Data-Driven Investigation of Gait Patterns in Individuals Affected by Normal Pressure Hydrocephalus
Abstract
:1. Introduction
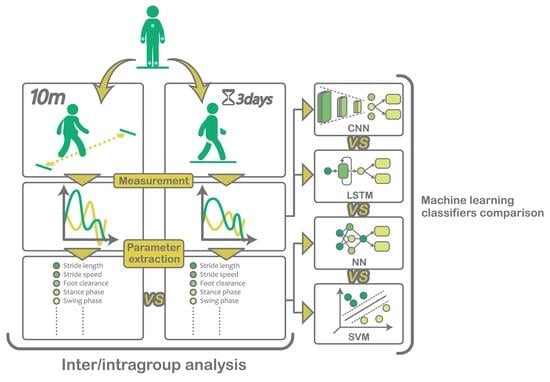
2. Materials and Methods
2.1. Sensors
2.2. Subjects
2.3. Protocol
2.4. Gait Analysis
2.5. Statistical Analysis
2.6. Data-Driven Approaches for Stride Classification
3. Results
3.1. Real-World versus Controlled Environment
3.1.1. Significant Differences between Environments for All Subjects Groups YHC, EHC and NPH
3.1.2. Significant Differences between Environments Only for EHC and NPH
3.1.3. Significant Differences between Environments Only for the NPH Group
3.2. Elderly Subjects versus NPH Patients
3.3. Stride Classification through Machine Learning
4. Discussion
4.1. Real-World versus Controlled Environment
4.2. Elderly Subjects versus NPH Patients
4.3. Stride Classification through Machine Learning
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abu-Faraj, Z.O.; Harris, G.F.; Smith, P.A.; Hassani, S. Human Gait and Clinical Movement Analysis, 2nd ed.; John Wiley & Sons, Inc.: New York, NY, USA, 2015; pp. 1–34. [Google Scholar] [CrossRef]
- Figueiredo, J.; Félix, P.; Costa, L.; Moreno, J.C.; Santos, C.P. Gait Event Detection in Controlled and Real-Life Situations: Repeated Measures from Healthy Subjects. IEEE Trans. Neural Syst. Rehabil. Eng. 2018, 26, 1945–1956. [Google Scholar] [CrossRef]
- Wang, W.; Adamczyk, P.G. Analyzing gait in the real world using wearable movement sensors and frequently repeated movement paths. Sensors 2019, 19, 1925. [Google Scholar] [CrossRef] [Green Version]
- Renggli, D.; Graf, C.; Tachatos, N.; Singh, N.; Meboldt, M.; Taylor, W.R.; Stieglitz, L.; Schmid Daners, M. Wearable Inertial Measurement Units for Assessing Gait in Real-World Environments. Front. Physiol. 2020, 11, 90. [Google Scholar] [CrossRef]
- Stolze, H.; Kuhtz-Buschbeck, J.P.; Drücke, H.; Jöhnk, K.; Illert, M.; Deuschl, G. Comparative analysis of the gait disorder of normal pressure hydrocephalus and Parkinson’s disease. J. Neurol. Neurosurg. Psychiatry 2001, 70, 289–297. [Google Scholar] [CrossRef] [Green Version]
- Gallia, G.L.; Rigamonti, D.; Williams, M.A. The diagnosis and treatment of idiopathic normal pressure hydrocephalus. Nat. Clin. Pract. Neurol. 2006, 2, 375–381. [Google Scholar] [CrossRef] [Green Version]
- Jaraj, D.; Rabiei, K.; Marlow, T.; Jensen, C.; Skoog, I.; Wikkelsø, C. Prevalence of idiopathic normal-pressure hydrocephalus. Neurology 2014, 82, 1449–1454. [Google Scholar] [CrossRef] [Green Version]
- König, N.; Singh, N.B.; von Beckerath, J.; Janke, L.; Taylor, W.R. Is gait variability reliable? An assessment of spatio-temporal parameters of gait variability during continuous overground walking. Gait Posture 2014, 39, 615–617. [Google Scholar] [CrossRef]
- König, N.; Singh, N.B.; Baumann, C.R.; Taylo, W.R. Can gait signatures provide quantitative measures for aiding clinical decision-making? A systematic meta-analysis of gait variability behavior in patients with Parkinson’s disease. Front. Hum. Neurosci. 2016, 10, 319. [Google Scholar] [CrossRef] [Green Version]
- Stolze, H.; Diercks, C.; Palmie, S.; Jo, K.; Dru, H.; Mehdorn, H.M.; Illert, M.; Deuschl, G. Gait analysis in idiopathic normal pressure hydrocephalus ± which parameters respond to the CSF tap test? Clin. Neurophysiol. 2000, 111, 1678–1686. [Google Scholar] [CrossRef]
- Relktin, N.; Marmarou, A.; Klinge, P.; Bergsneider, M.; Black, P.M.L. INPH guidelines, part II: Diagnosing idio-pathic normal-pressure hydrocephalus. Neurosurgery 2005, 57, 4–16. [Google Scholar] [CrossRef] [Green Version]
- Shrinivasan, A.; Maïté, B.P.; Barth, A.; Lach, J. Analysis of gait in patients with normal pressure hydrocephalus. In mHealthSys 2011, Proceedings of the 1st ACM Workshop on Mobile Systems, Applications, and Services for HealthCare-Co-Held with ACM SenSys, Seattle WA, USA, 1 November 2011; Association for Computing Machinery: New York, NY, USA, 2011. [Google Scholar] [CrossRef]
- Liao, R.; Makihara, Y.; Muramatsu, D.; Mitsugami, I.; Yagi, Y.; Yoshiyama, K.; Kazui, H.; Takeda, M. A video-based gait disturbance assessment tool for diagnosing idiopathic normal pressure hydrocephalus. IEEJ Trans. Electr. Electron. Eng. 2020, 15, 433–441. [Google Scholar] [CrossRef]
- Tunca, C.; Salur, G.; Ersoy, C. Deep Learning for Fall Risk Assessment with Inertial Sensors: Utilizing Domain Knowledge in Spatiooral Gait Parameters. IEEE J. Biomed. Health Inform. 2020, 24, 1994–2005. [Google Scholar] [CrossRef]
- Rampp, A.; Barth, J.; Schülein, S.; Gaßmann, K.G.; Klucken, J.; Eskofier, B.M. Inertial Sensor-Based Stride Parameter Calculation From Gait Sequences in Geriatric Patients. IEEE Trans. Biomed. Eng. 2015, 62, 1089–1097. [Google Scholar] [CrossRef]
- Benoussaad, M.; Sijobert, B.; Mombaur, K.; Coste, C.A. Robust foot clearance estimation based on the integration of foot-mounted IMU acceleration data. Sensors 2015, 16, 12. [Google Scholar] [CrossRef] [Green Version]
- Mathworks. Statistics and Machine Learning Toolbox™ User’s Guide R2019b. 2019. Available online: https://www.mathworks.com/products/statistics.html (accessed on 22 September 2021).
- Kühnast, C.; Neuhäuser, M. A note on the use of the non-parametric Wilcoxon-Mann- Whitney test in the analysis of medical studies. Ger. Med Sci. 2008, 6, 3–7. [Google Scholar]
- Mathworks. Deep Learning Toolbox™ User’s Guide R2019b. 2019. Available online: https://www.mathworks.com/products/deep-learning.html (accessed on 22 September 2021).
- Eyobu, O.S.; Han, D.S. Feature representation and data augmentation for human activity classification based on wearable IMU sensor data using a deep LSTM neural network. Sensors 2018, 18, 2892. [Google Scholar] [CrossRef] [Green Version]
- Buda, M.; Maki, A.; Mazurowski, M.A. A systematic study of the class imbalance problem in convolutional neural networks. Neural Netw. 2018, 106, 249–259. [Google Scholar] [CrossRef] [Green Version]
- Alcock, L.; Galna, B.; Lord, S.; Rochester, L. Characterisation of foot clearance during gait in people with early Parkinson’s disease: Deficits associated with a dual task. J. Biomech. 2016, 49, 2763–2769. [Google Scholar] [CrossRef] [Green Version]
- Tunca, C.; Pehlivan, N.; Ak, N.; Arnrich, B.; Salur, G.; Ersoy, C. Inertial sensor-based robust gait analysis in non-hospital settings for neurological disorders. Sensors 2017, 17, 825. [Google Scholar] [CrossRef] [Green Version]
- Weerdesteyn, V.; De Niet, M.; Van Duijnhoven, H.J.; Geurts, A.C. Falls in individuals with stroke. J. Rehabil. Res. Dev. 2008, 45, 1195–1214. [Google Scholar] [CrossRef]
- Del Din, S.; Godfrey, A.; Galna, B.; Lord, S.; Rochester, L. Free-living gait characteristics in ageing and Parkinson’s disease: Impact of environment and ambulatory bout length. J. NeuroEng. Rehabil. 2016, 13, 1–12. [Google Scholar] [CrossRef] [Green Version]




| No | Parameter | Unit | YHC—20 Subjects | EHC—20 Subjects | NPH—12 Subjects | |||
|---|---|---|---|---|---|---|---|---|
| Rel. Diff. | Wilcoxon | Rel. Diff. | Wilcoxon | Rel. Diff. | Wilcoxon | |||
| 1 | Stride length | m | −0.9% | 0.7333 | −5.6% | 0.0935 | −43.4% | 0.0073 |
| 2 | Sensor clearance | m | 0.3% | 0.8078 | 0.0% | 0.8496 | −24.9% | 0.0117 |
| 3 | Gait speed | m/s | −2.1% | 0.7990 | −12.1% | 0.0068 | −44.3% | 0.0073 |
| 4 | Outward rotation | rad | 19.4% | 0.0094 | 15.8% | 0.0018 | 16.6% | 0.0317 |
| 5 | Step width | m | 31.5% | 0.0010 | −17.7% | 0.0046 | 23.4% | 0.0293 |
| 6 | Hand distance | m | 5.7% | 0.1737 | 3.6% | 0.5377 | −6.8% | 0.4370 |
| 7 | Hand max. ang. velocity | rad/s | 7.3% | 0.0868 | −1.3% | 0.8373 | −16.4% | 0.6107 |
| 8 | Number of steps per | steps | 19.3% | 0.008 | 19.8% | 0.0016 | 6.3% | 0.3276 |
| 9 | Stance phase | % | 0.9% | 0.1982 | 0.6% | 0.5285 | 4.0% | 0.0073 |
| 10 | Swing phase | % | −1.5% | 0.3749 | −0.8% | 0.5606 | −7.6% | 0.0209 |
| 11 | Stance/swing ratio | % | 3.0% | 0.0868 | 2.5% | 0.5182 | 9.4% | 0.0098 |
| 12 | Double support phase | % | 6.8% | 0.0560 | 5.1% | 0.0926 | 16.8% | 0.0183 |
| 13 | Cadence | steps/min | −0.12% | 0.9477 | −6.0% | 0.0056 | −0.3% | 0.7339 |
| 14 | Stride time | s | −0.6% | 0.8414 | 4.8% | 0.0058 | −1.28% | 0.6906 |
| 15 | Variability stride time | % | 51.3% | 0.0013 | 57.6% | 0.0013 | −43.41% | 0.0073 |
| No | Parameter | Unit | Controlled—p-Value | Non-Controlled—p-Value |
|---|---|---|---|---|
| 1 | Stride length | m | <0.0001 | <0.0001 |
| 2 | Sensor clearance | m | 0.0005 | <0.0001 |
| 3 | Gait speed | m/s | <0.0001 | <0.0001 |
| 4 | Outward rotation | rad | 0.3811 | 0.3023 |
| 5 | Step width | m | 0.3603 | 0.4249 |
| 6 | Hand distance | m | 0.0068 | <0.0001 |
| 7 | Hand max. ang. velocity | rad/s | 0.0003 | <0.0001 |
| 8 | Number of steps per | steps | <0.0001 | 0.0001 |
| 9 | Stance phase | % | 0.2200 | 0.0006 |
| 10 | Swing phase | % | 0.0409 | 0.0002 |
| 11 | Stance/swing ratio | % | 0.0703 | 0.0003 |
| 12 | Double support phase | % | 0.1554 | 0.0006 |
| 13 | Cadence | steps/min | 0.0589 | 0.2739 |
| 14 | Walk-cycle time | (s) | 0.0562 | 0.1913 |
| 15 | Variability walk-cycle time | % | 0.0029 | 0.2201 |
| 16 | Variability stride length | % | 0.0005 | 0.0004 |
| 17 | Variability stance/swing ratio | % | 0.0001 | 0.0002 |
| Controlled environment | Real-world environment | ||||||
| True Class | EHC | 89.5% | 10.5% | True Class | EHC | 95.8% | 4.2% |
| NPH | 24.4% | 75.6% | NPH | 8.4% | 91.6% | ||
| EHC | NPH | EHC | NPH | ||||
| Predicted Class | Predicted Class | ||||||
| True Class | Subject | SVM Class Prediction | NN Class Prediction | LSTM Class Prediction | CNN Class Prediction | ||||
|---|---|---|---|---|---|---|---|---|---|
| EHC | NPH | EHC | NPH | EHC | NPH | EHC | NPH | ||
| EHC | 1 | 96.2% | 3.8% | 96.3% | 3.7% | 87.6% | 12.4% | 60.5% | 39.4% |
| 2 | 98.1% | 1.9% | 97.7% | 2.3% | 81.0% | 19.0% | 97.0% | 2.9% | |
| 3 | 90.0% | 10.0% | 89.1% | 10.9% | 79.5% | 20.5% | 88.7% | 11.2% | |
| 4 | 67.3% | 32.7% | 54.6% | 45.4% | 97.0% | 3.0% | 82.8% | 17.1% | |
| 5 | 96.6% | 3.4% | 96.4% | 3.6% | 59.0% | 41.0% | 94.0% | 5.9% | |
| 6 | 92.0% | 8.0% | 92.1% | 7.9% | 62.7% | 37.3% | 70.0% | 29.9% | |
| 7 | 66.0% | 34.0% | 43.0% | 57.0% | 90.6% | 9.4% | 74.6% | 25.3% | |
| 8 | 97.4% | 2.6% | 97.7% | 2.3% | 98.2% | 1.8% | 95.4% | 4.5% | |
| 9 | 98.3% | 1.7% | 98.6% | 1.4% | 97.7% | 2.3% | 95.5% | 4.4% | |
| 10 | 98.9% | 1.1% | 98.6% | 1.5% | 99.4% | 0.6% | 97.4% | 2.5% | |
| 11 | 98.8% | 1.2% | 99.0% | 1.0% | 98.9% | 1.1% | 99.2% | 0.7% | |
| 12 | 98.9% | 1.1% | 98.8% | 1.2% | 86.3% | 13.7% | 94.9% | 5.0% | |
| 13 | 98.4% | 1.6% | 98.8% | 1.2% | 97.9% | 2.1% | 98.6% | 1.3% | |
| 14 | 97.5% | 2.5% | 99.0% | 1.0% | 99.3% | 0.7% | 98.2% | 1.7% | |
| 15 | 98.3% | 1.7% | 98.0% | 2.0% | 88.9% | 11.1% | 91.4% | 8.5% | |
| 16 | 95.2% | 4.8% | 94.5% | 5.5% | 95.0% | 5.0% | 96.0% | 3.9% | |
| 17 | 95.9% | 4.1% | 96.1% | 3.9% | 92.3% | 7.7% | 90.2% | 9.7% | |
| 18 | 99.1% | 0.9% | 99.3% | 0.7% | 98.8% | 1.2% | 98.1% | 1.8% | |
| 19 | 99.1% | 0.9% | 99.9% | 0.1% | 96.2% | 3.8% | 97.6% | 2.3% | |
| 20 | 95.7% | 4.3% | 98.0% | 2.0% | 99.6% | 0.4% | 98.8% | 1.1% | |
| NPH | 1 | 11.0% | 89.0% | 10.4% | 89.6% | 45.3% | 54.7% | 57.2% | 42.7% |
| 2 | 60.4% | 39.6% | 51.5% | 48.5% | 54.0% | 45.9% | 74.7% | 25.2% | |
| 3 | 6.8% | 93.2% | 21.9% | 78.1% | 28.7% | 71.3% | 49.8% | 50.1% | |
| 4 | 29.7% | 70.3% | 7.4% | 92.6% | 13.6% | 86.4% | 44.1% | 55.8% | |
| 5 | 2.2% | 97.8% | 3.8% | 96.2% | 13.3% | 86.7% | 16.8% | 83.1% | |
| 6 | 61.7% | 38.3% | 63.1% | 36.9% | 76.4% | 23.6% | 85.2% | 14.7% | |
| 7 | 1.6% | 98.4% | 11.7% | 88.3% | 9.5% | 90.5% | 47.2% | 52.7% | |
| 8 | 12.8% | 87.2% | 34.9% | 65.1% | 44.2% | 55.8% | 52.1% | 47.8% | |
| 9 | 1.9% | 98.1% | 2.6% | 97.4% | 32.9% | 67.1% | 34.0% | 65.9% | |
| 10 | 1.0% | 99.0% | 0.8% | 99.2% | 10.1% | 89.9% | 11.0% | 88.9% | |
| 11 | 1.1% | 98.9% | 1.8% | 98.2% | 3.4% | 96.6% | 6.5% | 93.4% | |
| 12 | 9.9% | 90.1% | 18.6% | 81.4% | 19.5% | 80.5% | 25.9% | 74.0% | |
| Method | Classifier | Mean AUROC | std AUROC | Accuracy |
|---|---|---|---|---|
| Parameter representation | SVM-stride | 0.976 | 0.020 | 89.9 |
| NN-stride | 0.976 | 0.016 | 88.0 | |
| Time-series representation | LSTM | 0.903 | 0.076 | 83.0 |
| CNN | 0.876 | 0.084 | 78.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuruvithadam, K.; Menner, M.; Taylor, W.R.; Zeilinger, M.N.; Stieglitz, L.; Schmid Daners, M. Data-Driven Investigation of Gait Patterns in Individuals Affected by Normal Pressure Hydrocephalus. Sensors 2021, 21, 6451. https://doi.org/10.3390/s21196451
Kuruvithadam K, Menner M, Taylor WR, Zeilinger MN, Stieglitz L, Schmid Daners M. Data-Driven Investigation of Gait Patterns in Individuals Affected by Normal Pressure Hydrocephalus. Sensors. 2021; 21(19):6451. https://doi.org/10.3390/s21196451
Chicago/Turabian StyleKuruvithadam, Kiran, Marcel Menner, William R. Taylor, Melanie N. Zeilinger, Lennart Stieglitz, and Marianne Schmid Daners. 2021. "Data-Driven Investigation of Gait Patterns in Individuals Affected by Normal Pressure Hydrocephalus" Sensors 21, no. 19: 6451. https://doi.org/10.3390/s21196451