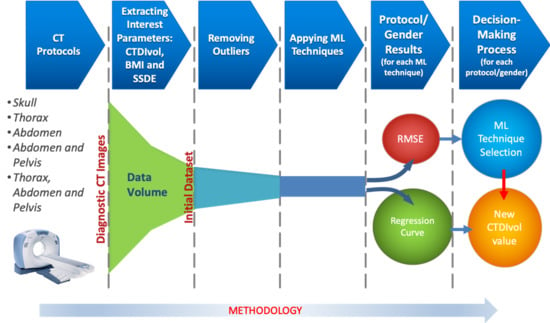
4.2.1. Removing Outliers
Once the data are gathered, those considered to be atypical (outliers); that is, values denoted as errors or unrepresentative are eliminated from the population (in this work, the sample values match the population). There are diverse techniques in order to do this [
24], which are applied according to the distribution of the sample and the data percentage to be removed.
If the data are treated as separated variables, outliers are eliminated while using the
univariate method.
Figure 2 shows the boxplot diagram that was proposed by Tukey in 1977 [
25], in which the distribution of a set of data is observed, and different regions are identified from their statistical information.
As
Figure 2 shows, we define the
Interquartile range (RIC) as the difference between Q3 (third quartile or 75th percentile) and Q1 (first quartile or 25th percentile). In this way, if we want to remove extreme values, those greater than Q3 + 3 *RIC and those less than Q1 − 3 *RIC will be eliminated. Even if we intend to be more restrictive, data that are higher than Q3 + 1.5 *RIC and lower than Q1 − 1.5 *RIC can be eliminated from our samples. However, the
univariate method has a clear drawback; the data removed are located at the ends of each variable and outside of these zones there could be undetected outliers.
Another way of eliminating outliers is to employ the multivariate method. This is ideal for internal areas with low data density; meaning the number of data is not very representative in the whole sample.
To detect and remove outliers, we apply the technique called the
density-based spatial clustering of applications with noise technique, DBSCAN [
26]. In this technique, the
epsilon (euclidean distance) and
MinPts (threshold) parameters must be previously defined, to later operate, as follows: for each particular data the number of neighbors in a certain
epsilon must be quantified. If that number exceeds the established threshold (
MinPts), the specific data and their neighbors are included in a cluster, as well as the neighbors of the previous data that fulfill the same condition. The iterative process continues until all of the data are checked and a cluster of connected data is established. On the other hand, if the number does not exceed the value of
MinPts, the specific data will be considered to be noise, and will therefore be eliminated from the sample (see
Figure 3).
The goal of both techniques is to eliminate the smallest number of data, trying not to exceed 5% of the total data of the sample. Once the atypical data have been removed, the regression techniques that are described in
Section 4.2.2 will be applied to the remaining data.
The data are separated into two groups, called training and test, in order to obtain a good regression model [
27]. The
cross-validation technique will be used, which divides the sample into
k groups of data. One of them will be used as a test and the rest for training.
This process is repeated during k iterations, with each time using a different group as test, without repetition. The value of k will vary depending on the number of available data. The value of 10 is the one that is selected for this work.
The regression algorithm will be fed with the number of data allotted to training, which will fit a curve with these data, obtaining the mathematical model as a result. For each iteration, the error
Ek will be estimated; the mean of all the errors results in the total error
E, as shown in
Figure 4.
After training the algorithm and obtaining the model, we will verify its performance by means of new data that have not been employed in the training process. We use the test data for this purpose.
If the error from the test data is much greater than the error committed by the training data, the model is overfitting the training data, diminishing the generality for the test set. The reason is the following: the algorithm extracts a large amount of information from the dataset to generate the training data, deriving a complex model that is capable of precisely adjusting its predictions. This model can include noise or random fluctuations due to the great number of data. When assessing new data (test), a minor amount of them are selected, which implies a low probability of noise/random data. The result is a clear deterioration in the performance of the model. This issue has a negative effect on the precision of the predictions, to the point of making it unfeasible for the problem contemplated here.
4.2.2. ML Techniques
Machine learning (ML) algorithms, as a subfield of artificial intelligence (AI), have been providing effective solutions in engineering applications and to scientific problems for many decades. The ML methods have the ability to adapt to new conditions and detect and estimate patterns. To this end, ML conceives self-learning algorithms to derive knowledge from data in order to carry out system predictions. ML provides a suitable solution that captures the awareness present in data and gradually enhances the performance of predictive models to build models that analyze a large amount of data. The main goal is to make the best decisions, or to take the best actions based on these predictions.
ML is divided into three categories: supervised learning, unsupervised learning, and reinforcement learning. In this work, we will focus our attention on supervised learning techniques, since they allow for a model to learn from training data to make predictions about unseen or future data. In supervised learning, the input data are defined by labels (such as, for instance, mail labels) or raw data. One of the subcategories of supervised learning is regression analysis, which addresses the prediction of continuous results from labels/raw data. Given a set of variables, x, named predictors, and their corresponding response variables, y, we can fit a curve graph (the simplest is a straight line) to these data that minimizes the distance between the sample points and the fitted linear/non-linear graph. The set unsupervised learning and regression analysis is adjusted to the requirements while observing the nature of the data to analyze in this work, allowing for us to predict the continuous outcomes of target variables.
In regression analysis techniques, the scientific literature presents different approaches that are useful in the massive analysis of data (Big Data). Furthermore, these techniques help in the forecasting of future doses to be received by patients, which is a distinctive objective of this work.
Regarding regression analysis models, we will concentrate our efforts on specifying (data selection), accommodating (eliminating outliers and anomalous points), and analyzing our large amount of CT exam data by using the following models:
1. Linear regression.
This technique consists of finding a line that fits a data set following a certain criterion. The most common criterion, which will also be employed in this work, is least squares adjustment [
28].
2. Decision Tree Learning.
This scheme breaks down our data by making decisions based on asking a series of questions. In particular, in the training phase, the decision tree model learns questions that are used to stamp class labels on the samples. As a tree model, the process starts at the root of the tree and then splits the data along its branches. The splitting procedure is repeated at each child node up to the leaves (of the tree). This means that the samples of each node belong to the same class. Note that the error is minimized if the tree is deep, but it can lead to overfitting. Thus, the usual procedure is to prune the tree, restricting its maximum depth. A better way to improve the results of the Decision Tree Learning algorithm is to employ a technique, called Bagged Decision Tree, which reduces the variance of a decision tree.
3. Bagged Decision Tree.
In this technique, multiple regression trees are constructed. In particular, several subsets of data are created from training samples, for each collection of them to be later used to train their own decision trees. The average derived from these different decision trees provides a more robust solution than a single decision tree. The use of several trees also reduces overfitting.
4.
Artificial Neural Networks [
29].
Our focus will be on analyzing the data for the training phase with a technique, called
Bayesian regularization [
30]. This algorithm allows for us to perform binary classification, and we will use the
Levenberg-Marquardt optimization [
31] to learn the weight coefficients of the model (in each iteration of the training phase, the coefficients are updated). Furthermore, it is possible to obtain the optimal weights employing cost functions, such as those called
Sum of Squared Errors (SSE). To find the predicted values, the solution involves connecting multiple single neurons to a
multi–layer feedforward neural network. This particular type of network is also called a
multi–layer perceptron (MLP), which consists of three layers (input, hidden, and output layers). Both techniques (
Bayesian regularization and
Levenberg–Marquardt optimization), together with an infrastructure MLP achieve an optimal model capable of generalizing the mathematical problem thanks to the minimization of a combination of weights and errors. This algorithm allows for overfitting to be reduced at the cost of longer execution time.
5.
Gaussian Process Regression [
32].
Parametric regression methods, for instance, linear/logistic regression, generate a line or a curve in the graph of inputs and outputs, replacing the training data. Accordingly, once the regression weights have been obtained, the original training data may be eliminated from the graph. On the other hand, non-parametric regression methods may retain the initial training data (also called latent variables) to be used as a significant element in generating a regressor function. To this end, test data are compared to the training data points; each output value of the test point is estimated via the distance of the test data input to the training data input. It is notable that non-parametric regression considers that data points with similar input values will be close in output space. The mathematical expressions include the covariance function formed of latent variables, which reflects the smoothness of the response. Covariance and mean functions were used in conjunction with a Gaussian likelihood for prediction, employing as an initial expression. In it, is a posterior distribution, X is a matrix of training inputs, y is a vector of training target, and is a matrix of test inputs.
To maximize this expression, we have carefully studied the mathematical model that was derived in [
33]. From this previous study, we opt as a useful equation for the problem here planned the following exponential function, which will, in turn, be employed as a kernel function:
where the parameter
is the standard deviation, while l
k is the scalar dimension for each predictor,
k is the number of evaluations to fulfill the maximization problem, and
x and
x’ are two near values.
6.
Support Vector Regression (
SVR) [
34].
In this case, we consider the following training data , where indicate the input space of the sample and its corresponding target value, respectively, and l denotes the size of the training data. Our objective is to find a function that has, at most, ε deviation from the obtained targets for all of the training data, and at the same time is as flat as possible. In other words, we do not care about errors because they are less than ε. Additionally, the results must avoid senseless predictions to find a function that returns the best fit.
Regarding the relationship between
and
it is approximately linear, which means that the model is represented as:
(
w represents coefficients and
b is an intercept). Therefore, this problem can be formulated as a convex optimization problem:
Here, our optimization problem is planned as a non-linear case. Keeping this in mind and, thanks to the support of the work [
35], the solution for (2) is the following Equation (3):
The constant C > 0 determines the trade-off between the flatness (this means that one seeks a small w value) of f and the amount up to which deviations that are larger than ε are tolerated. On the other hand, , , are Lagrange multipliers. Finally, is a Kernel function. A common kernel that is used for this model is the radial basis function:
A more detailed study of these aforementioned techniques can be found in the
Supplementary File 1 “Description of Machine Learning Techniques”. Finally,
Table 3 shows the parameters used in the previously defined algorithms: