Multispectral Image Analysis. Sensors, 19(2), 271. https://doi.org/10.3390/s19020271






